Published online Aug 6, 2021. doi: 10.12998/wjcc.v9.i22.6287
Peer-review started: April 25, 2021
First decision: May 24, 2021
Revised: May 29, 2021
Accepted: June 3, 2021
Article in press: June 3, 2021
Published online: August 6, 2021
Processing time: 93 Days and 13.3 Hours
Secreted protein acidic and rich in cysteine (SPARC) is an extracellular matrix-associated protein. Studies have revealed that SPARC is involved in the cell interaction and function including proliferation, differentiation, and apoptosis. However, the role of SPARC in cancer is controversial, as it was reported as the promoter or suppressor in different cancers. Further, the role of SPARC in lymphoma is unclear.
To identify the expression and significance of SPARC in lymphoma, especially in diffuse large B-cell lymphoma (DLBCL).
The expression analysis of SPARC in different cancers was evaluated with Oncomine. The Brune, Eckerle, Piccaluga, Basso, Compagno, Alizadeh, and Rosenwald datasets were included to evaluate the mRNA expression of SPARC in lymphoma. The Cancer Genome Atlas (TCGA)-DLBCL was used to analyze the diagnostic value of SPARC in DLBCL. The Compagno and Brune DLBCL datasets were used for validation. Then, the diagnostic value was evaluated with the receiver operating characteristic (ROC) curve. The Kaplan-Meier plot was conducted with TCGA-DLBCL, and the ROC analysis was performed based on the survival time. Further, the overall survival analysis based on the level of SPARC expression was performed with the GSE4475 and E-TABM-346. The Gene Set Enrichment Analyses (GSEA) was performed to make the underlying mechanism-regulatory networks.
The pan-cancer analysis of SPARC showed that SPARC was highly expressed in the brain and central nervous system, breast, colon, esophagus, stomach, head and neck, pancreas, and sarcoma, especially in lymphoma. The overexpression of SPARC in lymphoma, especially DLBCL, was confirmed in several datasets. The ROC analysis revealed that SPARC was a valuable diagnostic biomarker. More importantly, compared with DLBCL patients with low SPARC expression, those with higher SPARC expression represented a higher overall survival rate. The ROC analysis showed that SPARC was a favorable prognostic biomarker for DLBCL. Results of the GSEA confirmed that the high expression of SPARC was closely associated with focal adhesion, extracellular matrix receptor interaction, and leukocyte transendothelial migration, which suggested that SPARC may be involved in the regulation of epithelial-mesenchymal transition, KRAS, and myogenesis in DLBCL.
SPARC was highly expressed in DLBCL, and the overexpression of SPARC showed sound diagnostic value. More interestingly, the overexpression of SPARC might be a favorable prognostic biomarker for DLBCL, suggesting that SPARC might be an inducible factor in the development of DLBCL, and inducible SPARC was negative in some oncogenic pathways. All the evidence suggested that inducible SPARC might be a good diagnostic and prognostic biomarker for DLBCL.
Core Tip: In this study, the expression and significance of secreted protein acidic and rich in cysteine (SPARC) in the diffuse large B-cell lymphoma (DLBCL) were evaluated. The overexpression of SPARC can be an efficient diagnostic and prognostic biomarker for DLBCL, suggesting that SPARC has a potential value in future clinical application.
- Citation: Pan PJ, Liu JX. Diagnostic and prognostic value of secreted protein acidic and rich in cysteine in the diffuse large B-cell lymphoma. World J Clin Cases 2021; 9(22): 6287-6299
- URL: https://www.wjgnet.com/2307-8960/full/v9/i22/6287.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i22.6287
Secreted protein acidic and rich in cysteine (SPARC) is a secreted protein, which regulates various biological activities, including proliferation, migration, adhesion, differentiation, and apoptosis[1,2]. Recently, it was reported that SPARC could control the extracellular matrix (ECM) turnover, which plays a significant role in the re
The expression of SPARC in different cancers is ragged. According to the latest reports, the expression of SPARC could be controlled by many environmental factors. Studies showed that SPARC expression can be induced remarkably with the development of lung, esophageal, pancreatic, and prostate cancer, suggesting that SPARC could be a tumor promoter[5-9]. In detail, mechanism studies revealed that SPARC regulated the tumor growth factor β (TGF-β) signaling and then promoted the epithelial-mesenchymal transition (EMT), which participates in the cancer metastasis[5,10,11]. SPARC was a tumor promoter to activate the phosphatidylinositol 3-phosphate kinase (PI3K)/AKT pro-oncogenic pathway[12,13]. Additionally, SPARC was an anti-apoptotic factor, inhibiting the caspase activity[14].
However, other reports revealed SPARC was a tumor suppressor. In the colon, prostate, ovarian, and cervical cancers, it was reported that the expression of SPARC was closely associated with negative regulation, indicating that SPARC was a favorable prognostic factor[15]. Sailaja et al[16] reported that SPARC could increase the PTEN expression, which was a negative molecule of AKT suppressor. Additionally, SPARC was reported to induce endoplasmic reticulum (ER) stress. Moreover, SPARC inhibited cell proliferation by inducing the G2/M cell cycle arrest[17]. Taken together, the above evidence suggested that SPARC might play a dual role in different cancers. However, the role of SPARC is still unclear in some cancers, including diffuse large B-cell lymphoma (DLBCL), the most common clinical lymphoma. Thus, this study aimed to evaluate the significance of SPARC in the DLBCL.
In this study, we evaluated the pan-cancer expression of SPARC, and then con
The pan-cancer expression of SPARC was confirmed in the Oncomine database[18]. The threshold for P value was set below 0.05, the fold change was over 2 times, and the top 1% gene rank was included in the results. The included datasets were shown in Table 1[19-26], including the detailed sample number and the reporter platform.
The TCGA-DLBCL database was evaluated as the training dataset. Firstly, the difference of SPARC expression was confirmed, and the ROC analysis was conducted based on the expression results. The area under curve (AUC) was calculated to evaluate the diagnostic value, and an AUC value from 0.5 to 1 was considered to be statistically significant. The closer the AUC value was to 1, the better diagnostic effect[27,28]. The Compagno and Brune DLBCL datasets were confirmed as validation datasets.
The patients from TCGA-DLBCL with information on survival status were included in the overall survival analysis for the training test. The log-rank method was used to analyze the difference between SPARC high- and low-expression groups, and hazard ratio (HR) with 95% confidence interval (CI) was set to evaluate the prognostic value of SPARC. The value of HR was below 1.0, indicating that the high-expression group predicted a favorable prognosis[29,30]. The ROC analysis based on the survival rate was conducted in 12, 36, and 60 mo, respectively. The AUC value was calculated to evaluate the statistical difference, with an AUC value over 0.5 considered significant in the prognostic prediction. The GSE4475[31] and E-TABM-346[32] datasets were included for validation tests to evaluate the prognoses of DLBCL patients. In the GSE4475 and E-TABM-346 datasets, the median expression value of SPARC was set as the cut-off, and the survival time of censored data was the time from the start event to the truncation point.
