Published online Jun 26, 2021. doi: 10.12998/wjcc.v9.i18.4480
Peer-review started: January 8, 2021
First decision: March 27, 2021
Revised: April 7, 2021
Accepted: April 23, 2021
Article in press: April 23, 2021
Published online: June 26, 2021
Processing time: 153 Days and 15 Hours
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the most recent global health threat, is spreading throughout the world with worrisome speed, and the current wave of coronavirus disease 2019 (COVID-19) seems to have no mercy. While this mysterious virus challenges our ability to control viral infections, our opportunities to control the COVID-19 pandemic are gradually fading. Currently, pandemic management relies on preventive interventions. Although prevention is a good strategy to mitigate SARS-CoV-2 transmission, it still cannot be considered an absolute solution to eliminate this pandemic. Currently, developing a potent immunity against this viral infection seems to be the most promising strategy to drive down this ongoing global tragedy. However, with the emergence of new challenges in the context of immune responses to COVID-19, the road to control this devastating pandemic seems bumpier; thus, it is pivotal to characterize the dynamics of host immune responses to COVID-19, in order to develop efficient prophylactic and therapeutic tools. This begs the question of whether the effector mechanisms of the immune system are indeed potent or a possible contributing factor to developing more severe and lethal forms of COVID-19. In this review, the possible role of the immunopathologic phenomena including antibody-dependent enhancement, cytokine storm, and original antigenic sin in severity and mortality of COVID-19 will be discussed.
Core Tip: This study provides an overview on the possible role of immunopathologic phenomena including antibody-dependent enhancement, cytokine storm, and original antigenic sin in severity and mortality of coronavirus disease 2019 (COVID-19). With the emergence of new challenges in the context of immunity to COVID-19, it is pivotal to characterize the dynamics of host immune responses to COVID-19, in order to develop efficient prophylactic and therapeutic tools. This begs the question of whether the effector mechanisms of the immune system are indeed potent or a possible contributing factor to developing more severe forms of COVID-19.
- Citation: Taherkhani R, Taherkhani S, Farshadpour F. Dynamics of host immune responses to SARS-CoV-2. World J Clin Cases 2021; 9(18): 4480-4490
- URL: https://www.wjgnet.com/2307-8960/full/v9/i18/4480.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i18.4480
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative pathogen of the ongoing coronavirus disease 2019 (COVID-19) pandemic, is an enveloped positive-stranded RNA virus, belonging to the family Coronaviridae[1-5]. Based on phylogenetic analyses, coronaviruses are classified into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus[6-9]. There are seven coronavirus species capable of causing human infection: 229E, OC43, NL63, and HKU1 are endemic seasonal coronaviruses, causing the common cold and SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV) and SARS-CoV-2 are responsible for large epidemics or worldwide outbreaks of severe respiratory syndrome in humans[2,6,10-13]. SARS-CoV-2 appears to be less virulent but more contagious than SARS-CoV and MERS-CoV[1,10]. Moreover, the recent mutation in the receptor-binding domain (RBD) of spike protein makes it more transmissible[8,14,15].
SARS-CoV-2 is a novel Betacoronavirus in the subgenus Sarbecovirus and is grouped as SARS-related coronaviruses. SARS-CoV-2 shares 79.6% and about 96% sequence identity to SARS-CoV and a bat coronavirus (RaTG13), respectively[2]. Considering the high similarities between SARS-CoV-2 and the bat coronavirus, bats are considered natural reservoir hosts, and pangolins (Manis javanica) are introduced as intermediate hosts. However, there are uncertainties about the origin of SARS-CoV-2[2,16,17]. RNA recombination among coronaviruses is most likely responsible for the emergence of SARS-CoV-2, enabling events of cross-species transmissions[7,16,18,19].
COVID-19 is preliminary a pneumonia-like disease with a wide spectrum of clinical presentations from asymptomatic to mild or severe disease, which spontaneously clear or progress to acute respiratory distress syndrome (ARDS), pulmonary edema, tissue damage, multiple organ dysfunction, and eventually death[1,20]. However, progression to life-threatening clinical illness has mostly been reported in older patients and those with underlying problems and co-morbidities such as chronic respiratory conditions, diabetes, cancer, hypertension, cardiovascular and cerebrovascular disease, requiring hospital care[2,21-24].
SARS-CoV-2 is predominantly transmitted through exposure to infective respiratory droplets and contact with contaminated surfaces[2,25-27]. Currently, reducing exposure to SARS-CoV-2 through public awareness, personal preventive actions, and social distancing as well as the routine screening of population and quarantining of infected subjects seems to be the best preventive measures to mitigate SARS-CoV-2 transmission[26,28-30]. However, poor self-quarantine strategies and asymptomatic transmission, as well as elongated social isolation and lockdown and subsequently the growing economic crisis are the main challenges in the management of the COVID-19 pandemic[1,31].
Considering these challenges, developing a potent immunity against this viral infection seems to be the best option to drive down this ongoing global tragedy. However, with the emergence of new challenges in the context of immunity to COVID-19, the road to control this pandemic seems bumpier; thus, it is pivotal to characterize the dynamics of host immune responses to COVID-19, in order to develop efficient prophylactic and therapeutic tools. This begs the question of whether the effector mechanisms of the immune system are indeed potent or a possible contributing factor to developing more severe forms of COVID-19. In this review, the possible role of immunopathologic phenomena including antibody-dependent enhancement, cytokine storm, and original antigenic sin in severity and mortality of COVID-19 will be discussed.
Upon infection, several types of antibodies are produced. These antibodies are able to provide protection or promote inflammation to maintain health and pathogenesis. IgM emerges rapidly after infection and promotes the pro-inflammatory responses through activation of the complement pathway, resulting in the elimination of the pathogen. IgG is formed later and neutralizes pathogens by inhibiting host cell infections and provides protective immunity or causes antibody-dependent enhancement of infection and acute inflammation[32,33].
