Published online Aug 26, 2021. doi: 10.12998/wjcc.v9.i24.7139
Peer-review started: February 21, 2021
First decision: May 6, 2021
Revised: May 23, 2021
Accepted: June 7, 2021
Article in press: June 7, 2021
Published online: August 26, 2021
Processing time: 183 Days and 14.6 Hours
During meiosis, the recombination of homologous chromosomes produces some new heritable mutations, which are the basis of biological evolution and diversity. However, when there is pericentric inversion of chromosomes, unbalanced gametes will be formed in the process of germ cell meiosis.
A 23-year-old pregnant woman at 25 wk of gestation wanted to terminate her pregnancy due to fetal chromosomal abnormalities. She had no exposure to toxic or hazardous substances before and during pregnancy, no history of medication usage during pregnancy, and she underwent cystectomy of ovarian cysts in 2017. On the second day of the 16th week of gestation, non-invasive prenatal testing showed chromosome 8 copy number variation. Following genetic counseling, her pregnancy was terminated.
Recombinant offspring chromosome is rarely seen when the inversion segment is shorter than one-third of the chromosome length. In terms of the mechanism of chromosome 8 duplication/deletion occurrence, attention should be paid to the production of unbalanced gametes by the pairing of homologous chromosome during meiosis, and the possibility of mitotic recombination exchange as well.
Core Tip: The mechanism of partial deletion/duplication at the end of chromosome 8 involves two prevailing theories: Parental chromosome 8 inversion producing un
- Citation: Jiang Y, Tang S, He F, Yuan JX, Zhang Z. New mechanism of partial duplication and deletion of chromosome 8: A case report. World J Clin Cases 2021; 9(24): 7139-7145
- URL: https://www.wjgnet.com/2307-8960/full/v9/i24/7139.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i24.7139
The occurrence of duplicated and deleted offspring chromosome ends is commonly triggered by the joint pairing of pericentric inversion of chromosomes and homo
A 23-year-old pregnant woman at 25 wk of gestation wanted to terminate her preg