The TCGA-DLBCL data was subjected to the Gene Set Enrichment Analysis (GSEA), and the KEGG and HALLMARK modules were adopted in the SangerBox tool (http://sangerbox.com/Index). The detailed setting was as follows: weighted manner enrichment statistic; Signal2Noise was subjected to the metric for ranking genes; the number of permutations was 1000; the number of markers was above 100; the minimum gene number sets was over 15.
Results were shown in mean ± SEM, and the data was processed with Graphpad Prism 8.0 software. The difference between two groups was analyzed with Student’s t test. The AUC value of ROC over 0.50 was considered statistically significant. The survival analysis was conducted with log-rank and Cox methods with CI, respectively. P < 0.05 was considered statistically significant.
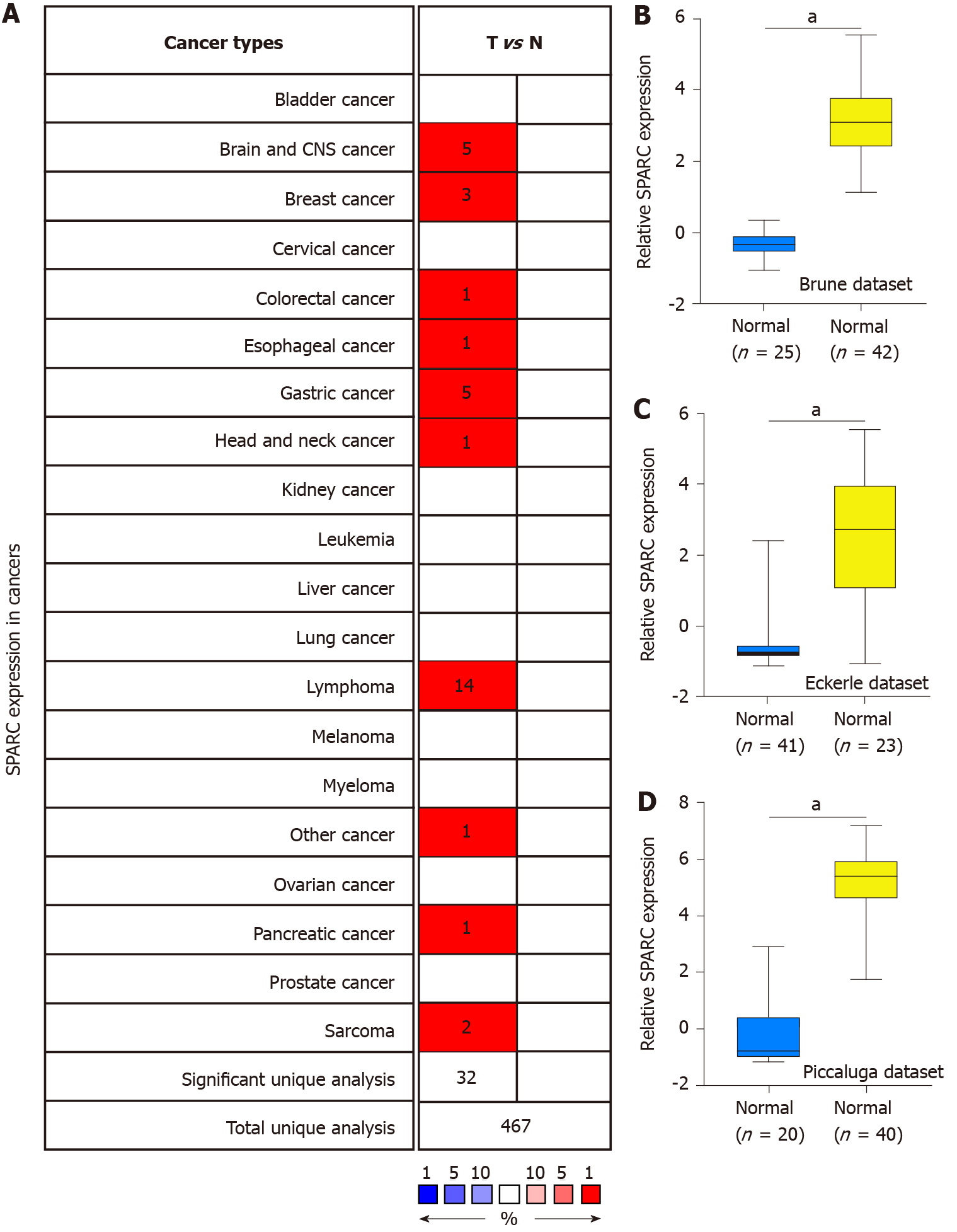
Previous studies showed that SPARC might play a dual role in some cancers[33]. To further confirm the expression of SPARC in cancers, the Oncomine database with different expression datasets was used. As is shown in Figure 1A, the differences of SPARC expression between tumors and normal tissues were confirmed, and the results showed that SPARC was highly expressed in the brain and central nervous system cancer, breast cancer, cervical cancer, colon cancer, esophageal cancer, stomach cancer, head and neck cancer, pancreatic cancer, lymphoma, and sarcoma. The most significant overexpressing cancer was the lymphoma. Considering the expression of SPARC in lymphoma was controversial, this study focused on the potential role of SPARC in lymphoma. To fully understand the role of SPARC in the lymphoma, more lymphoma datasets were included to study the expression manners. As is shown in Figure 1B-D, three independent datasets revealed that SPARC was overexpressed in lymphoma cells compared with normal control cells. Further study is warranted to understand the significance of SPARC in lymphoma.