In the case of COVID-19, anti-SARS-CoV-2 IgM and IgG antibodies appear between 6 and 15 d post-disease onset. IgM level increases during the acute phase of infection, and then decreases at 4 wk and becomes undetectable in most patients after 5-7 wk. Meanwhile, the IgG level peaks at approximately day 25 and remains high after 4 wk of infection with unknown longevity. Overall, the antibody response to SARS-CoV-2 in most patients is detected between day 10 and day 21 after the onset of illness. Nevertheless, the detection of antibodies may take a longer time, 4 wk or more, in those cases with mild manifestations, and antibody response is not detectable in a small number of cases[34-36].
Neutralizing antibodies contribute to pathogen clearance by inhibiting the interaction of the virus with the host cell receptor as well as by interacting with complement, natural killer cells, and phagocytes[33]. The most efficient SARS-CoV-2-specific neutralizing antibodies target the RBD and the heptad repeat 2 domain of the viral spike protein, blocking viral docking on angiotensin-converting enzyme 2 (ACE2) receptor on the host cell surface and subsequently inhibiting fusion between the viral and host cell membranes and viral entry[33]. Neutralizing anti-RBD antibody is species-specific. Some antibodies are produced against regions outside the RBD. These antibodies are cross-reactive but not neutralizing[37].
In a recent study, patients were in the active and severe course of COVID-19 despite developing anti-nucleoprotein and anti-RBD specific antibodies[38]. Of note, Zhao et al[34] found that higher titers of antibodies against SARS-CoV-2 correlate with worse disease outcomes. Similarly, the results of Liu et al[39] study showed that patients with severe presentations of COVID-19 have vigorous antibody responses. While in the Zhu et al[11] study, COVID-19 patients with mild symptoms had high antibody titers against RBD, S1, and S2 domains of the spike. In addition to disease severity, the titer of antibody is reportedly correlated with age, so that older patients develop higher titers of antibodies than younger patients[11,37,40]. Therefore, the development of antibodies in the course of COVID-19 might not be indicative of protective immunity and viral clearance, and the antibody-mediated immune response might not work through ordinary immune pathways.
Several factors such as specificity, affinity, concentration, and isotype of antibody may dictate potential outcomes of antibody response to infection. Antibodies recognize specific targeting domains. Experimental studies with SARS-CoV report that antibodies specific for the RBD of the spike protein promote stronger neutralization and subsequently better protection, whereas antibodies reactive to S protein epitopes outside the RBD and N protein can induce more severe clinical illness. Therefore, specific antibodies capable of targeting receptor binding and blocking viral entry tend to elicit neutralization. Moreover, the virus-neutralizing effect correlates with antibody concentration and affinity, so that antibodies with higher affinity and concentration induce stronger neutralization and subsequently better protection[33].
The effector functions of antibodies are dependent on antibody isotype as well. IgG subclasses display different biological functions due to their varied affinities to Fcγ receptors (FcγRs). In SARS-CoV infection, ectopic expression of FcγRIIa and FcγRIIb induce more severe disease[10,33]. FcγRIIb is an inhibitory FcγR that signals through an immunoreceptor tyrosine-based inhibition motif on its cytoplasmic tail. The other FcγRs, FcγRI, FcγRIIa, FcγRIIc and FcγRIIIa, activate signaling pathways through immunoreceptor tyrosine-based activation motifs (ITAMs)[41,42]. FcγR polymorphism is another effector factor in this context. The FcγRIIa allelic polymorphism alters the affinity and specificity of FcγRIIa for different IgG subclasses, affecting the severity and susceptibility of infections. The low-affinity allele (FcγRIIa-R131) is capable of binding to both IgG1 and IgG2 and is associated with SARS severity[10,33]. This polymorphic variant is frequently observed in deceased and hospitalized COVID-19 patients; whereas, the high-affinity allele (FcγRIIa-H131) is associated with reduced severity and mortality of SARS-CoV infection[42].
