Published online Sep 18, 2025. doi: 10.13105/wjma.v13.i3.111111
Revised: July 10, 2025
Accepted: August 25, 2025
Published online: September 18, 2025
Processing time: 79 Days and 1.5 Hours
The causative agent for dengue is dengue virus (DENV), with humans and mos
Core Tip: The dengue disease, caused by the dengue virus, is mosquito-borne. Recent epidemiological studies of this highly contagious disease revealed its increasing incidence and mortality rates. Despite these concerns, approved therapeutic and prophylactic measures are still lacking. The ocean houses organisms that are prolific producers of specialized metabolites with pharmacological importance. Marine natural products have potential as drug molecules with anti-dengue virus activity.
- Citation: Gbadebo OS, Oke ED. Therapeutic potential of marine natural products against dengue virus. World J Meta-Anal 2025; 13(3): 111111
- URL: https://www.wjgnet.com/2308-3840/full/v13/i3/111111.htm
- DOI: https://dx.doi.org/10.13105/wjma.v13.i3.111111
The dengue virus (family = Flaviviridae), the causative agent of dengue, is a positive-stranded RNA virus with four serotypes, DENV-1, DENV-2, DENV-3, and DENV-4[1]. The dengue disease is currently the most contagious mosquito-borne disease worldwide, transmitted by Aedes aegypti and Aedes albopictus[2]. Following infection, the virus incubates for four to seven days, after which the full-blown disease takes a triad phase – febrile, critical, and recovery phases[3]. Monophasic courses also occur[3]. About 75% of the disease cases are asymptomatic while other clinical presentations could be mild or severe. Severe presentations include hemorrhage, organ impairment-associated shock, and death[3]. As of 2023, over 80 countries were reported to have been affected and since then, over 6.5 million cases and 7300 deaths associated with dengue have been reported[4]. Stanaway et al[5] estimated 9221 mortalities per year between 1990 and 2013, ranging from a low of 8227 in 1992 and peaking in 2010 with 11302 deaths. The increasing trend in dengue incidence and consequent mortality can be linked to globalization and urbanization, causing the emergence of breeding sites for mosquitoes. These factors and a rising global population increase the accumulation of stagnant water and the deposition of waste that either house the mosquitoes or foster their development[6]. Increasing temperatures due to climate change may further worsen the situation, especially in the currently low-risk regions of the world. All these contribute to the predicted global risk population of 63% in 2080[7].
Specific treatment options for dengue are currently lacking, as the disease is only managed by either improving the patient’s immunity or controlling the symptoms[3]. Encoded in the viral genome are three structural proteins and seven non-structural proteins, some of which may serve as potential anti-dengue drug targets[8]. The capsid, a structural protein, has been targeted as a retroviral inhibition approach, where the viral capsid protein is fused with a ligand that inhibits the virus[9]. This capsid-targeted viral inactivation approach elicited moderate anti-dengue effect in vitro, and the prophylactic model also decreased the viral infectious titers by 103 to 104-fold[10]. Viral growth involves steps like viral attachment, viral cell fusion, reverse transcription, integration, and translation, which are potential targets for antiviral drug molecules[11]. Crucial enzymes involved in the synthesis of DNA, RNA, and glycoproteins may also be targeted[12]. The knowledge of the structure and replication cycle of the DENV is a vital tool in the development of new anti-dengue drug molecules. While macromolecules like fusion proteins have been developed, small molecules also have potential as antivirals.
Lately, the marine environment has been a hub for the discovery of specialized metabolites with diverse bioactivities[12]. The MarinLit database, a collection of marine natural products contains 42237 compounds from marine organisms, especially invertebrates and microorganisms. A lot of these compounds have made it to the clinical trial phase while several of them have obtained approval for commercial distribution. The anti-HSV drug, vidarabine, was developed from the lead, spongouridine, which was first isolated from marine sponges[13]. The microbial diversity in the seawater presents great potential for drug development. Marine cyanobacteria, actinomycetes, bacteria, and fungi are prolific producers of alkaloids, terpenes, peptides, polyketides, shikimates, etc, with antitumor, antibacterial, and antiviral activities[12]. While the cyanobacterial-derived anti-HIV cyanovirin N was involved in preclinical studies, the marine microbiomes still lack extensive exploration as sources of anti-viral agents for clinical studies[14,15]. Algae have also been a reservoir for antiviral specialized metabolites. Algal-derived compounds such as sulfated polysaccharides, carrageenan, lectin, fucoidan, and sulfated galactan have been reported to inhibit different stages of viral replication[16]. Their impressive potential as sources of bioactive molecules can be utilized for anti-dengue drug discovery.