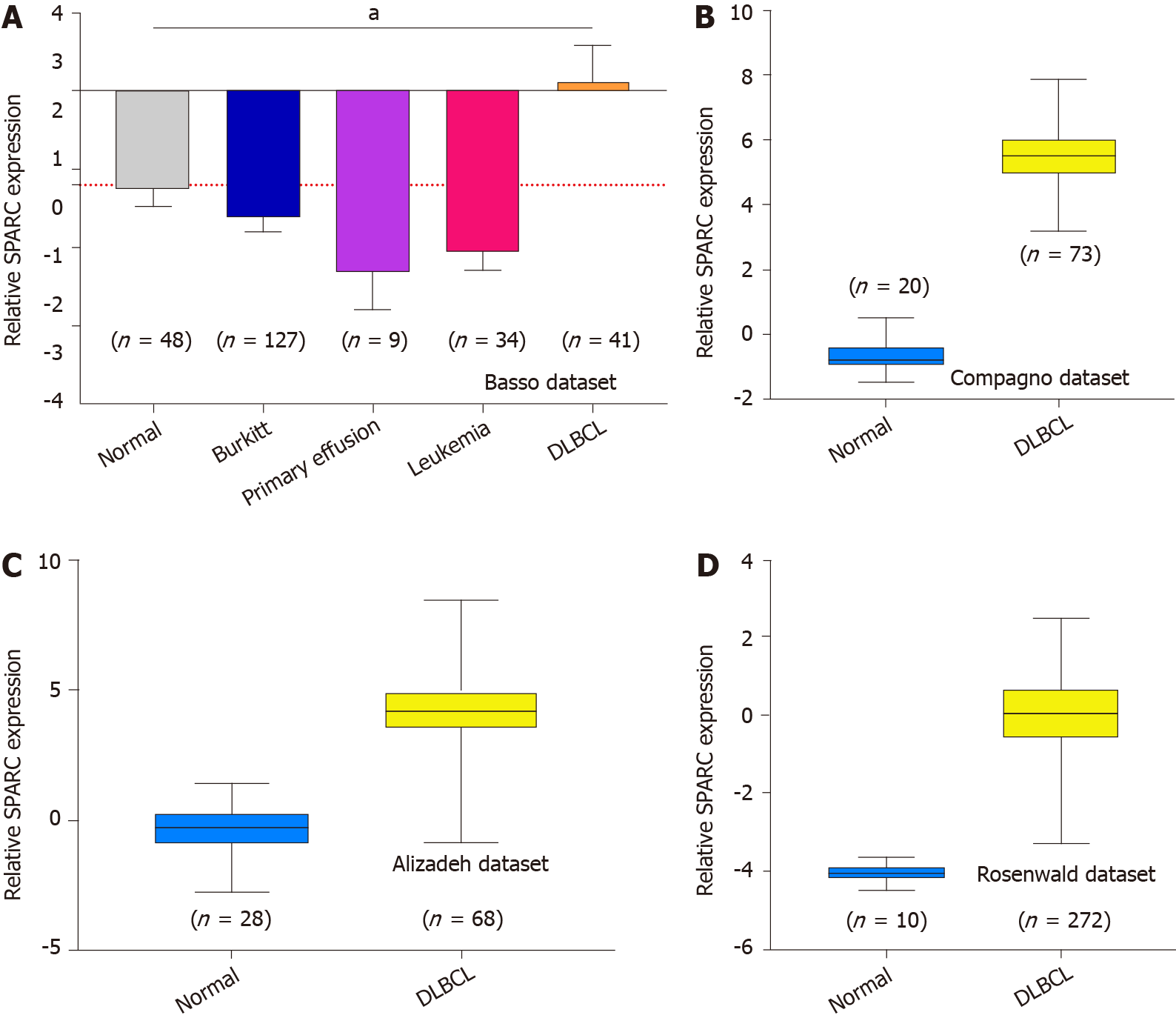
Figure 1 shows the overexpression of SPARC in lymphoma. Due to the significant effect of tumor heterogeneity and molecular typing on the development of lymphoma, this study analyzed the SPARC expression in different types of lymphoma. As is shown in Figure 2A, there was no significant difference in the SPARC expression in Burkitt lymphoma and primary effusion lymphoma. However, we found a remarkable difference in DLBCL. Leukemia was included to test the SPARC expression as a blood tumor; however, no significant difference in the SPARC expression was found. These results suggest that SPARC was highly expressed in the lymphoma, mainly in DLBCL. The overexpression of SPARC in the DLBCL was confirmed with three independent datasets, including the Compagno dataset, Alizadeh dataset, and Rosenwald dataset (Figure 2B-D). Together, these data indicate the overexpression of SPARC in lymphoma was primarily in DLBCL, suggesting that SPARC might be closely associated with certain molecular types of lymphoma.
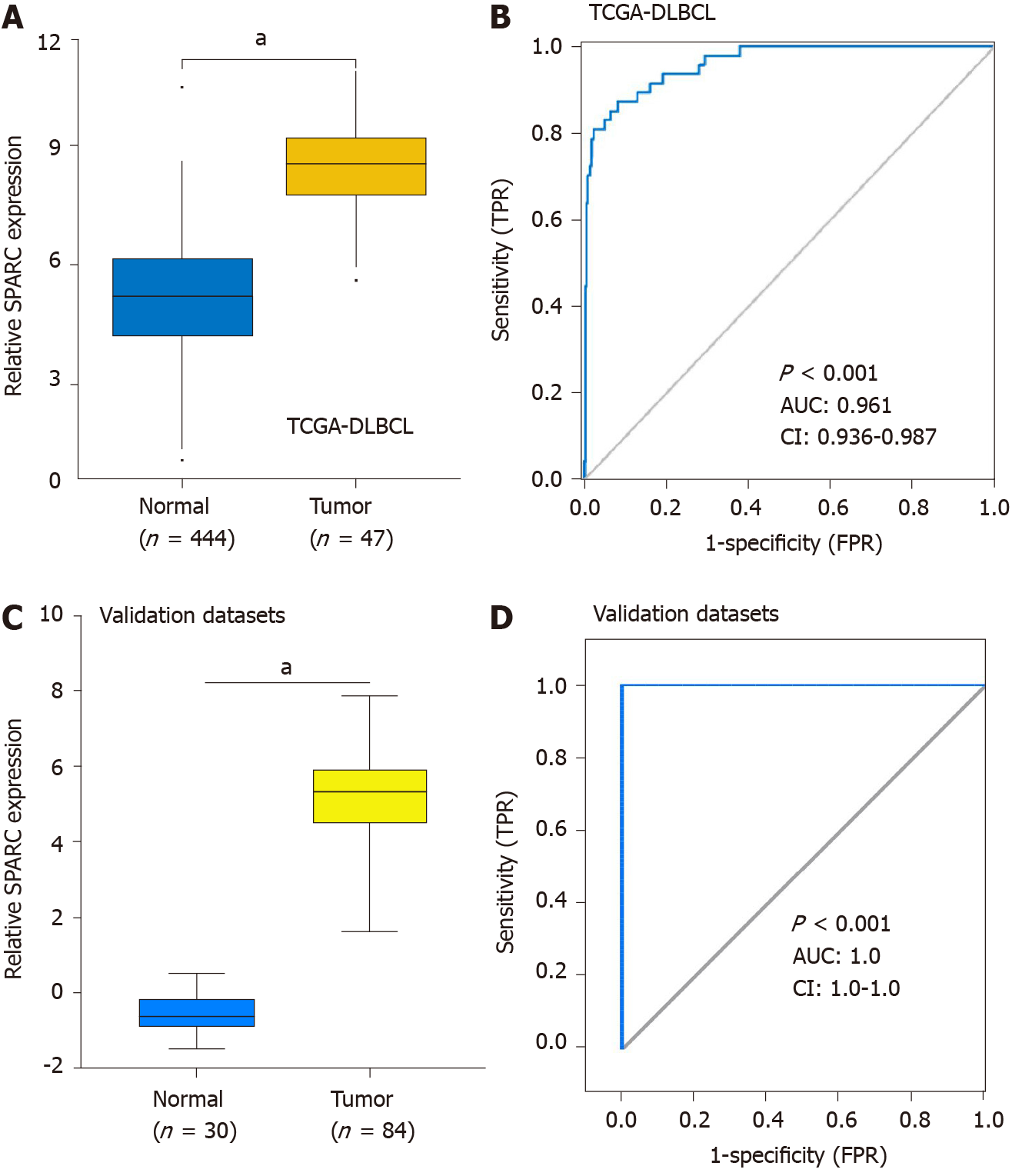
Because SPARC was specifically overexpressed in DLBCL, we hypothesized that the overexpression of SPARC had diagnostic potential for DLBCL. To validate the hypothesis, tumor and normal control cells from DLBCL patients and healthy donors were included for the ROC analysis. Firstly, the overexpression of SPARC was reconfirmed in the TCGA-DLBCL database (Figure 3A). As is shown in Figure 3B, the ROC results of TCGA-DLBCL suggested high diagnostic potential, with the AUC value of up to 0.961 (P < 0.001). Furthermore, similar results were validated in Compagno and Brune DLBCL datasets, with the AUC value close to 1.0 (P < 0.001). These results indicate that SPARC is a diagnostic marker with a high true positive rate (TPR).