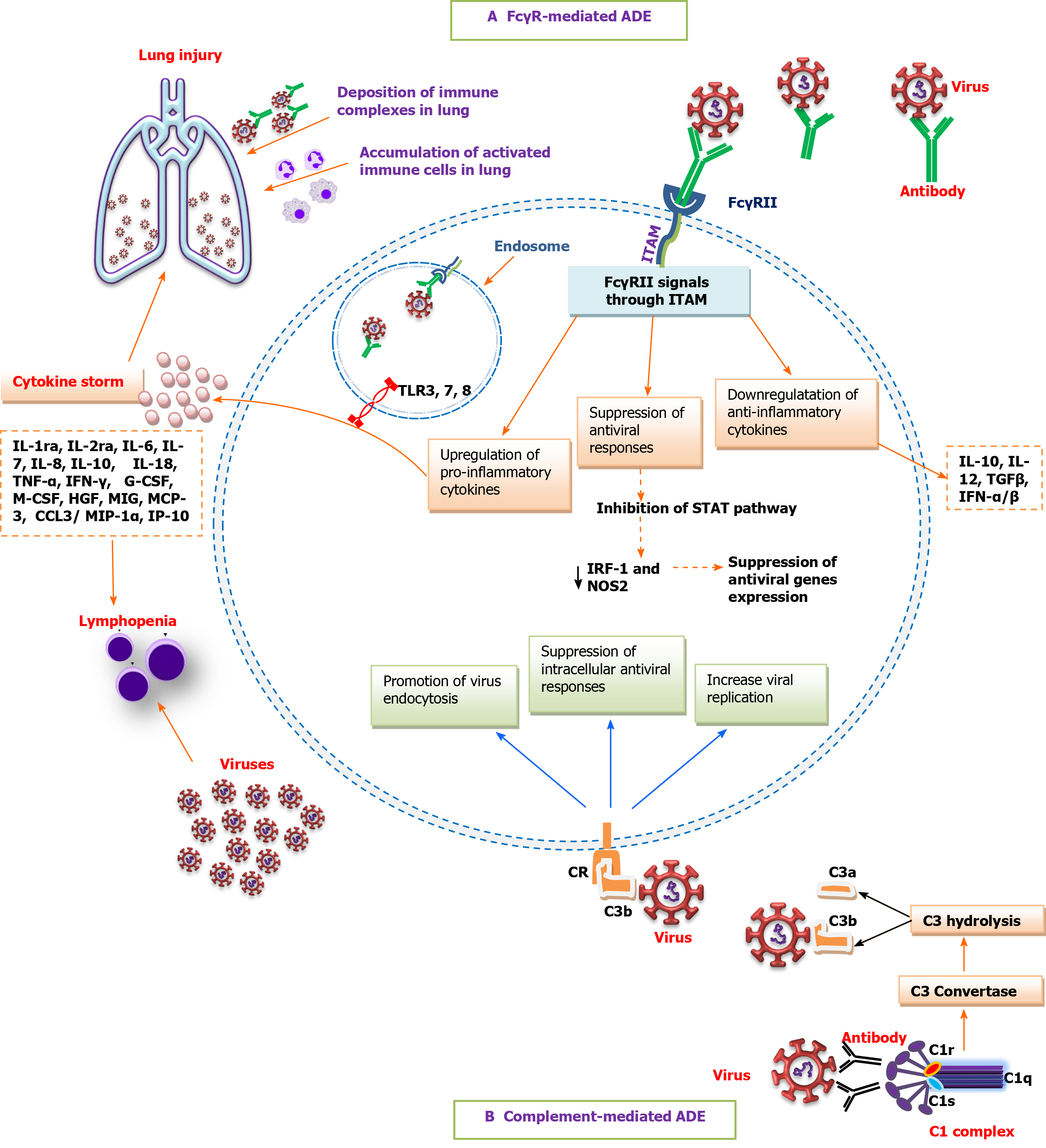
Although antibody-effector functions are commonly beneficial and protective, some viral-induced antibodies might not neutralize the infection, and conversely, lead to enhanced virus infectivity and virulence[10,43]. Antibody-mediated enhancement of infection (ADE) is a well-characterized phenomenon whereby sub-neutralizing concentrations of antibodies or non-neutralizing antibodies bind to virus particles and enhance viral cell entry through interaction with FcγRs or complement receptors (CR) on susceptible cells[1,42,44-46]. In FcγR-mediated ADE, upon binding of virus-antibody complexes to FcγRII on antigen-presenting cells or B cells, FcγRII signals through an ITAM on its cytoplasmic tail to upregulate pro-inflammatory cytokines, downregulate anti-inflammatory cytokines, and suppress antiviral genes expression by inhibiting the signal transducer and activator of transcription (commonly known as STAT) pathway and subsequent decrease in antiviral transcription factors such as interferon regulatory factor 1 (IRF-1) and nitric oxide synthase 2 (NOS2), resulting in increased viral replication due to suppression of the intracellular antiviral responses. On the other hand, following internalization, virus-antibody immune complexes and viral RNA in the endosomes initiate signaling through toll-like receptor 3 (TLR3), TLR7, and TLR8 to activate immune cells. Subsequently, the accumulation of activated myeloid cells in the lung promotes inflammation, acute lung injury, and vasculitis[1,33]. In complement-mediated antibody-dependent enhancement (ADE), virus-antibody complexes activate the complement cascade, resulting in the formation of complement-coated virions and subsequent interaction with complement receptor (CR) on CR-bearing cells. The activated CR initiates intracellular signaling to promote endocytosis of the virus, suppress intracellular antiviral responses, and increase viral replication[46,47]. Furthermore, the formation and deposition of immune complexes in the lung can lead to cell lysis and tissue damage due to complement activation[48,49]. Taken together, ADE can modulate host immune responses toward an inflammatory profile, leading to cytokine storm, lymphopenia, and tissue damage (Figure 1)[1,49,50].
One of the hallmarks of the ADE phenomenon is the excessive expression of inflammatory mediators and cytokines, known as a cytokine storm. Cytokine storm is a form of uncontrolled systemic inflammatory responses characterized by increased levels of interleukin 1 receptor antagonist (IL-1ra), IL-2ra, IL-6, IL-7, IL-8, IL-10, IL-18, tumor necrosis factor alpha (TNF-α), interferon gamma (IFN-γ), granulocyte colony-stimulating factor (G-CSF), macrophage CSF (M-CSF), hepatocyte growth factor, monokine-induced gamma IFN, C-X-C motif chemokine ligand 10 (CXCL10), C-C motif chemokine ligand 2 (CCL2)/monocyte chemoattractant protein 1 (MCP-1), MCP-3, CCL3/macrophage inflammatory protein 1 alpha, IFN-γ-induced protein 10, cutaneous T cell-attracting cytokine, RANTES (CCL5); and decreased levels of IL-10, IL-12, and TGFβ and IFN-α/β. Furthermore, high expression of chemokines receptors, including CCR5 (CCL3 receptor) and CCR2 (CCL2 receptor) has been reported[1,2,23,50,51]. Among them, IL-6 and GM-CSF play major roles in triggering inflammatory events by monocyte activation and differentiation to inflammatory macrophages.
Uncontrolled proliferation of inflammatory macrophages, commonly known as macrophage activation syndrome, plays a key role in driving cytokine storm and is closely linked to lymphopenia, hyperferritinemia, liver insufficiency, acute lung injury, fulminant myocarditis, neurologic symptoms, coagulopathy, and thrombotic events[51,52].