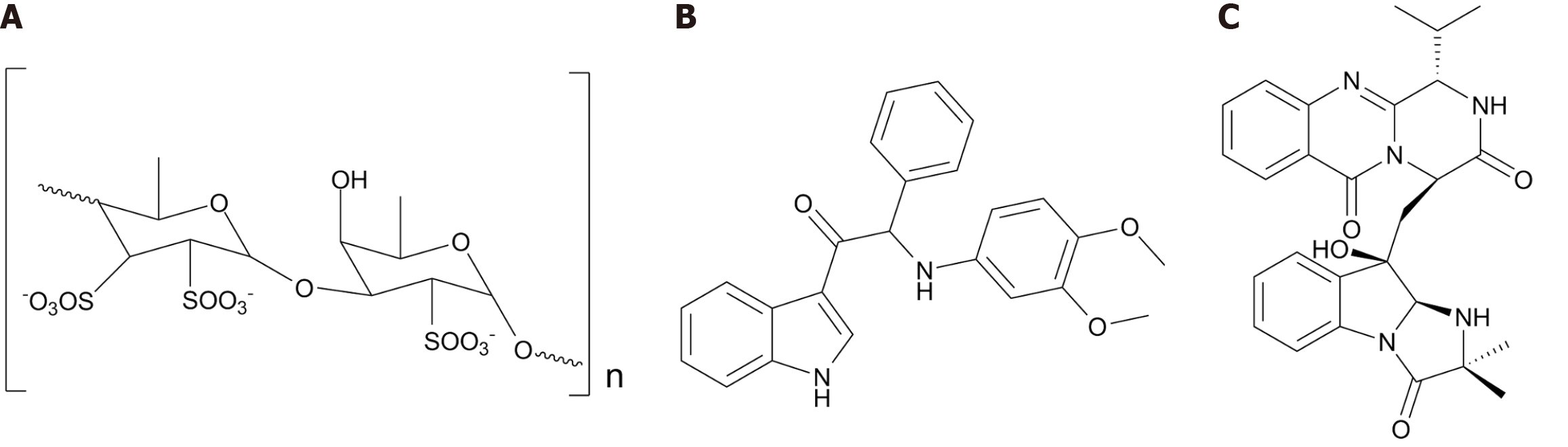
Despite the potential of the ocean and drug source, only a few marine-sourced anti-dengue specialized metabolites, including fucoidan, scequinadoline A, and indole derivatives, have been reported (Figure 1). Fucoidan, found in Sargassum spp. polysaccharide extract elicited anti-dengue activity by preventing viral attachment to the host cell[17]. Fucoidan is a sulfated polysaccharide composed of two main monosaccharides, fucose and glucuronic acid. Structure diversity is a major strength of sulfated polysaccharides as antiviral agents[17]. They elicit antiviral activity by blocking viral attachment, adsorption, and reproduction. A rich source of sulfated polysaccharides is marine algae[17]. These polysaccharides have been studied for activity against influenza A, mumps, measles, HSV-1, HIV-1, and human cytomegalovirus[18]. Carrageenan is another polysaccharide from red algae, which moderately reduced viral replication to varying extents in the four dengue serotypes[19]. Scequinadoline A, a specialized metabolite from the marine-derived fungus Dichotomomyces cejpii, inhibited DENV serotype 2 viral production by plaque assay[20]. Scequinadoline A is an alkaloid made up of the pyrazino[2,1-b]quinazoline-3,6-dione and 3,3a-dihydroimidazo[1,2-a]indol-1-one moieties. The two chiral centers on the pyrazino[2,1-b]quinazoline-3,6-dione moiety may be important for activity[20]. Indole alkaloids have also been studied for activity against DENV. Bardiot et al[21] performed a medium-throughput cytopathic effect reduction assay using the CD3 (Center for Drug Design and Discovery, KU Leuven) compound library. This led to the discovery of 2-[(3,4-dimethoxyphenyl)amino]-1-(1H-indol-3-yl)-2-phenylethan-1-one, a novel anti-dengue molecule. Other indole derivatives with significant viral inhibitory activity were also identified[21]. These compounds prevented viral replication by blocking the interaction between NS3 and NS4B viral proteins in vitro[22,23]. A follow-up lead optimization effort yielded two compounds with improved spectrum of activity, ADME, and pharmacokinetic properties[24].