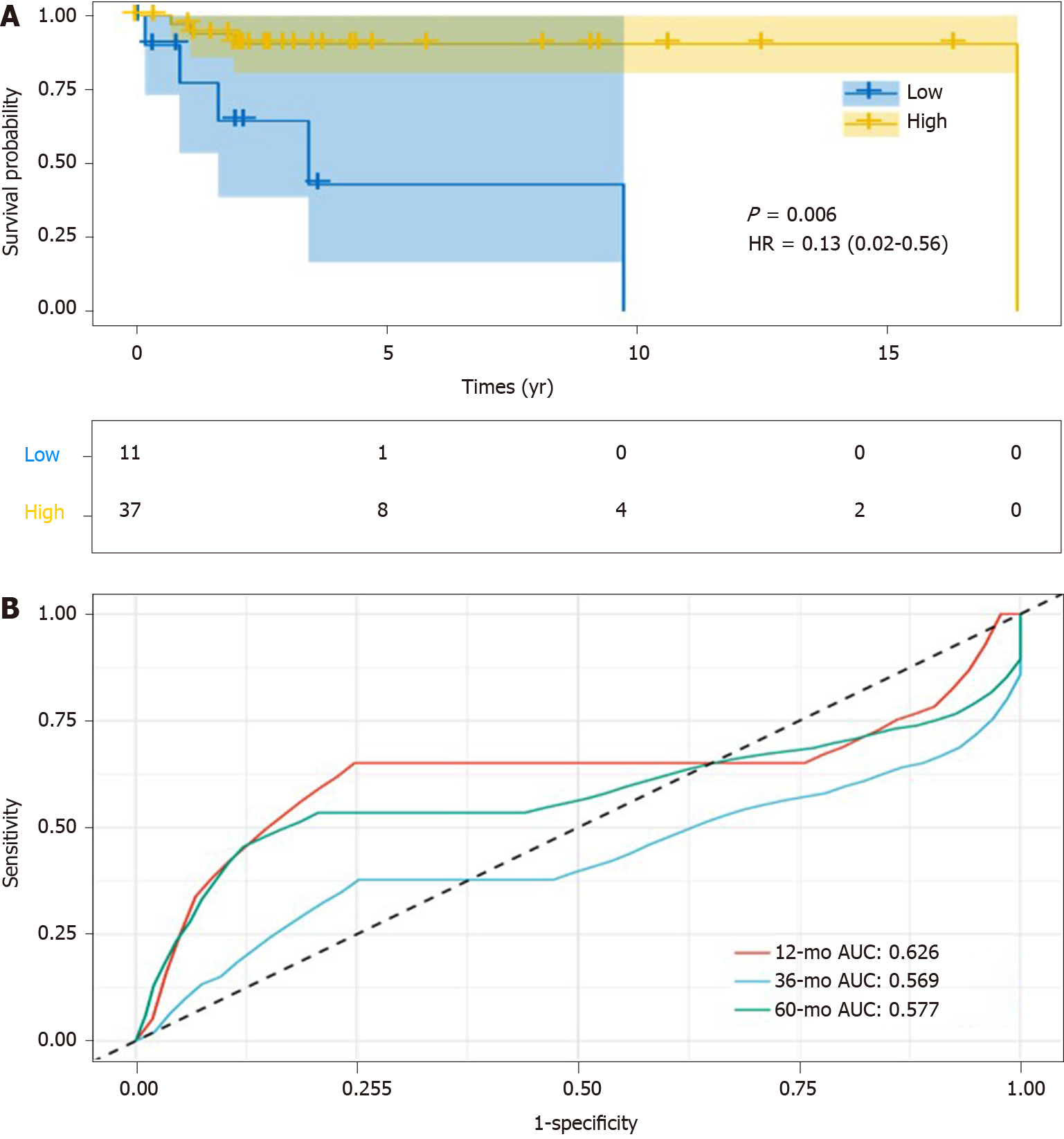
To elucidate the question of whether the overexpression of SPARC could be used as a potential diagnostic biomarker for DLBCL, the overall survival analysis was conducted. As is shown in Figure 4A, DLBCL patients with higher SPARC expression displayed longer survival time (median, 17.6 years), compared with lower SPARC levels (median, 3.4 years). The prognostic evaluation analysis was included with ROC analysis, which revealed that SPARC expression could act as a prognostic biomarker for the DLBCL patients, with the AUC value of 0.626, 0.569, and 0.577 for survival rate analysis at 1, 3, and 5 years, respectively (Figure 4B).