Lymphopenia, a condition defined by a drastic decrease in T cell counts, notably CD8+ T cells, is frequently observed in severe cases of COVID-19 with a high mortality rate[53]. Although the exact underlying etiology has not been identified, several factors have been proposed. First, SARS-CoV-2 entry into lymphocytes through ACE2 may induce lymphocyte lysis[54,55]. Second, high expression of inflammatory cytokines especially IL-6 and TNF-α may promote lymphocyte apoptosis and atrophy of lymphoid organs together with the spleen. Massive lymphocyte death and atrophy of lymphoid organs have been observed in the autopsy of those patients who succumbed to COVID-19[53,54]. Third, hypercytokinemia and coexisting lactic acid acidosis may inhibit lymphocyte proliferation and expansion[54]. Although downregulation of those genes involved in lymphocyte proliferation and activation, including mitogen-activated protein kinase kinase 7 and SOS1 has been reported in T lymphocytes of severe cases of COVID-19, more investigations are required to understand how SARS-CoV-2 infection affects T-cell expansion and function[53,56]. Furthermore, impairment of the function of T helper and regulatory T cells during COVID-19 infection may eventually result in the exhaustion of cytotoxic CD8+ T-cells. Increased cell surface expression of markers of T-cell exhaustion, including programmed cell death protein 1 and T cell immunoglobulin and mucin domain 3, has been reported in severe COVID-19 patients. Although some studies exclude the role of regulatory T cells in the development of T-cell exhaustion, the possible underlying causes remain unknown[53,57]. Nevertheless, an association between a high level of IL-6 and impairment of the cytotoxic activity of CD8+ T-cells has been reported in COVID-19 patients[58].
In 1964, Hawkes[59] first reported the possible role of ADE in the enhanced infectivity of some members of the Flaviviridae family. ADE has been observed in Dengue virus, Ebola virus, Zika virus, yellow fever virus, and human immunodeficiency virus infections and during influenza pandemics, and has been highlighted in the course of vaccine development against feline infectious peritonitis virus, respiratory syncytial virus, and Dengue virus[44-47,60]. ADE has been experimentally described for SARS-CoV and MERS-CoV in animal models and HL-CZ human promonocyte cell line, in which it can be mediated by non-neutralizing IgG antibodies to the viral spike protein[44,51,61,62]. However, ADE in coronaviruses infection differs fundamentally from that of the other viruses. For example, according to in vitro experimental observations, ADE mediated by Dengue virus antibodies is dependent on activating FcγRs such as FcγRIIa (CD32a) and leads to productive viral infection in FcγRII-expressing cells such as macrophages. Whereas SARS-CoV, despite having high-affinity entry receptor ACE2, employs low-affinity FcγRs such as inhibitory FcγRIIb to infect non-permissive leukocytes preferably B cell lines within the lung microenvironment, leading to an abortive viral infection[42]. However, SARS-CoV entry into monocytes, macrophages, and B-cells through FcγRIIa has also been reported. SARS-CoV infection is abortive in these cells[61,63]. Taken together, ADE mediated by SARS-CoV and MERS-CoV antibodies is unlikely to cause productive viral replication as revealed by the Dengue virus. Instead, FcγRII-mediated coronavirus entry can promote hypercytokinemia, inflammation, and acute lung injury by activating immune cells[33,64].
Considering the high genetic homology between SARS-CoV and SARS-CoV-2 as well as epidemiological similarities between dengue fever and COVID-19, ADE has been presumptively proposed as a possible contributing factor in the pathogenesis of COVID-19. Nonetheless, the underlying pathomechanisms remain unknown[33,51,65]. One possible speculation is that the antibodies obtained from primary infections with circulating seasonal human coronaviruses or SARS-CoV-2 not only fail to completely neutralize secondary infection with SARS-CoV-2 but also trigger the ADE-FcγRII mechanism[10,21,32,44,65]. This speculation might also explain mild or the relative absence of clinical symptoms of COVID-19 in children and young people as compared with adults and the elderly. A possible explanation is the relative lack of immune memory to closely related coronaviruses in younger ages. A history of repeated confrontations with seasonal coronaviruses in the lifetime is prone to induce ADE[66]. Overall, children develop life-threatening complications less frequently than the elderly. Nevertheless, the pediatric infection risk is reported to be similar to adults but mostly remain undiagnosed due to diagnostic focusing on symptomatic cases. Moreover, the association between advanced age and severity and mortality of COVID-19 is suggestive of dengue fever and SARS epidemic[67].
Another speculation is the original antigenic sin phenomenon, in which primary infection or immunization with a pathogen influences the following immune responses to the infections with closely related pathogens. This phenomenon has already been observed in infection with influenza virus, Zika virus, Dengue virus, and coronaviruses. According to this phenomenon, the host body prefers utilizing memory B cells instead of establishing new immunity against the new antigenically related pathogens[60]. In this context, it is hypothesized that in those COVID-19 patients with a history of previous exposure to seasonal coronaviruses, the immune system produces early and high-titer recall antibodies to related coronaviruses instead of developing antibodies to SARS-CoV-2. These recall antibodies not only fail to provide protection against COVID-19, but also boost the disease severity. Hereby, rapid production of these cross-reactive memory IgG antibodies can trigger the ADE-FcγRII mechanism and immune complex deposition, resulting in hyper inflammation and tissue injury[66]. The early appearance of IgG and relatively long incubation period before symptomatic disease onset in some COVID-19 patients are indicative of immunological recall effects. Whereby, a seroconversion of IgG earlier than IgM, which is expected in memory immune responses, has an enhancing role in clinically overt infection. Interestingly, COVID-19 severity is strongly associated with early seroconversion and high titers of antibodies, suggesting the possible role of original antigenic sin in the development of ADE[66,68].