Advances in bioinformatic tools and the application of molecular biology to natural product drug discovery give room for rational natural product discovery endeavors. Unlike the traditional trial-and-error and bioassay-guided fractionation approaches, the genomes of marine microbes can be mined for biosynthetic gene clusters responsible for specialized metabolite production. Molecular network generation has also been used to identify metabolites with important pharmacological activities from extracts of marine microbial cultures[25]. Computational methods can also be applied to anti-DENV small molecules from the ocean.
Computational methods are increasingly important for identifying potential treatment options for the DENV. By leveraging computational tools, medicinal chemists can identify potential drug targets, perform virtual screening of large chemical libraries, and evaluate the binding and activity of potential lead compounds, thus accelerating the discovery of newer and more effective therapeutic options for the DENV.
The highly conserved serine NS2B/NS3 protease, which functions in both the viral replication process and immune system invasion, is considered the major therapeutic target for anti-flavivirus agents[26-28]. Crystal structures of the DENV NS2B/NS3 protease reveal a negatively charged active site, which favors molecular interactions with positively charged residues such as arginine and lysine. Its flat surface and preference for binding positively charged residues present a significant challenge for developing effective inhibitors[29]. Hence, allosteric sites within the protease are being explored for inhibition. A diverse array of synthetic compounds has been identified as orthosteric and allosteric inhibitors of the DENV NS2B/NS3 protease[28]. Moreover, the synthetic compound NITD-88 is currently being developed in clinical trials as a drug candidate for dengue. NITD-88 acts as an NS4B/NS3 inhibitor, disrupting critical interactions between these two protein counterparts[30].
The potential of plant-based natural products as inhibitors of the DENV protease has been elucidated through molecular docking and molecular dynamics simulation studies. Compounds including rosmarinic acid, niloticin, luteolin, methoxyflavones, diterpenes, diterpenoids, and their derivatives have been shown to inhibit DENV’s non-structural proteins[31-35]. Databases of phytochemicals have also been screened against the dengue viral proteases NS1, NS3/NS2B, and NS5, generating novel scaffolds for drug development against the DENV[36].
A virtual screening of the Comprehensive Marine Natural Products Database, containing 31561 marine compounds, against the DENV NS1 generated hit compounds with good binding affinities, some of which can be further optimized to serve as lead molecules for the development of potential drug candidates for DENV[37]. Based on the conservation of the NS2B/NS3 protease in the flavivirus genus, a multi-target drug development approach is more commonly explored. However, as part of the drug discovery and development process, a more focused approach for the selective targeting of dengue viral protease should also be considered. This can be achieved by investigating other less conserved proteases or exploring novel allosteric sites with less conservation among the flaviviruses.