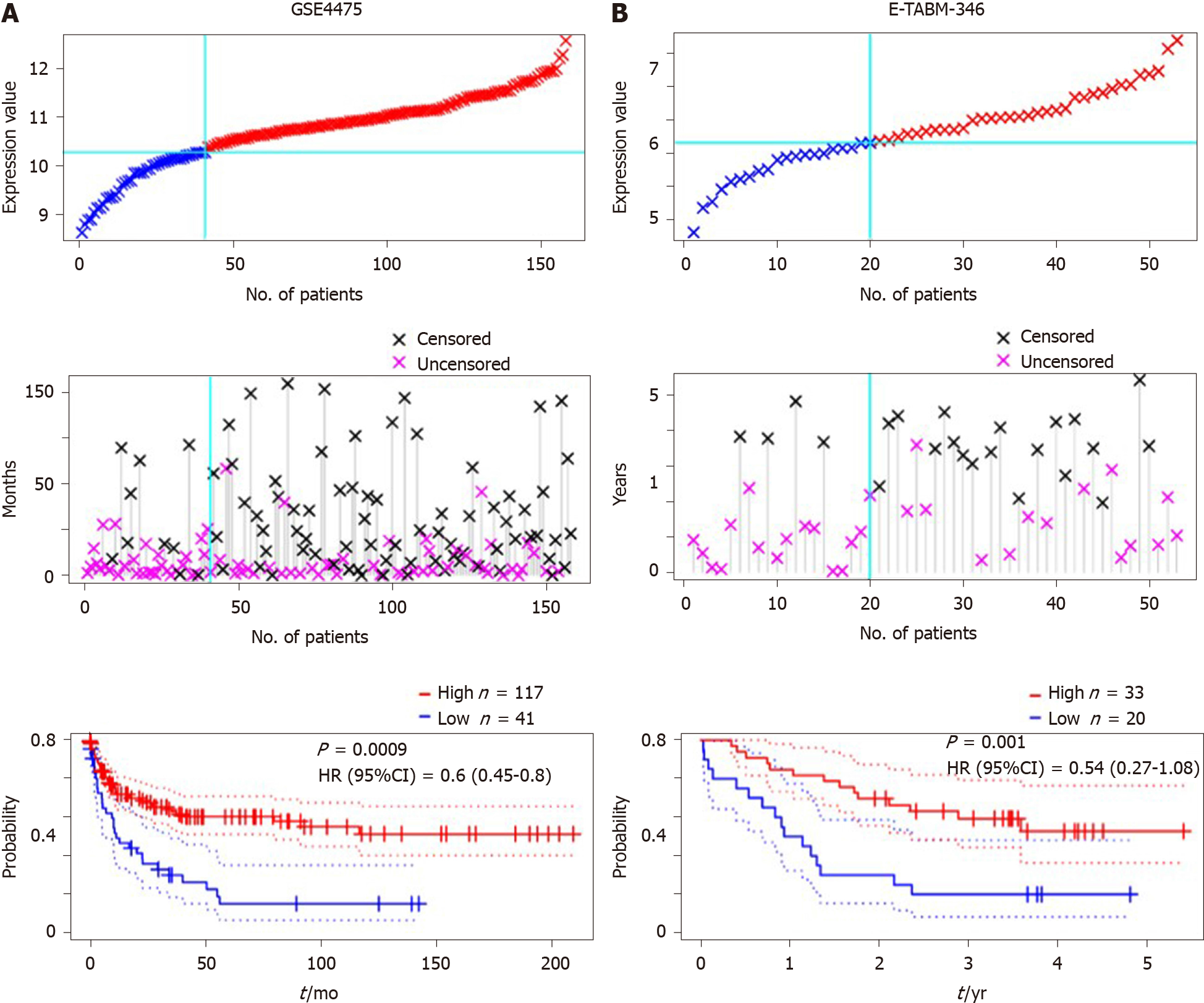
Due to the small samples of DLBCL patients with detailed clinical information, other DLBCL datasets were included to validate the prognostic value of SPARC. The GSE4475 and E-TABM-346 datasets were included, and the detailed information is presented in Table 2. The overall survival analysis was conducted based on the SPARC expression. As is shown in Figure 5A, DLBCL patients with higher SPARC expression presented a higher survival rate than those with low SPARC levels [P = 0.0009, HR = 0.6 (0.45-0.8)]. Similar results were confirmed in the E-TABM-346 dataset [P = 0.001, HR = 0.54 (0.27-1.08), Figure 5B]. Furthermore, the Cox analysis was conducted in Table 2, and the Cox analysis of SPARC was confirmed in the GSE4475 as a prognostic biomarker. However, due to the small samples, the Cox analysis of the E-TABM-346 dataset showed no statistical significance (P = 0.08).
| Features | GSE4475 | E-TABM-346 |
| Number | 158 | 53 |
| P value | 0.000024 | 0.001367 |
| Adj. P value | 0.000984 | 0.033088 |
| Cox P value | 0.000532 | 0.082877 |
| HR (95%CI) | 0.60 (0.45-0.80) | 0.54 (0.27-1.08) |
| Sex, n (%) | ||
| Female | 68 (43.0) | 26 (49.1) |
| Male | 90 (57.0) | 27 (50.9) |
| Event, n (%) | ||
| 0 | 83 (52.5) | 23 (43.4) |
| 1 | 75 (47.5) | 30 (56.6) |
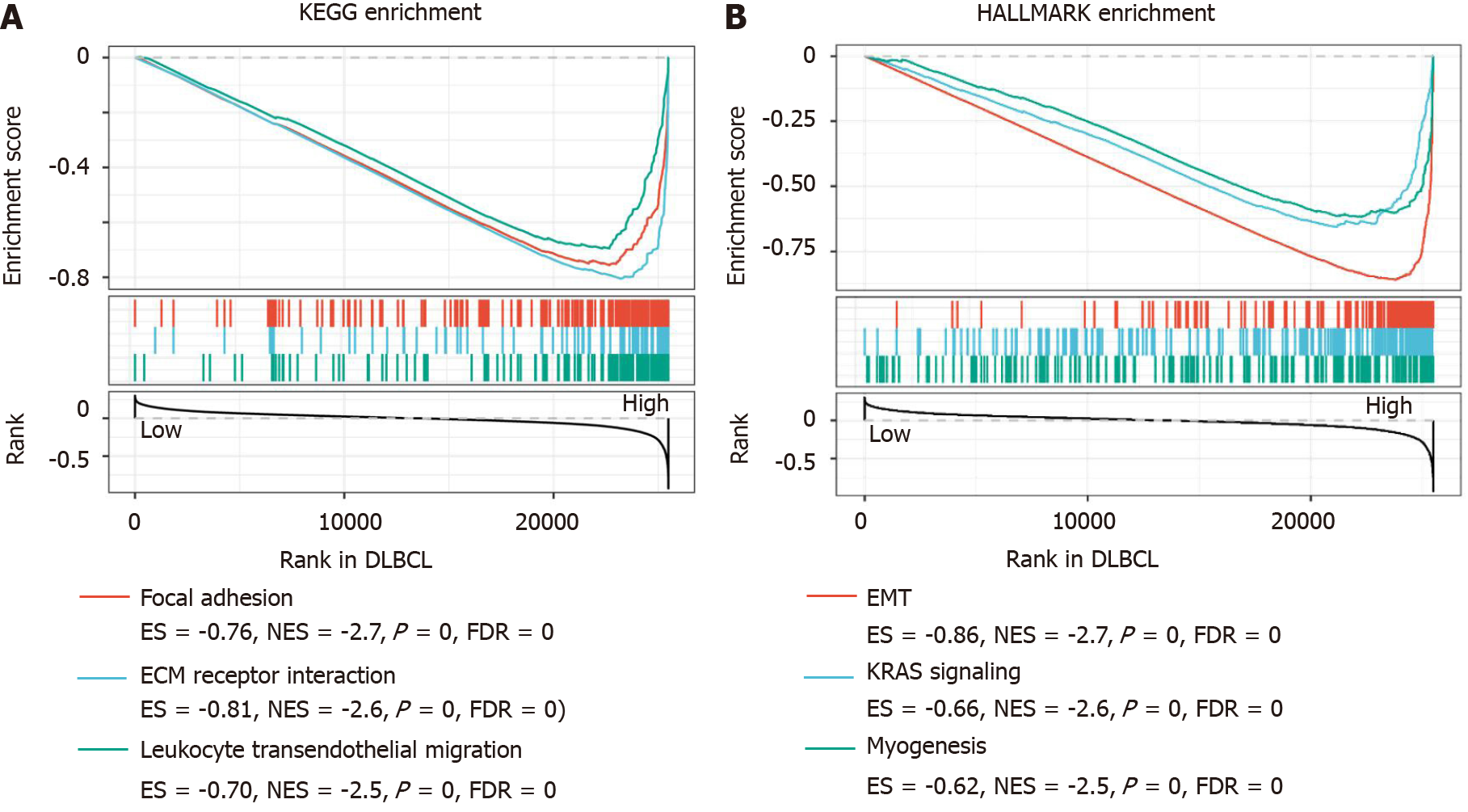
The overexpression of SPARC might act as a favorable prognostic biomarker for DLBCL. To investigate the potential pathways mediated by SPARC in DLBCL, the GSEA analysis was performed and revealed that SPARC was implicated in focal adhesion (P = 0, FDR = 0, NES = -2.7), ECM receptor interaction (P = 0, FDR = 0, NES = -2.6), and leukocyte transendothelial migration (P = 0, FDR = 0, NES = -2.5). Furthermore, as is shown in Figure 6B, SPARC in DLBCL was enriched in EMT (P = 0, FDR = 0, NES = -2.7), KRAS signaling (P = 0, FDR = 0, NES = -2.6), and myogenesis (P = 0, FDR = 0, NES = -2.5). These negative correlation results with some pro-oncogenic pathways, combined with the prognostic analysis, suggest that SPARC might work as an onco-promoter in DLBCL.