On the second day of the 16th week of gestation, non-invasive prenatal testing (NIPT) showed chromosome 8 copy number variation.
The patient underwent a cystectomy for benign ovarian cysts in 2017.
The patient had no special personal and family history.
The pregnant woman’s uterine height was 23 cm, abdominal circumference was 84 cm and blood pressure was 114/64 mmHg.
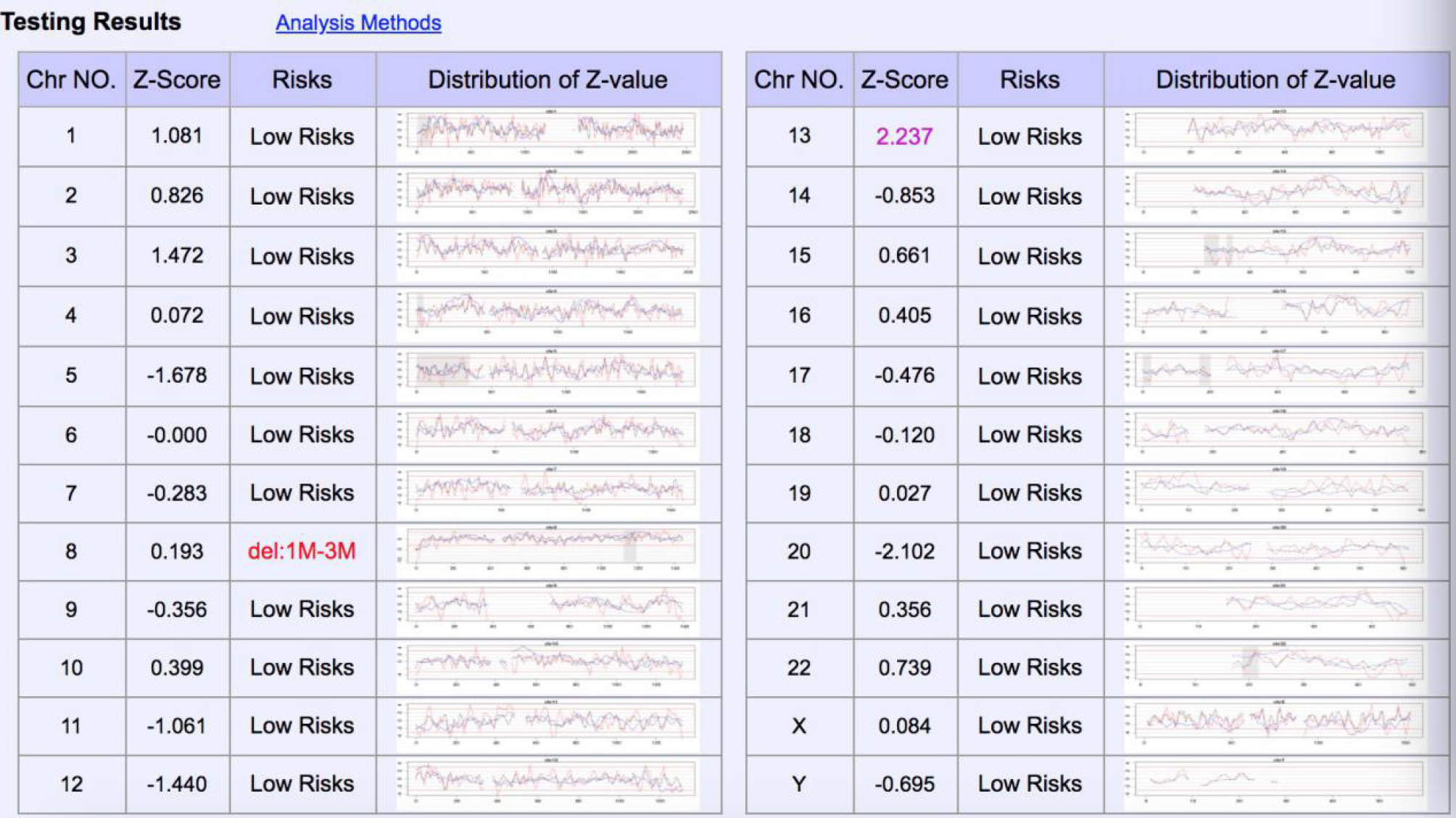
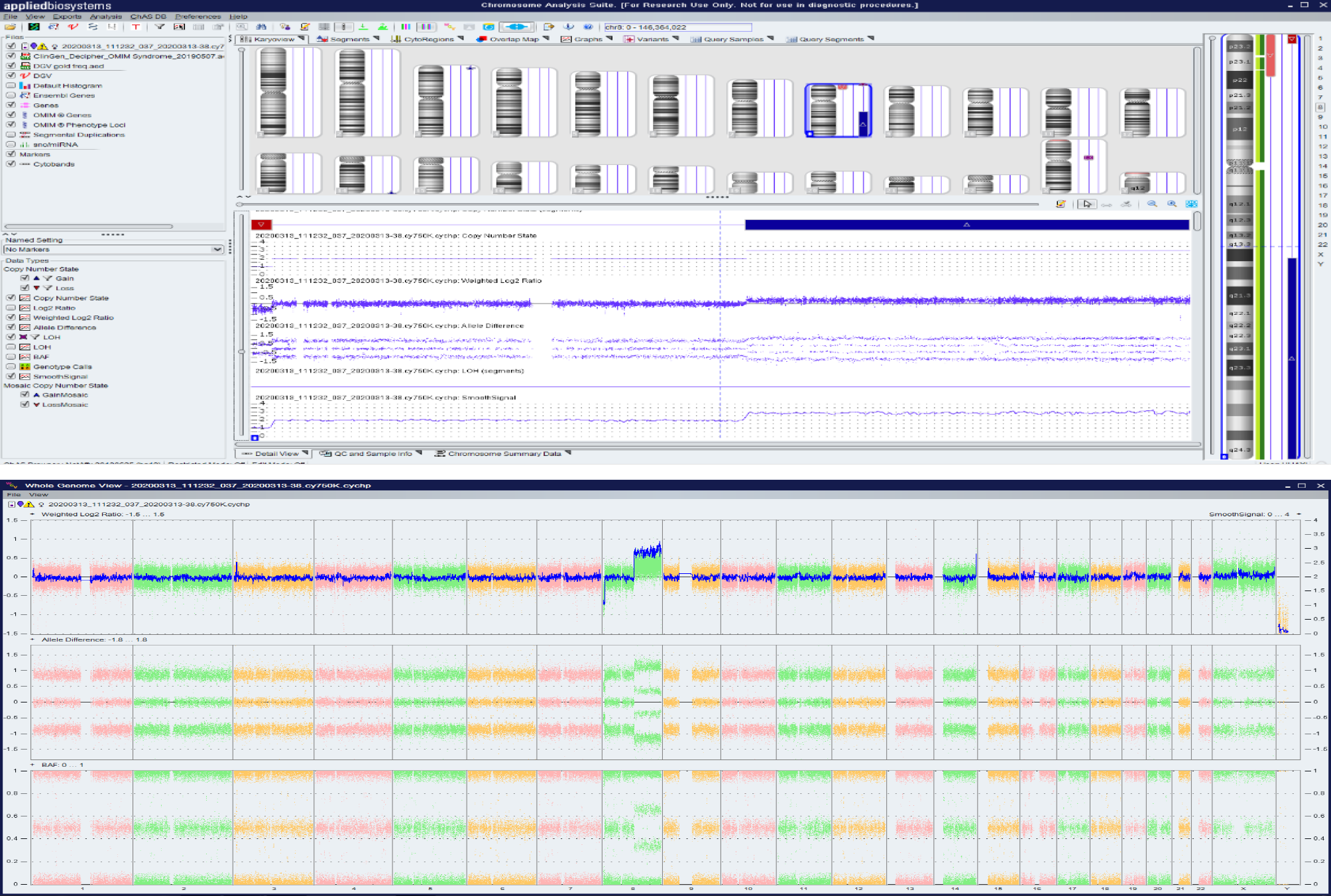
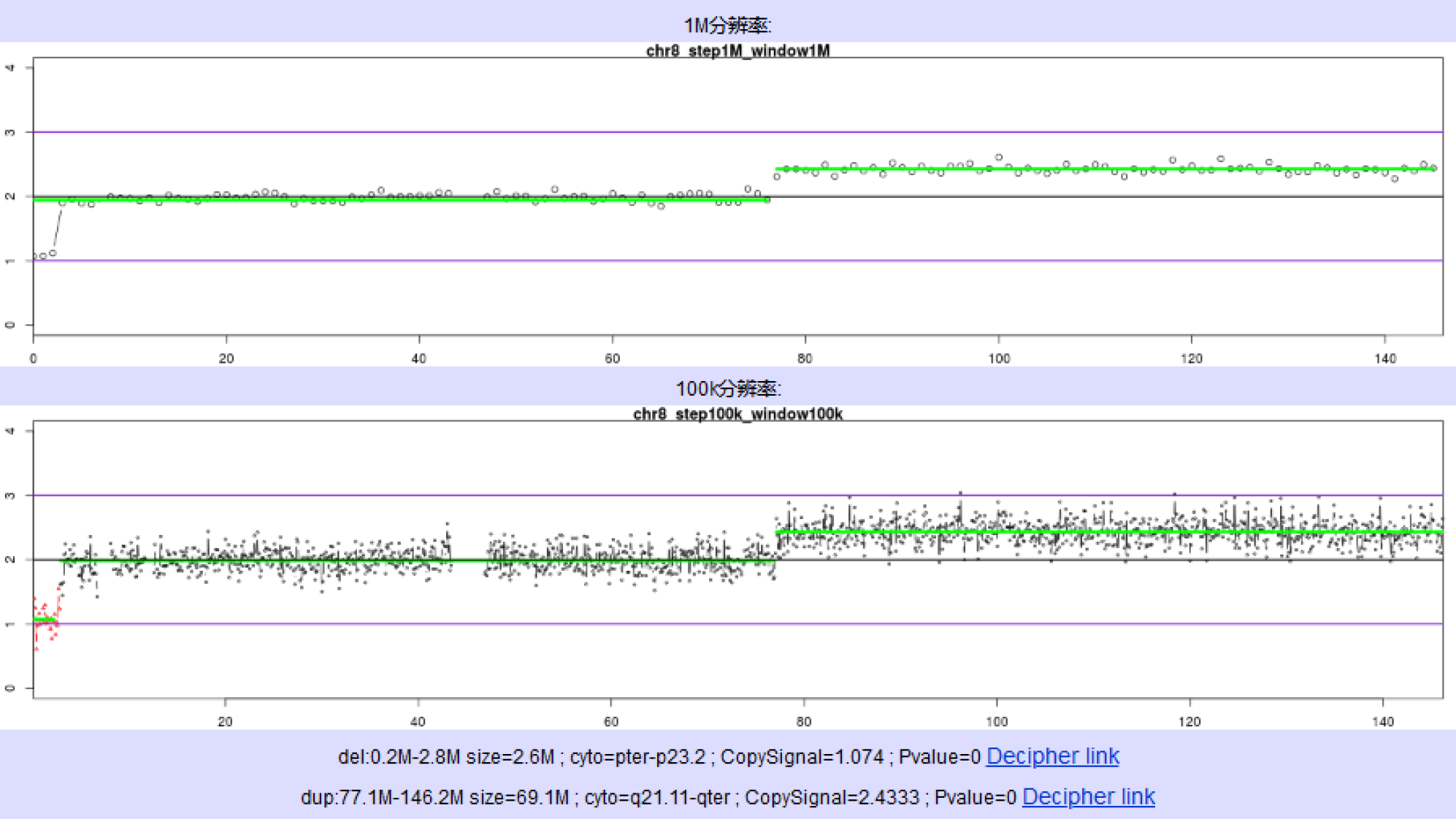
NIPT showed that there was low-risk syndrome of chromosome 13, 18 and 21 and high risk of the end of the short arm of chromosome 8 missing about 3 Mb (Figure 1). Amniocentesis chromosome microarray analysis showed: arr[GRCH37]8p23.3p23.2 (158048-3220759)x1,8q21.11q24.3(77115706-146295771)x3 (Figure 2).
Systematic ultrasonography showed that the fetal ventricles were widened bilaterally, and the measured value of the septum pellucidum was smaller than the normal. Cardiac ultrasound suggested fetal venous catheter occlusion or absence.