Although ADE has so far not been confirmed for SARS-CoV-2, cytokine storm and the subsequent hyper-inflammation have been observed in severe COVID-19 patients requiring hospitalization[22,69,70]. Notably, IL-6 and GM-CSF are the main hallmarks of SARS-CoV-2-induced hyper-inflammation and play a major role in the onset of pediatric inflammatory multisystemic syndrome, an autoimmune inflammatory disease related to SARS-CoV-2[51,71]. Furthermore, IL-6 has a key role in driving metabolic inflammation due to obesity and diabetes and, therefore, increases the severity of COVID-19 in diabetic patients[51,72]. On the other hand, this exuberant inflammatory response associated with infiltration and accumulation of inflammatory macrophages in various organs may promote microvascular vessel obstructive thrombo-inflammatory syndrome and ARDS that are characteristic of severe COVID-19, resulting in multiple-organ dysfunction and death[51]. Specifically, the risk of ARDS development during the COVID-19 course is associated with lymphopenia. Furthermore, a high level of IL-6 is a key factor in the development of lymphopenia and intensive care requirement in COVID-19 patients[53,54]. Therefore, blockade of cytokine signaling pathways has become an effective therapeutic option to inhibit the cytokine storm. In this light, blockade of the IL-6 receptor signaling pathway with tocilizumab has been shown to limit hyper-inflammation, boost the number of circulatory lymphocytes and decrease lethality in severe cases of COVID-19[51,70]. Moreover, CCR5 antagonist leronlimab, IL-1 receptor antagonist anakinra, GM-CSF blockade as well as therapeutic depletion of IL-1, IL-6, and macrophages at disease onset are some other therapeutic blockade options with promising results[1,49,51].
The clinical relevance of the ADE mechanism in SARS-CoV-2 pathogenesis, if confirmed, would be the greatest concern in COVID-19 vaccine development and would affect the safety and efficacy of antibody-based therapies. Considering our limited knowledge in this regard, further thoughtful and rigorous research works are urgently required to explore the possible role of ADE in the severity and mortality of COVID-19. Knowledge of this immunopathologic phenomenon will help to understand the viral pathogenesis as well as to guide vaccine design and public health policy for better management of the pandemic.
We would like to extend our sincere gratitude to the warriors who devote their lives to fighting against COVID-19.
| 1. | Siracusano G, Pastori C, Lopalco L. Humoral Immune Responses in COVID-19 Patients: A Window on the State of the Art. Front Immunol. 2020;11:1049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 2. | Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, Tan KS, Wang DY, Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020;7:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1854] [Cited by in RCA: 2029] [Article Influence: 338.2] [Reference Citation Analysis (0)] |
| 3. | Liu W, Liu L, Kou G, Zheng Y, Ding Y, Ni W, Wang Q, Tan L, Wu W, Tang S, Xiong Z, Zheng S. Evaluation of Nucleocapsid and Spike Protein-Based Enzyme-Linked Immunosorbent Assays for Detecting Antibodies against SARS-CoV-2. J Clin Microbiol. 2020;58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 377] [Cited by in RCA: 455] [Article Influence: 75.8] [Reference Citation Analysis (0)] |
| 4. | Yoshimoto FK. The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19. Protein J. 2020;39:198-216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 387] [Cited by in RCA: 363] [Article Influence: 60.5] [Reference Citation Analysis (0)] |
| 5. | Satarker S, Nampoothiri M. Structural Proteins in Severe Acute Respiratory Syndrome Coronavirus-2. Arch Med Res. 2020;51:482-491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 258] [Cited by in RCA: 253] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 6. | Ugurel OM, Ata O, Turgut-Balik D. An updated analysis of variations in SARS-CoV-2 genome. Turk J Biol. 2020;44:157-167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 7. | Kumar S, Maurya VK, Prasad AK, Bhatt MLB, Saxena SK. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV). Virusdisease. 2020;31:13-21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 149] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 8. | Alanagreh L, Alzoughool F, Atoum M. The Human Coronavirus Disease COVID-19: Its Origin, Characteristics, and Insights into Potential Drugs and Its Mechanisms. Pathogens. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 209] [Cited by in RCA: 166] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 9. | Yin Y, Wunderink RG. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23:130-137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 632] [Cited by in RCA: 651] [Article Influence: 72.3] [Reference Citation Analysis (0)] |
| 10. | Ulrich H, Pillat MM, Tárnok A. Dengue Fever, COVID-19 (SARS-CoV-2), and Antibody-Dependent Enhancement (ADE): A Perspective. Cytometry A. 2020;97:662-667. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 11. | Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W; China Novel Coronavirus Investigating and Research Team. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382:727-733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18987] [Cited by in RCA: 17890] [Article Influence: 2981.7] [Reference Citation Analysis (2)] |
| 12. | Chen B, Tian EK, He B, Tian L, Han R, Wang S, Xiang Q, Zhang S, El Arnaout T, Cheng W. Overview of lethal human coronaviruses. Signal Transduct Target Ther. 2020;5:89. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 190] [Cited by in RCA: 222] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 13. | Shokri S, Mahmoudvand S, Taherkhani R, Farshadpour F. Modulation of the immune response by Middle East respiratory syndrome coronavirus. J Cell Physiol. 2019;234:2143-2151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 67] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 14. | Shah M, Ahmad B, Choi S, Woo HG. Mutations in the SARS-CoV-2 spike RBD are responsible for stronger ACE2 binding and poor anti-SARS-CoV mAbs cross-neutralization. Comput Struct Biotechnol J. 2020;18:3402-3414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 60] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 15. | Ou J, Zhou Z, Dai R, Zhang J, Lan W, Zhao S, Wu J, Seto D, Cui L, Zhang G, Zhang Q. Emergence of RBD mutations in circulating SARS-CoV-2 strains enhancing the structural stability and human ACE2 receptor affinity of the spike protein. 2020 Preprint. Available from: bioRxiv. [DOI] [Full Text] |