The currently high incidence and mortality rates of the DENV require urgent public health attention. There is yet no approved drug for the treatment and prophylaxis of the viral infection. Therefore, endeavors to discover new anti-DENV drugs need to be heightened. With the ocean being a known source of bioactive specialized metabolites, marine natural products have high potential as sources of these novel drug molecules. Bioinformatic and computational advancements may aid and accelerate these discoveries.
| 1. | Sinha S, Singh K, Ravi Kumar YS, Roy R, Phadnis S, Meena V, Bhattacharyya S, Verma B. Dengue virus pathogenesis and host molecular machineries. J Biomed Sci. 2024;31:43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 49] [Reference Citation Analysis (0)] |
| 2. | Haider N, Hasan MN, Onyango J, Billah M, Khan S, Papakonstantinou D, Paudyal P, Asaduzzaman M. Global dengue epidemic worsens with record 14 million cases and 9000 deaths reported in 2024. Int J Infect Dis. 2025;158:107940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 35] [Reference Citation Analysis (0)] |
| 3. | Witte P, Venturini S, Meyer H, Zeller A, Christ M. Dengue Fever—Diagnosis, Risk Stratification, and Treatment. Dtsch Arztebl Int. 2024;121:773-778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 4. | Haider N, Hasan MN, Onyango J, Asaduzzaman M. Global landmark: 2023 marks the worst year for dengue cases with millions infected and thousands of deaths reported. IJID Reg. 2024;13:100459. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 29] [Reference Citation Analysis (0)] |
| 5. | Stanaway JD, Shepard DS, Undurraga EA, Halasa YA, Coffeng LE, Brady OJ, Hay SI, Bedi N, Bensenor IM, Castañeda-Orjuela CA, Chuang TW, Gibney KB, Memish ZA, Rafay A, Ukwaja KN, Yonemoto N, Murray CJL. The global burden of dengue: an analysis from the Global Burden of Disease Study 2013. Lancet Infect Dis. 2016;16:712-723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 567] [Cited by in RCA: 744] [Article Influence: 74.4] [Reference Citation Analysis (0)] |
| 6. | Rahman MS, Faruk MO, Tanjila S, Sabbir NM, Haider N, Chowdhury S. Entomological survey for identification of Aedes larval breeding sites and their distribution in Chattogram, Bangladesh. Beni-Suef Univ J Basic Appl Sci. 2021;10:32. [RCA] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 7. | Messina JP, Brady OJ, Golding N, Kraemer MUG, Wint GRW, Ray SE, Pigott DM, Shearer FM, Johnson K, Earl L, Marczak LB, Shirude S, Davis Weaver N, Gilbert M, Velayudhan R, Jones P, Jaenisch T, Scott TW, Reiner RC Jr, Hay SI. The current and future global distribution and population at risk of dengue. Nat Microbiol. 2019;4:1508-1515. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 763] [Cited by in RCA: 768] [Article Influence: 109.7] [Reference Citation Analysis (0)] |
| 8. | Perera R, Kuhn RJ. Structural proteomics of dengue virus. Curr Opin Microbiol. 2008;11:369-377. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 345] [Cited by in RCA: 319] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 9. | Hozáková L, Vokatá B, Ruml T, Ulbrich P. Targeting the Virus Capsid as a Tool to Fight RNA Viruses. Viruses. 2022;14:174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 10. | Qin CF, Qin ED. Capsid-targeted viral inactivation can destroy dengue 2 virus from within in vitro. Arch Virol. 2006;151:379-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 11. | Novikova M, Zhang Y, Freed EO, Peng K. Multiple Roles of HIV-1 Capsid during the Virus Replication Cycle. Virol Sin. 2019;34:119-134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 77] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 12. | Yi M, Lin S, Zhang B, Jin H, Ding L. Antiviral potential of natural products from marine microbes. Eur J Med Chem. 2020;207:112790. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 13. | Gogineni V, Schinazi RF, Hamann MT. Role of Marine Natural Products in the Genesis of Antiviral Agents. Chem Rev. 2015;115:9655-9706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 81] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 14. | Yasuhara-Bell J, Lu Y. Marine compounds and their antiviral activities. Antiviral Res. 2010;86:231-240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 135] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 15. | Barzkar N, Tamadoni Jahromi S, Poorsaheli HB, Vianello F. Metabolites from Marine Microorganisms, Micro, and Macroalgae: Immense Scope for Pharmacology. Mar Drugs. 2019;17:464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 78] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 16. | Kumar A, Soratur A, Kumar S, Venmathi Maran BA. A Review of Marine Algae as a Sustainable Source of Antiviral and Anticancer Compounds. Macromol. 2025;5:11. [DOI] [Full Text] |
| 17. | Panwong S, Phinyo K, Duangjan K, Sattayawat P, Pekkoh J, Tragoolpua Y, Yenchitsomanus PT, Panya A. Inhibition of dengue virus infection in vitro by fucoidan and polysaccharide extract from marine alga Sargassum spp. Int J Biol Macromol. 2024;276:133496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 18. | Pradhan B, Nayak R, Patra S, Bhuyan PP, Behera PK, Mandal AK, Behera C, Ki JS, Adhikary SP, MubarakAli D, Jena M. A state-of-the-art review on fucoidan as an antiviral agent to combat viral infections. Carbohydr Polym. 2022;291:119551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 53] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 19. | Talarico LB, Pujol CA, Zibetti RG, Faría PC, Noseda MD, Duarte ME, Damonte EB. The antiviral activity of sulfated polysaccharides against dengue virus is dependent on virus serotype and host cell. Antiviral Res. 2005;66:103-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 203] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 20. | Wu DL, Li HJ, Smith DR, Jaratsittisin J, Xia-Ke-Er XF, Ma WZ, Guo YW, Dong J, Shen J, Yang DP, Lan WJ. Polyketides and Alkaloids from the Marine-Derived Fungus Dichotomomyces cejpii F31-1 and the Antiviral Activity of Scequinadoline A against Dengue Virus. Mar Drugs. 2018;16:229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 21. | Bardiot D, Koukni M, Smets W, Carlens G, McNaughton M, Kaptein S, Dallmeier K, Chaltin P, Neyts J, Marchand A. Discovery of Indole Derivatives as Novel and Potent Dengue Virus Inhibitors. J Med Chem. 2018;61:8390-8401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 22. | Kaptein SJF, Goethals O, Kiemel D, Marchand A, Kesteleyn B, Bonfanti JF, Bardiot D, Stoops B, Jonckers THM, Dallmeier K, Geluykens P, Thys K, Crabbe M, Chatel-Chaix L, Münster M, Querat G, Touret F, de Lamballerie X, Raboisson P, Simmen K, Chaltin P, Bartenschlager R, Van Loock M, Neyts J. A pan-serotype dengue virus inhibitor targeting the NS3-NS4B interaction. Nature. 2021;598:504-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 134] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 23. | Goethals O, Kaptein SJF, Kesteleyn B, Bonfanti JF, Van Wesenbeeck L, Bardiot D, Verschoor EJ, Verstrepen BE, Fagrouch Z, Putnak JR, Kiemel D, Ackaert O, Straetemans R, Lachau-Durand S, Geluykens P, Crabbe M, Thys K, Stoops B, Lenz O, Tambuyzer L, De Meyer S, Dallmeier K, McCracken MK, Gromowski GD, Rutvisuttinunt W, Jarman RG, Karasavvas N, Touret F, Querat G, de Lamballerie X, Chatel-Chaix L, Milligan GN, Beasley DWC, Bourne N, Barrett ADT, Marchand A, Jonckers THM, Raboisson P, Simmen K, Chaltin P, Bartenschlager R, Bogers WM, Neyts J, Van Loock M. Blocking NS3-NS4B interaction inhibits dengue virus in non-human primates. Nature. 2023;615:678-686. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 81] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 24. | Kesteleyn B, Bardiot D, Bonfanti JF, De Boeck B, Goethals O, Kaptein SJF, Stoops B, Coesemans E, Fortin J, Muller P, Doublet F, Carlens G, Koukni M, Smets W, Raboisson P, Chaltin P, Simmen K, Van Loock M, Neyts J, Marchand A, Jonckers THM. Discovery of Acyl-Indole Derivatives as Pan-Serotype Dengue Virus NS4B Inhibitors. J Med Chem. 2023;66:8808-8821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 25. | Morehouse NJ, Clark TN, McMann EJ, van Santen JA, Haeckl FPJ, Gray CA, Linington RG. Annotation of natural product compound families using molecular networking topology and structural similarity fingerprinting. Nat Commun. 2023;14:308. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 48] [Reference Citation Analysis (0)] |