The fetus had an abnormal copy number of chromosome 8 and restricted placental mosaicism.
The pregnancy was terminated after genetic counseling.
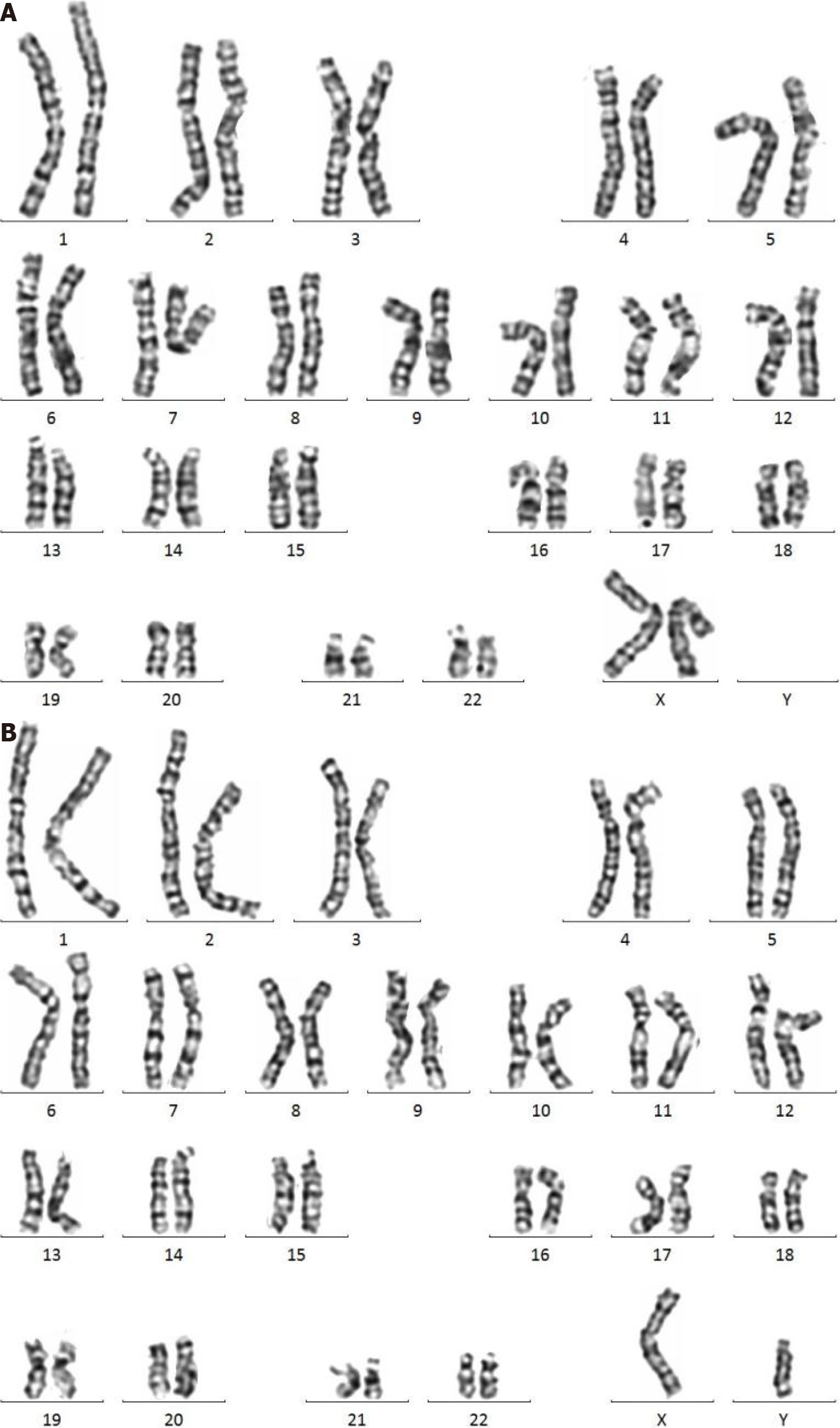
The couple underwent peripheral blood karyotype examination, and no significant abnormalities were seen in the G-dominant band (400 bands). They have no plans for another pregnancy.
In the present case, NIPT showed that the fetus may have a terminal deletion of chromosome 8p (Figure 1), and amniocentesis chromosome microarray analysis show
All the chromosomes, mostly chromosomes 2 and 8, are known to be involved in pericentric inversions[8]. Carriers of these inversions can produce a significant per
Chromosome 8p is especially prone to various genomic rearrangements mainly due to the existence of the two olfactory receptor gene clusters (REPD and REPP) of 8p23.1[10-12].
In the present case, the chromosome microarray analysis indicated a deletion of 8p and a duplication of 8q, and pericentric inversions of chromosome 8 were not found in the couple’s G-dominant band (400 bands) of chromosomal karyotype (Figure 4). CNV-seq of the placenta indicated a deletion of 8p and a duplication of 8q with 40% mosaicism (Figure 3). All the above data indicated that the short-arm deletion and long-arm duplication of fetal chromosome 8 were new mutations. The deletion of chromosome 8p is presumed to have a high possibility of a deletion in the meiotic homologous chromosome synapsis and exchange, which is consistent with the high recombination rate of the terminal arm of chromosome 8 based on the database of recombination rates of human homologous chromosomes[3]. Cases of terminal de
Recombinant offspring chromosomes are rarely seen when the inversion segment is shorter than one-third of the chromosome length. The extent of the genetic imbalance of these recombinants depends on the relative size of the inversion segment. In terms of the mechanism of chromosome 8 duplication/deletion occurrence, attention should be paid to the production of unbalanced gametes by the pairing of homologous chromosome during meiosis, and the possibility of mitotic recombination exchange as well.