| 16. | Bibiana S O F, Vargas-Pinilla P, Amorim CEG, Sortica VA, Bortolini MC. ACE2 diversity in placental mammals reveals the evolutionary strategy of SARS-CoV-2. Genet Mol Biol. 2020;43:e20200104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 17. | Lam TT, Jia N, Zhang YW, Shum MH, Jiang JF, Zhu HC, Tong YG, Shi YX, Ni XB, Liao YS, Li WJ, Jiang BG, Wei W, Yuan TT, Zheng K, Cui XM, Li J, Pei GQ, Qiang X, Cheung WY, Li LF, Sun FF, Qin S, Huang JC, Leung GM, Holmes EC, Hu YL, Guan Y, Cao WC. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1019] [Cited by in RCA: 1215] [Article Influence: 202.5] [Reference Citation Analysis (0)] |
| 18. | Ji W, Wang W, Zhao X, Zai J, Li X. Homologous recombination within the spike glycoprotein of the newly identified coronavirus may boost cross-species transmission from snake to human. J Med Virol. 2020;92. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 518] [Cited by in RCA: 534] [Article Influence: 89.0] [Reference Citation Analysis (0)] |
| 19. | Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562-569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1933] [Cited by in RCA: 2253] [Article Influence: 375.5] [Reference Citation Analysis (1)] |
| 20. | Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, Levantovsky R, Malle L, Moreira A, Park MD, Pia L, Risson E, Saffern M, Salomé B, Esai Selvan M, Spindler MP, Tan J, van der Heide V, Gregory JK, Alexandropoulos K, Bhardwaj N, Brown BD, Greenbaum B, Gümüş ZH, Homann D, Horowitz A, Kamphorst AO, Curotto de Lafaille MA, Mehandru S, Merad M, Samstein RM; Sinai Immunology Review Project. Immunology of COVID-19: Current State of the Science. Immunity. 2020;52:910-941. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1336] [Cited by in RCA: 1195] [Article Influence: 199.2] [Reference Citation Analysis (0)] |
| 21. | Cegolon L, Pichierri J, Mastrangelo G, Cinquetti S, Sotgiu G, Bellizzi S, Pichierri G. Hypothesis to explain the severe form of COVID-19 in Northern Italy. BMJ Glob Health. 2020;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 22. | Chowdhury MA, Hossain N, Kashem MA, Shahid MA, Alam A. Immune response in COVID-19: A review. J Infect Public Health. 2020;13:1619-1629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 274] [Cited by in RCA: 228] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 23. | Nikolich-Zugich J, Knox KS, Rios CT, Natt B, Bhattacharya D, Fain MJ. SARS-CoV-2 and COVID-19 in older adults: what we may expect regarding pathogenesis, immune responses, and outcomes. Geroscience. 2020;42:505-514. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 356] [Cited by in RCA: 347] [Article Influence: 57.8] [Reference Citation Analysis (0)] |
| 24. | Hou Y, Zhao J, Martin W, Kallianpur A, Chung MK, Jehi L, Sharifi N, Erzurum S, Eng C, Cheng F. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020;18:216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 279] [Cited by in RCA: 325] [Article Influence: 54.2] [Reference Citation Analysis (0)] |
| 25. | Meselson M. Droplets and Aerosols in the Transmission of SARS-CoV-2. N Engl J Med. 2020;382:2063. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 220] [Cited by in RCA: 231] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 26. | Javed M, Javed F, Ergin HE, Maung TZ, Khan S. Do COVID-19 and SARS Gene Complexities and Variations Help Overcome the Knowledge Gap? Cureus. 2020;12:e8439. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 27. | Chen D, Xu W, Lei Z, Huang Z, Liu J, Gao Z, Peng L. Recurrence of positive SARS-CoV-2 RNA in COVID-19: A case report. Int J Infect Dis. 2020;93:297-299. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 231] [Cited by in RCA: 222] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 28. | Kirkcaldy RD, King BA, Brooks JT. COVID-19 and Postinfection Immunity: Limited Evidence, Many Remaining Questions. JAMA. 2020;323:2245-2246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 163] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 29. | Taherkhani R, Farshadpour F. Lurking epidemic of hepatitis C virus infection in Iran: A call to action. World J Hepatol. 2017;9:1040-1042. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (1)] |
| 30. | Taherkhani R, Farshadpour F. Global elimination of hepatitis C virus infection: Progresses and the remaining challenges. World J Hepatol. 2017;9:1239-1252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (1)] |
| 31. | Nicola M, Alsafi Z, Sohrabi C, Kerwan A, Al-Jabir A, Iosifidis C, Agha M, Agha R. The socio-economic implications of the coronavirus pandemic (COVID-19): A review. Int J Surg. 2020;78:185-193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2872] [Cited by in RCA: 2719] [Article Influence: 453.2] [Reference Citation Analysis (0)] |
| 32. | Kumar R, Gupta N, Kodan P, Mittal A, Soneja M, Wig N. Is there antibody-dependent enhancement in SARS Coronavirus 2? J Family Med Prim Care. 2020;9:2589-2590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 33. | Iwasaki A, Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol. 2020;20:339-341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 416] [Cited by in RCA: 384] [Article Influence: 64.0] [Reference Citation Analysis (0)] |
| 34. | Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, Wang X, Yuan J, Li T, Li J, Qian S, Hong C, Wang F, Liu Y, Wang Z, He Q, Li Z, He B, Zhang T, Fu Y, Ge S, Liu L, Zhang J, Xia N, Zhang Z. Antibody Responses to SARS-CoV-2 in Patients With Novel Coronavirus Disease 2019. Clin Infect Dis. 2020;71:2027-2034. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1590] [Cited by in RCA: 1855] [Article Influence: 309.2] [Reference Citation Analysis (0)] |