| 26. | Miao J, Yuan H, Rao J, Zou J, Yang K, Peng G, Cao S, Chen H, Song Y. Identification of a small compound that specifically inhibits Zika virus in vitro and in vivo by targeting the NS2B-NS3 protease. Antiviral Res. 2022;199:105255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 27. | Norshidah H, Leow CH, Ezleen KE, Wahab HA, Vignesh R, Rasul A, Lai NS. Assessing the potential of NS2B/NS3 protease inhibitors biomarker in curbing dengue virus infections: In silico vs. In vitro approach. Front Cell Infect Microbiol. 2023;13:1061937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 25] [Reference Citation Analysis (0)] |
| 28. | Starvaggi J, Previti S, Zappalà M, Ettari R. The Inhibition of NS2B/NS3 Protease: A New Therapeutic Opportunity to Treat Dengue and Zika Virus Infection. Int J Mol Sci. 2024;25:4376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 29. | Erbel P, Schiering N, D'Arcy A, Renatus M, Kroemer M, Lim SP, Yin Z, Keller TH, Vasudevan SG, Hommel U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol. 2006;13:372-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 459] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 30. | Wang Y, Sun L, Fernandes L, Wang YH, Zou J, Franklin SJ, Hu Y, Palmer LK, Yeung J, Barriga D, Russell WK, Moquin SA, Shi PY, Skepper C, Xie X. Mechanistic insights into dengue virus inhibition by a clinical trial compound NITD-688. Proc Natl Acad Sci USA. 2025;122:e2426922122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 31. | Sujitha S, Murugesan R. Rosmarinic acid and dengue virus: Computational insights into antiviral potential. LabMed Discovery. 2025;2:100042. [DOI] [Full Text] |
| 32. | Stalin A, Han J, Daniel Reegan A, Ignacimuthu S, Liu S, Yao X, Zou Q. Exploring the antiviral inhibitory activity of Niloticin against the NS2B/NS3 protease of Dengue virus (DENV2). Int J Biol Macromol. 2024;277:133791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 33. | Hossain MS, Hasnat S, Akter S, Mim MM, Tahcin A, Hoque M, Sutradhar D, Keya MAA, Sium NR, Hossain S, Masuma R, Rakib SH, Islam MA, Islam T, Bhattacharya P, Hoque MN. Computational identification of Vernonia cinerea-derived phytochemicals as potential inhibitors of nonstructural protein 1 (NSP1) in dengue virus serotype-2. Front Pharmacol. 2024;15:1465827. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 34. | Mustafa NF, Cheng KK, Razali SA, Wahab HA, Salin NH, Zakaria II, Nadri MH. Evaluation of methoxyflavones as dengue NS2B-NS3 protease inhibitors: an in silico and in vitro studies. Mol Divers. 2025;29:1175-1187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 35. | Khan RA, Hossain R, Siyadatpanah A, Al-Khafaji K, Khalipha ABR, Dey D, Asha UH, Biswas P, Saikat ASM, Chenari HA, Wilairatana P, Islam MT. Diterpenes/Diterpenoids and Their Derivatives as Potential Bioactive Leads against Dengue Virus: A Computational and Network Pharmacology Study. Molecules. 2021;26:6821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 36. | Tahir Ul Qamar M, Maryam A, Muneer I, Xing F, Ashfaq UA, Khan FA, Anwar F, Geesi MH, Khalid RR, Rauf SA, Siddiqi AR. Computational screening of medicinal plant phytochemicals to discover potent pan-serotype inhibitors against dengue virus. Sci Rep. 2019;9:1433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 87] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 37. | Bhat BA, Algaissi A, Khamjan NA, Dar TUH, Dar SA, Varadharajan V, Qasir NA, Lohani M. Exploration of comprehensive marine natural products database against dengue viral non-structural protein 1 using high-throughput computational studies. J Biomol Struct Dyn. 2025;43:3276-3285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/