| 1. | Caer E, Perrin A, Douet-Guilbert N, Amice V, De Braekeleer M, Morel F. Differing mechanisms of meiotic segregation in spermatozoa from three carriers of a pericentric inversion of chromosome 8. Fertil Steril. 2008;89:1637-1640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 2. | Vermeesch JR, Thoelen R, Salden I, Raes M, Matthijs G, Fryns JP. Mosaicism del(8p)/inv dup(8p) in a dysmorphic female infant: a mosaic formed by a meiotic error at the 8p OR gene and an independent terminal deletion event. J Med Genet. 2003;40:e93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 3. | Giorda R, Ciccone R, Gimelli G, Pramparo T, Beri S, Bonaglia MC, Giglio S, Genuardi M, Argente J, Rocchi M, Zuffardi O. Two classes of low-copy repeats comediate a new recurrent rearrangement consisting of duplication at 8p23.1 and triplication at 8p23.2. Hum Mutat. 2007;28:459-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 38] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 4. | Yin AH, Peng CF, Zhao X, Caughey BA, Yang JX, Liu J, Huang WW, Liu C, Luo DH, Liu HL, Chen YY, Wu J, Hou R, Zhang M, Ai M, Zheng L, Xue RQ, Mai MQ, Guo FF, Qi YM, Wang DM, Krawczyk M, Zhang D, Wang YN, Huang QF, Karin M, Zhang K. Noninvasive detection of fetal subchromosomal abnormalities by semiconductor sequencing of maternal plasma DNA. Proc Natl Acad Sci USA. 2015;112:14670-14675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 100] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 5. | Wang Q, Zeng XL, Yu L, Zhao SY. Clinical research progress of non-invasive prenatal detection for fetal chromosome microdeletion/microrepetition. Shiyong Fuke Neifenmi Dianzi Zazhi. 2020;13:22-24. [DOI] [Full Text] |
| 6. | Ye XQ, Gao Y, Song XW, Wu XJ, Chen JY, Wang JY, Yan HC, Chen M. Evaluation of Detection Efficiency for Fetal Chromosome Copy Number Varia- tion Measured by Low-Depth Sequencing Noninvasive Prenatal Test. Shiyong Fuchanke Zazhi. 2020;36:380-384. |
| 7. | Yu D, Zhang K, Han M, Pan W, Chen Y, Wang Y, Jiao H, Duan L, Zhu Q, Song X, Hong Y, Chen C, Wang J, Hui F, Huang L, Du Y. Noninvasive prenatal testing for fetal subchromosomal copy number variations and chromosomal aneuploidy by low-pass whole-genome sequencing. Mol Genet Genomic Med. 2019;7:e674. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 8. | Jaarola M, Martin RH, Ashley T. Direct evidence for suppression of recombination within two pericentric inversions in humans: a new sperm-FISH technique. Am J Hum Genet. 1998;63:218-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 53] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 9. | Vera-Carbonell A, López-González V, Bafalliu JA, Piñero-Fernández J, Susmozas J, Sorli M, López-Pérez R, Fernández A, Guillén-Navarro E, López-Expósito I. Pre- and postnatal findings in a patient with a novel rec(8)dup(8q)inv(8)(p23.2q22.3) associated with San Luis Valley syndrome. Am J Med Genet A. 2013;161A:2369-2375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 10. | Hollox EJ, Barber JC, Brookes AJ, Armour JA. Defensins and the dynamic genome: what we can learn from structural variation at human chromosome band 8p23.1. Genome Res. 2008;18:1686-1697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 75] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 11. | Cooke SL, Northup JK, Champaige NL, Zinser W, Edwards PA, Lockhart LH, Velagaleti GV. Molecular cytogenetic characterization of a unique and complex de novo 8p rearrangement. Am J Med Genet A. 2008;146A:1166-1172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 12. | Shimokawa O, Miyake N, Yoshimura T, Sosonkina N, Harada N, Mizuguchi T, Kondoh S, Kishino T, Ohta T, Remco V, Takashima T, Kinoshita A, Yoshiura K, Niikawa N, Matsumoto N. Molecular characterization of del(8)(p23.1p23.1) in a case of congenital diaphragmatic hernia. Am J Med Genet A. 2005;136:49-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 56] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 13. | Liu FR, Hao SJ, Zhang C, Zhou BB, Wang X, Zheng L. Cytogenetic and molecular genetic study of duplication deletion of 8p in a new case. Linchuang Erke Zazhi. 2020;38:707-709. [DOI] [Full Text] |
| 14. | Han X, Zhang JM, Jiang WT, Hu Q, Tao J. Cytogenetic and molecular genetic study of a case with 8p inverted duplication deletion syndrome. Zhonghua Yixue Yichuanxue Zazhi. 2010;27:361-366. [RCA] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/Licenses/by-nc/4.0/
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bolshakova GB, soleimanian S S-Editor: Gao CC L-Editor: Webster JR P-Editor: Yuan YY