DLBCL is the most common type of lymphoma. In clinical practice, the pathological characteristics of DLBCL were in a highly heterogeneous status[34,35]. Thus, there was great variation in response to the treatment and prognoses of DLBCL patients. With the development of the next-generation sequencing, the discovery of novel prognosis biomarkers has achieved great progress. C-myc, Bcl-2, Ki-67, and CD5 were identified as independent prognostic markers for DLBCL, the overexpression of which predicts the poor prognosis of DLBCL[34,36]. However, many factors can regulate the expressions of these genes, and the prognostic value is inaccurate[37,38]. Thus, it is important to screen new biomarkers for DLBCL prognosis.
Interestingly, this study revealed that SPARC was specifically overexpressed in DLBCL, rather than other types of lymphomas (Figures 1 and 2). Previous studies showed that SPARC was overexpressed in some cancers, including melanoma, breast cancer, pancreatic cancer, lung cancer, and liver cancer, and the overexpression of SPARC might act as a tumor promoter[3,33]. In glioma cells, SPARC could activate the PI3K/AKT pathway, and inhibit the apoptotic pathway[13]. However, the molecular functions of SPARC in other cancers are completely the opposite. In neuroblastoma, SPARC induces ER stress and suppresses the AKT activity, suggesting that SPARC might play a dual role in different cancers[16]. Additionally, SPARC was reported to be downregulated in T-cell lymphoma, and acted as a tumor suppressor through the inhibition of cell proliferation and metastasis[39]. This study confirmed the overexpression and the diagnostic potential of SPARC in the DLBCL. The results showed an excellent TPR of SPARC as a diagnostic biomarker for the DLBCL (Figure 3). Considering the complex functions of SPARC in cancers, this study evaluated the significance of SPARC overexpression in the DLBCL. This study revealed that the overexpression of SPARC might represent a favorable prognosis for DLBCL patients (Figures 4 and 5), suggesting that SPARC might function as a tumor suppressor in DLBCL. Meyer et al[40] reported the positive SPARC in stromal cells in DLBCL displayed a higher survival rate than those with negative SPARC. Our study further confirmed the clinical significance of SPARC in DLBCL. The survival rate analysis and the potential mechanism network were included in this study. The negative results were associated with the biological processes, including focal adhesion, ECM receptor interaction, and leukocyte transendothelial migration (Figure 6A). Focal adhesion and ECM receptor interaction were fully studied in the migration of cancer cells[41], and leukocyte transendothelial migration had a significant effect on the endothelial barrier function[42]. The three biological processes were closely associated with the tumor development, and the negative association between SPARC and focal adhesion, ECM receptor interaction, and leukocyte transendothelial migration indicated the potential suppressing role of SPARC in DLBCL. Moreover, SPARC-associated pathways were enriched in EMT, KRAS, and myogenesis, which were implicated as tumor promoters in many cancers[43]. Thus, the analysis of SPARC-associated pathways confirmed the negative regulation of SPARC on some pro-oncogenic pathways.
This study confirmed the overexpression of SPARC in lymphoma, and its overexpression is specific for DLBCL. More importantly, the overexpression of SPARC can be a diagnostic and prognostic biomarker for the DLBCL with high clinical potential.
Secreted protein acidic and rich in cysteine (SPARC), is a protein related to the extracellular matrix. Studies have shown that SPARC regulate cell interactions and display multi-functions, including proliferation, differentiation and apoptosis. However, the role of SPARC in cancer is controversial because it is reported to be a promoter or inhibitor in different cancers. In addition, the role of SPARC in lymphoma is unclear.
The role of SPARC in different cancers is controversial, and the expression and clinical application of SPARC in lymphoma is unclear.
This study aimed to explore the expression and clinical value of SPARC in lymphoma, especially in the diffuse large B-cell lymphoma (DLBCL).
The expression of SPARC in pan-cancer was conducted in Oncomine database. The Gene Expression Omnibus including Brune, Eckerle, Piccaluga, Basso, Compagno, Alizadeh, and Rosenwald datasets were subjected to confirm the expression of SPARC. The diagnostic value of SPARC was conducted in the Cancer Genome Atlas (TCGA)-DLBCL. The validated datasets were included with Compagno and Brune DLBCL datasets. Receiver operating characteristic (ROC) curve was applied to test the diagnostic value. The survival rate was conducted with Kaplan-Meier plot in TCGA-DLBCL database. The effect of SPARC on the overall survival was also confirmed in GSE4475 and E-TABM-346. The potential signaling pathways of SPARC in DLBCL was conducted with The Gene Set Enrichment Analyses (GSEA) software.
SPARC was highly expressed in pan-cancers, including brain and central nervous system cancer, breast cancer, colorectal cancer, esophageal cancer, gastric cancer, head and neck cancer, pancreatic cancer and sarcoma, most significantly in lymphoma. The overexpression of SPARC in lymphoma was confirmed by validated datasets. This study also identified that the overexpression of SPARC occurred significantly in DLBCL. And the overexpression of SPARC in DLBCL was tested in TCGA-DLBCL, and the ROC result showed a significant value of SPARC as a biomarker for DLBCL. Furthermore, the validation datasets including Compagno and Brune datasets confirmed the excellent diagnostic value of SPARC for DLBCL. In further prognostic analysis, DLBCL patients with high SPARC expression represented a favorable survival rate, and the ROC analysis of SPARC also demonstrated that SPARC as a favorable prognostic biomarker. The results of GSEA also revealed that SPARC was closely associated with focal adhesion, extracellular matrix receptor interaction and leukocyte transendothelial migration, which was involved in the regulation of epithelial-mesenchymal transition, KRAS and myogenesis signaling pathways in DLBCL.
SPARC was overexpressed in DLBCL, showing an excellent diagnostic value. Furthermore, the overexpression of SPARC could be a favorable prognostic biomarker. The inducible SPARC was also negatively correlated with some oncogenic pathways. Overall, the inducible SPARC could serve as a good diagnostic and prognostic biomarker for DLBCL.
This study identified that SPARC as a novel biomarker for the diagnosis and prognosis of DLBCL. Also, the inducible SPARC might be negatively correlated with some oncogenic pathways, suggesting that the inducible SPARC in the development of DLBCL could guide the clinical practice of DLBCL. However, this study was based on expression level, SPARC as a secreted protein, the serum level in DLBCL patients could be included in the further study.