| 35. | Long QX, Liu BZ, Deng HJ, Wu GC, Deng K, Chen YK, Liao P, Qiu JF, Lin Y, Cai XF, Wang DQ, Hu Y, Ren JH, Tang N, Xu YY, Yu LH, Mo Z, Gong F, Zhang XL, Tian WG, Hu L, Zhang XX, Xiang JL, Du HX, Liu HW, Lang CH, Luo XH, Wu SB, Cui XP, Zhou Z, Zhu MM, Wang J, Xue CJ, Li XF, Wang L, Li ZJ, Wang K, Niu CC, Yang QJ, Tang XJ, Zhang Y, Liu XM, Li JJ, Zhang DC, Zhang F, Liu P, Yuan J, Li Q, Hu JL, Chen J, Huang AL. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26:845-848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2411] [Cited by in RCA: 2065] [Article Influence: 344.2] [Reference Citation Analysis (0)] |
| 36. | Kellam P, Barclay W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J Gen Virol. 2020;101:791-797. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 269] [Cited by in RCA: 262] [Article Influence: 43.7] [Reference Citation Analysis (1)] |
| 37. | Gorse GJ, Donovan MM, Patel GB. Antibodies to coronaviruses are higher in older compared with younger adults and binding antibodies are more sensitive than neutralizing antibodies in identifying coronavirus-associated illnesses. J Med Virol. 2020;92:512-517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 112] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 38. | To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, Yip CC, Cai JP, Chan JM, Chik TS, Lau DP, Choi CY, Chen LL, Chan WM, Chan KH, Ip JD, Ng AC, Poon RW, Luo CT, Cheng VC, Chan JF, Hung IF, Chen Z, Chen H, Yuen KY. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565-574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2296] [Cited by in RCA: 2273] [Article Influence: 378.8] [Reference Citation Analysis (4)] |
| 39. | Liu X, Wang J, Xu X, Liao G, Chen Y, Hu CH. Patterns of IgG and IgM antibody response in COVID-19 patients. Emerg Microbes Infect. 2020;9:1269-1274. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 166] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 40. | van der Heide V. Neutralizing antibody response in mild COVID-19. Nat Rev Immunol. 2020;20:352. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 41. | Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8:34-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1948] [Cited by in RCA: 2330] [Article Influence: 129.4] [Reference Citation Analysis (0)] |
| 42. | Bournazos S, Gupta A, Ravetch JV. The role of IgG Fc receptors in antibody-dependent enhancement. Nat Rev Immunol. 2020;20:633-643. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 467] [Article Influence: 77.8] [Reference Citation Analysis (0)] |
| 43. | Wan Y, Shang J, Sun S, Tai W, Chen J, Geng Q, He L, Chen Y, Wu J, Shi Z, Zhou Y, Du L, Li F. Molecular Mechanism for Antibody-Dependent Enhancement of Coronavirus Entry. J Virol. 2020;94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 335] [Cited by in RCA: 452] [Article Influence: 75.3] [Reference Citation Analysis (0)] |
| 44. | Negro F. Is antibody-dependent enhancement playing a role in COVID-19 pathogenesis? Swiss Med Wkly. 2020;150:w20249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 45. | Yager EJ. Antibody-dependent enhancement and COVID-19: Moving toward acquittal. Clin Immunol. 2020;217:108496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 46. | Smatti MK, Al Thani AA, Yassine HM. Viral-Induced Enhanced Disease Illness. Front Microbiol. 2018;9:2991. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 96] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 47. | Willey S, Aasa-Chapman MM, O'Farrell S, Pellegrino P, Williams I, Weiss RA, Neil SJ. Extensive complement-dependent enhancement of HIV-1 by autologous non-neutralising antibodies at early stages of infection. Retrovirology. 2011;8:16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 68] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 48. | Burton DR, Walker LM. Rational Vaccine Design in the Time of COVID-19. Cell Host Microbe. 2020;27:695-698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 100] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 49. | Felsenstein S, Herbert JA, McNamara PS, Hedrich CM. COVID-19: Immunology and treatment options. Clin Immunol. 2020;215:108448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 428] [Cited by in RCA: 412] [Article Influence: 68.7] [Reference Citation Analysis (0)] |
| 50. | Jafarzadeh A, Chauhan P, Saha B, Jafarzadeh S, Nemati M. Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID-19: Lessons from SARS and MERS, and potential therapeutic interventions. Life Sci. 2020;257:118102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 190] [Cited by in RCA: 244] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 51. | Gómez-Rial J, Rivero-Calle I, Salas A, Martinón-Torres F. Role of Monocytes/Macrophages in Covid-19 Pathogenesis: Implications for Therapy. Infect Drug Resist. 2020;13:2485-2493. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 85] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 52. | Lerkvaleekul B, Vilaiyuk S. Macrophage activation syndrome: early diagnosis is key. Open Access Rheumatol. 2018;10:117-128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 100] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 53. | Tavakolpour S, Rakhshandehroo T, Wei EX, Rashidian M. Lymphopenia during the COVID-19 infection: What it shows and what can be learned. Immunol Lett. 2020;225:31-32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 158] [Cited by in RCA: 238] [Article Influence: 39.7] [Reference Citation Analysis (0)] |
| 54. | Terpos E, Ntanasis-Stathopoulos I, Elalamy I, Kastritis E, Sergentanis TN, Politou M, Psaltopoulou T, Gerotziafas G, Dimopoulos MA. Hematological findings and complications of COVID-19. Am J Hematol. 2020;95:834-847. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 936] [Cited by in RCA: 1225] [Article Influence: 204.2] [Reference Citation Analysis (0)] |