Manuscript source: Unsolicited manuscript
Specialty type: Hematology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Shahjaman M, Sopo SM S-Editor: Gong ZM L-Editor: Filipodia P-Editor: Liu JH
| 1. | Brekken RA, Sage EH. SPARC, a matricellular protein: at the crossroads of cell-matrix communication. Matrix Biol. 2001;19:816-827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 391] [Cited by in RCA: 434] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 2. | Wong SL, Sukkar MB. The SPARC protein: an overview of its role in lung cancer and pulmonary fibrosis and its potential role in chronic airways disease. Br J Pharmacol. 2017;174:3-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 93] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 3. | Li L, Zhu Z, Zhao Y, Zhang Q, Wu X, Miao B, Cao J, Fei S. FN1, SPARC, and SERPINE1 are highly expressed and significantly related to a poor prognosis of gastric adenocarcinoma revealed by microarray and bioinformatics. Sci Rep. 2019;9:7827. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 111] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 4. | Melouane A, Carbonell A, Yoshioka M, Puymirat J, St-Amand J. Implication of SPARC in the modulation of the extracellular matrix and mitochondrial function in muscle cells. PLoS One. 2018;13:e0192714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 5. | Sun W, Feng J, Yi Q, Xu X, Chen Y, Tang L. SPARC acts as a mediator of TGF-β1 in promoting epithelial-to-mesenchymal transition in A549 and H1299 lung cancer cells. Biofactors. 2018;44:453-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 6. | Gong L, Mao W, Chen Q, Jiang Y, Fan Y. Analysis of SPARC and TUBB3 as predictors for prognosis in esophageal squamous cell carcinoma receiving nab-paclitaxel plus cisplatin neoadjuvant chemotherapy: a prospective study. Cancer Chemother Pharmacol. 2019;83:639-647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 7. | Khetan K, Baloda V, Sahoo RK, Vishnubhathla S, Yadav R, Saraya A, Sharma A, Gupta SD, Das P. SPARC expression in desmoplastic and non desmoplastic pancreatic carcinoma and cholangiocarcinoma. Pathol Res Pract. 2019;215:152685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 8. | Liu T, Qiu X, Zhao X, Yang R, Lian H, Qu F, Li X, Guo H. Hypermethylation of the SPARC promoter and its prognostic value for prostate cancer. Oncol Rep. 2018;39:659-666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (1)] |
| 9. | López-Moncada F, Torres MJ, Castellón EA, Contreras HR. Secreted protein acidic and rich in cysteine (SPARC) induces epithelial-mesenchymal transition, enhancing migration and invasion, and is associated with high Gleason score in prostate cancer. Asian J Androl. 2019;21:557-564. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 10. | Chang CH, Yen MC, Liao SH, Hsu YL, Lai CS, Chang KP. Secreted Protein Acidic and Rich in Cysteine (SPARC) Enhances Cell Proliferation, Migration, and Epithelial Mesenchymal Transition, and SPARC Expression is Associated with Tumor Grade in Head and Neck Cancer. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 11. | Aghamaliyev U, Gaitantzi H, Thomas M, Simon-Keller K, Gaiser T, Marx A, Yagublu V, Araos J, Cai C, Valous NA, Halama N, Kiesslich T, Ebert M, Grützmann R, Rückert F, Breitkopf-Heinlein K. Downregulation of SPARC Is Associated with Epithelial-Mesenchymal Transition and Low Differentiation State of Biliary Tract Cancer Cells. Eur Surg Res. 2019;60:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 12. | Jing Y, Jin Y, Wang Y, Chen S, Zhang X, Song Y, Wang Z, Pu Y, Ni Y, Hu Q. SPARC promotes the proliferation and metastasis of oral squamous cell carcinoma by PI3K/AKT/PDGFB/PDGFRβ axis. J Cell Physiol. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 13. | Thomas SL, Alam R, Lemke N, Schultz LR, Gutiérrez JA, Rempel SA. PTEN augments SPARC suppression of proliferation and inhibits SPARC-induced migration by suppressing SHC-RAF-ERK and AKT signaling. Neuro Oncol. 2010;12:941-955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 55] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 14. | Tang MJ, Tai IT. A novel interaction between procaspase 8 and SPARC enhances apoptosis and potentiates chemotherapy sensitivity in colorectal cancers. J Biol Chem. 2007;282:34457-34467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 64] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 15. | Aoi W, Naito Y, Takagi T, Tanimura Y, Takanami Y, Kawai Y, Sakuma K, Hang LP, Mizushima K, Hirai Y, Koyama R, Wada S, Higashi A, Kokura S, Ichikawa H, Yoshikawa T. A novel myokine, secreted protein acidic and rich in cysteine (SPARC), suppresses colon tumorigenesis via regular exercise. Gut. 2013;62:882-889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 239] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 16. | Sailaja GS, Bhoopathi P, Gorantla B, Chetty C, Gogineni VR, Velpula KK, Gondi CS, Rao JS. The secreted protein acidic and rich in cysteine (SPARC) induces endoplasmic reticulum stress leading to autophagy-mediated apoptosis in neuroblastoma. Int J Oncol. 2013;42:188-196. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 17. | Chetty C, Dontula R, Ganji PN, Gujrati M, Lakka SS. SPARC expression induces cell cycle arrest via STAT3 signaling pathway in medulloblastoma cells. Biochem Biophys Res Commun. 2012;417:874-879. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 18. | Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2463] [Cited by in RCA: 2842] [Article Influence: 129.2] [Reference Citation Analysis (0)] |
| 19. | Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, Hurt EM, Zhao H, Averett L, Yang L, Wilson WH, Jaffe ES, Simon R, Klausner RD, Powell J, Duffey PL, Longo DL, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Montserrat E, López-Guillermo A, Grogan TM, Miller TP, LeBlanc M, Ott G, Kvaloy S, Delabie J, Holte H, Krajci P, Stokke T, Staudt LM; Lymphoma/Leukemia Molecular Profiling Project. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937-1947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2802] [Cited by in RCA: 2717] [Article Influence: 113.2] [Reference Citation Analysis (0)] |
| 20. | Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, Bertoni F, Ponzoni M, Scandurra M, Califano A, Bhagat G, Chadburn A, Dalla-Favera R, Pasqualucci L. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717-721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 890] [Cited by in RCA: 873] [Article Influence: 51.4] [Reference Citation Analysis (0)] |
| 21. | Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7015] [Cited by in RCA: 6384] [Article Influence: 245.5] [Reference Citation Analysis (10)] |