| 55. | Violetis OA, Chasouraki AM, Giannou AM, Baraboutis IG. COVID-19 Infection and Haematological Involvement: a Review of Epidemiology, Pathophysiology and Prognosis of Full Blood Count Findings. SN Compr Clin Med. 2020;1-5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 56. | Ouyang Y, Yin J, Wang W, Shi H, Shi Y, Xu B, Qiao L, Feng Y, Pang L, Wei F, Guo X, Jin R, Chen D. Downregulated Gene Expression Spectrum and Immune Responses Changed During the Disease Progression in Patients With COVID-19. Clin Infect Dis. 2020;71:2052-2060. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 105] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 57. | Diao B, Wang C, Tan Y, Chen X, Liu Y, Ning L, Chen L, Li M, Wang G, Yuan Z, Feng Z, Zhang Y, Wu Y, Chen Y. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front Immunol. 2020;11:827. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1687] [Cited by in RCA: 1791] [Article Influence: 298.5] [Reference Citation Analysis (0)] |
| 58. | Mazzoni A, Salvati L, Maggi L, Capone M, Vanni A, Spinicci M, Mencarini J, Caporale R, Peruzzi B, Antonelli A, Trotta M, Zammarchi L, Ciani L, Gori L, Lazzeri C, Matucci A, Vultaggio A, Rossi O, Almerigogna F, Parronchi P, Fontanari P, Lavorini F, Peris A, Rossolini GM, Bartoloni A, Romagnani S, Liotta F, Annunziato F, Cosmi L. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J Clin Invest. 2020;130:4694-4703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 312] [Cited by in RCA: 384] [Article Influence: 64.0] [Reference Citation Analysis (0)] |
| 59. | Hawkes RA. Enhancement of the infectivity of arboviruses by specific antisera produced in domestic fowls. Aust J Exp Biol Med Sci. 1964;42:465-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 123] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 60. | Kadkhoda K. COVID-19: are neutralizing antibodies neutralizing enough? Transfusion. 2020;60:1602-1603. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 61. | Jaume M, Yip MS, Cheung CY, Leung HL, Li PH, Kien F, Dutry I, Callendret B, Escriou N, Altmeyer R, Nal B, Daëron M, Bruzzone R, Peiris JS. Anti-severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH- and cysteine protease-independent FcγR pathway. J Virol. 2011;85:10582-10597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 263] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 62. | Yeh C-S, Yang J-Y, Liu W-T, Huang JC, Chen Y-MA, Wang S-F. SARS coronavirus has antibody-dependent enhancement (ADE) effect through the autologous antibodies against envelope spikes on Fcγ receptor expressing cells. J Virus Erad. 2016;2:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 63. | Yip MS, Leung NH, Cheung CY, Li PH, Lee HH, Daëron M, Peiris JS, Bruzzone R, Jaume M. Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virol J. 2014;11:82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 186] [Cited by in RCA: 195] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 64. | Yip MS, Leung HL, Li PH, Cheung CY, Dutry I, Li D, Daëron M, Bruzzone R, Peiris JS, Jaume M. Antibody-dependent enhancement of SARS coronavirus infection and its role in the pathogenesis of SARS. Hong Kong Med J. 2016;22:25-31. [PubMed] |
| 65. | Tetro JA. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect. 2020;22:72-73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 295] [Cited by in RCA: 290] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 66. | Fierz W, Walz B. Antibody Dependent Enhancement Due to Original Antigenic Sin and the Development of SARS. Front Immunol. 2020;11:1120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 67. | Felsenstein S, Hedrich CM. COVID-19 in children and young people. Lancet Rheumatol. 2020;2:e514-e516. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 68. | Wang Y, Zhang L, Sang L, Ye F, Ruan S, Zhong B, Song T, Alshukairi AN, Chen R, Zhang Z, Gan M, Zhu A, Huang Y, Luo L, Mok CKP, Al Gethamy MM, Tan H, Li Z, Huang X, Li F, Sun J, Zhang Y, Wen L, Li Y, Chen Z, Zhuang Z, Zhuo J, Chen C, Kuang L, Wang J, Lv H, Jiang Y, Li M, Lin Y, Deng Y, Tang L, Liang J, Huang J, Perlman S, Zhong N, Zhao J, Malik Peiris JS. Kinetics of viral load and antibody response in relation to COVID-19 severity. J Clin Invest. 2020;130 (10):5235-5244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 364] [Cited by in RCA: 421] [Article Influence: 70.2] [Reference Citation Analysis (0)] |
| 69. | Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ; HLH Across Speciality Collaboration; UK. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033-1034. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6366] [Cited by in RCA: 6842] [Article Influence: 1140.3] [Reference Citation Analysis (1)] |
| 70. | Zhang C, Wu Z, Li JW, Zhao H, Wang GQ. Cytokine release syndrome in severe COVID-19: interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int J Antimicrob Agents. 2020;55:105954. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1228] [Cited by in RCA: 1224] [Article Influence: 204.0] [Reference Citation Analysis (0)] |
| 71. | Whittaker E, Bamford A, Kenny J, Kaforou M, Jones CE, Shah P, Ramnarayan P, Fraisse A, Miller O, Davies P, Kucera F, Brierley J, McDougall M, Carter M, Tremoulet A, Shimizu C, Herberg J, Burns JC, Lyall H, Levin M; PIMS-TS Study Group and EUCLIDS and PERFORM Consortia. Clinical Characteristics of 58 Children With a Pediatric Inflammatory Multisystem Syndrome Temporally Associated With SARS-CoV-2. JAMA. 2020;324:259-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1345] [Cited by in RCA: 1362] [Article Influence: 227.0] [Reference Citation Analysis (0)] |
| 72. | Sindhu S, Thomas R, Shihab P, Sriraman D, Behbehani K, Ahmad R. Obesity Is a Positive Modulator of IL-6R and IL-6 Expression in the Subcutaneous Adipose Tissue: Significance for Metabolic Inflammation. PLoS One. 2015;10:e0133494. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 181] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/Licenses/by-nc/4.0/
Manuscript source: Invited manuscript
Specialty type: Infectious diseases
Country/Territory of origin: Iran
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Vorobjova T S-Editor: Zhang H L-Editor: Filipodia P-Editor: Wang LL