| 22. | Rosenwald A, Alizadeh AA, Widhopf G, Simon R, Davis RE, Yu X, Yang L, Pickeral OK, Rassenti LZ, Powell J, Botstein D, Byrd JC, Grever MR, Cheson BD, Chiorazzi N, Wilson WH, Kipps TJ, Brown PO, Staudt LM. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194:1639-1647. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 844] [Cited by in RCA: 783] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 23. | Piccaluga PP, Agostinelli C, Califano A, Rossi M, Basso K, Zupo S, Went P, Klein U, Zinzani PL, Baccarani M, Dalla Favera R, Pileri SA. Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets. J Clin Invest. 2007;117:823-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 231] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 24. | Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1017] [Cited by in RCA: 941] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 25. | Brune V, Tiacci E, Pfeil I, Döring C, Eckerle S, van Noesel CJ, Klapper W, Falini B, von Heydebreck A, Metzler D, Bräuninger A, Hansmann ML, Küppers R. Origin and pathogenesis of nodular lymphocyte-predominant Hodgkin lymphoma as revealed by global gene expression analysis. J Exp Med. 2008;205:2251-2268. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 254] [Cited by in RCA: 265] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 26. | Eckerle S, Brune V, Döring C, Tiacci E, Bohle V, Sundström C, Kodet R, Paulli M, Falini B, Klapper W, Chaubert AB, Willenbrock K, Metzler D, Bräuninger A, Küppers R, Hansmann ML. Gene expression profiling of isolated tumour cells from anaplastic large cell lymphomas: insights into its cellular origin, pathogenesis and relation to Hodgkin lymphoma. Leukemia. 2009;23:2129-2138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 105] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 27. | Cao L, Cheng H, Jiang Q, Li H, Wu Z. APEX1 is a novel diagnostic and prognostic biomarker for hepatocellular carcinoma. Aging (Albany NY). 2020;12:4573-4591. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 28. | Mo H, Guan J, Yuan ZC, Lin X, Wu ZJ, Liu B, He JL. Expression and predictive value of miR-489 and miR-21 in melanoma metastasis. World J Clin Cases. 2019;7:2930-2941. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 10] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 29. | Rungsakulkij N, Suragul W, Mingphruedhi S, Tangtawee P, Muangkaew P, Aeesoa S. Prognostic role of alpha-fetoprotein response after hepatocellular carcinoma resection. World J Clin Cases. 2018;6:110-120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 26] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 30. | Liu Y, Wu L, Ao H, Zhao M, Leng X, Liu M, Ma J, Zhu J. Prognostic implications of autophagy-associated gene signatures in non-small cell lung cancer. Aging (Albany NY). 2019;11:11440-11462. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 116] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 31. | Richter J, Schlesner M, Hoffmann S, Kreuz M, Leich E, Burkhardt B, Rosolowski M, Ammerpohl O, Wagener R, Bernhart SH, Lenze D, Szczepanowski M, Paulsen M, Lipinski S, Russell RB, Adam-Klages S, Apic G, Claviez A, Hasenclever D, Hovestadt V, Hornig N, Korbel JO, Kube D, Langenberger D, Lawerenz C, Lisfeld J, Meyer K, Picelli S, Pischimarov J, Radlwimmer B, Rausch T, Rohde M, Schilhabel M, Scholtysik R, Spang R, Trautmann H, Zenz T, Borkhardt A, Drexler HG, Möller P, MacLeod RA, Pott C, Schreiber S, Trümper L, Loeffler M, Stadler PF, Lichter P, Eils R, Küppers R, Hummel M, Klapper W, Rosenstiel P, Rosenwald A, Brors B, Siebert R; ICGC MMML-Seq Project. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet. 2012;44:1316-1320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 316] [Cited by in RCA: 331] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 32. | Jais JP, Haioun C, Molina TJ, Rickman DS, de Reynies A, Berger F, Gisselbrecht C, Brière J, Reyes F, Gaulard P, Feugier P, Labouyrie E, Tilly H, Bastard C, Coiffier B, Salles G, Leroy K; Groupe d'Etude des Lymphomes de l'Adulte. The expression of 16 genes related to the cell of origin and immune response predicts survival in elderly patients with diffuse large B-cell lymphoma treated with CHOP and rituximab. Leukemia. 2008;22:1917-1924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 85] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 33. | Camacho D, Jesus JP, Palma AM, Martins SA, Afonso A, Peixoto ML, Pelham CJ, Moreno E, Gogna R. SPARC-p53: The double agents of cancer. Adv Cancer Res. 2020;148:171-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 34. | Cucco F, Barrans S, Sha C, Clipson A, Crouch S, Dobson R, Chen Z, Thompson JS, Care MA, Cummin T, Caddy J, Liu H, Robinson A, Schuh A, Fitzgibbon J, Painter D, Smith A, Roman E, Tooze R, Burton C, Davies AJ, Westhead DR, Johnson PWM, Du MQ. Distinct genetic changes reveal evolutionary history and heterogeneous molecular grade of DLBCL with MYC/BCL2 double-hit. Leukemia. 2020;34:1329-1341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 88] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 35. | Miao Y, Medeiros LJ, Li Y, Li J, Young KH. Genetic alterations and their clinical implications in DLBCL. Nat Rev Clin Oncol. 2019;16:634-652. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 146] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 36. | Huang H, Li Z, Huang C, Rao J, Xie Q, Cui W, Tou F, Zheng Z. CD5 and CD43 Expression are Associate with Poor Prognosis in DLBCL Patients. Open Med (Wars). 2018;13:605-609. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 37. | Abdulla M, Laszlo S, Triumf J, Hedström G, Berglund M, Enblad G, Amini RM. A population-based study of cellular markers in R-CHOP treated diffuse large B-cell lymphoma patients. Acta Oncol. 2016;55:1126-1131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 38. | McDermott MSJ, Sharko AC, Munie J, Kassler S, Melendez T, Lim CU, Broude EV. CDK7 Inhibition is Effective in all the Subtypes of Breast Cancer: Determinants of Response and Synergy with EGFR Inhibition. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 39. | Yan J, Zhang J, Zhang X, Li X, Li L, Li Z, Chen R, Zhang L, Wu J, Wang X, Sun Z, Fu X, Chang Y, Nan F, Yu H, Wu X, Feng X, Li W, Zhang M. SPARC is down-regulated by DNA methylation and functions as a tumor suppressor in T-cell lymphoma. Exp Cell Res. 2018;364:125-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 40. | Meyer PN, Fu K, Greiner T, Smith L, Delabie J, Gascoyne R, Ott G, Rosenwald A, Braziel R, Campo E, Vose J, Lenz G, Staudt L, Chan W, Weisenburger DD. The stromal cell marker SPARC predicts for survival in patients with diffuse large B-cell lymphoma treated with rituximab. Am J Clin Pathol. 2011;135:54-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 61] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 41. | Namani A, Liu K, Wang S, Zhou X, Liao Y, Wang H, Wang XJ, Tang X. Genome-wide global identification of NRF2 binding sites in A549 non-small cell lung cancer cells by ChIP-Seq reveals NRF2 regulation of genes involved in focal adhesion pathways. Aging (Albany NY). 2019;11:12600-12623. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 42. | Schimmel L, Heemskerk N, van Buul JD. Leukocyte transendothelial migration: A local affair. Small GTPases. 2017;8:1-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 43. | Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology. 2013;145:1215-1229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1070] [Cited by in RCA: 1010] [Article Influence: 77.7] [Reference Citation Analysis (6)] |