Published online Apr 16, 2021. doi: 10.12998/wjcc.v9.i11.2458
Peer-review started: December 22, 2020
First decision: January 10, 2021
Revised: January 12, 2021
Accepted: February 1, 2021
Article in press: February 1, 2021
Published online: April 16, 2021
Processing time: 101 Days and 1.5 Hours
Colorectal cancer (CRC) is common in elderly patients. Mismatch repair (MMR) protein deletion is one of the causes of CRC. The RAS (KRAS/NRAS), BRAF, and PIK3CA genes are important gene targets in CRC treatment and are closely related to the prognosis and survival of patients. However, little is known regarding the relationship between the expression of MMR, RAS, BRAF, PIK3CA and the clinicopathological features in CRC patients.
To analyze the relationship between the expression of MMR, RAS, BRAF, PIK3CA and the clinicopathological features in CRC.
A total of 327 elderly patients with CRC were enrolled, and immuno-histochemistry was used to detect the MMR protein. Real-time quantitative polymerase chain reaction was used to detect the RAS (KRAS/NRAS), BRAF, and PIK3CA genes. The clinicopathological data of the patients were recorded and analyzed by SPSS 19.0 statistical software.
In 327 elderly patients with CRC, the rate of MMR protein loss was 9.79% (32/327), and the deletion rate of four MMR proteins (MSH2, MSH6, MLH1, PMS2) was 1.83% (6/327), 3.06% (10/327), 7.65% (25/327), and 7.65% (25/327), respectively. There were no significant differences between MMR protein deletion and sex, pathological type, tumor morphology, differentiation degree or lymph node metastasis (P > 0.05), but there was a significant difference between MMR protein deletion and tumor diameter and tumor location (P = 0.048/P = 0.000). The mutation rates of the KRAS, NRAS, BRAF and PIK3CA genes in elderly CRC patients were 44.95% (147/327), 2.45% (8/327), 3.36% (11/327) and 2.75% (9/327), respectively; the KRAS gene mutation was closely related to tumor morphology (P = 0.002) but not to other clinicopathological features (P > 0.05), and there were no significant differences between NRAS gene mutation and clinicopathological features (P > 0.05). The BRAF gene mutation showed a significant difference in pathological type, tumor location, differentiation degree and lymph node metastasis (P < 0.05), but was not correlated with sex, tumor size and tumor morphology (P > 0.05). The PIK3CA gene mutation showed no significant differences in the above clinicopathological characteristics (P > 0.05). Significant differences were observed between MMR protein deletion and KRAS, BRAF, and PIK3CA gene mutations in elderly CRC patients (P = 0.044, P = 0.000, P = 0.003, respectively), but there was no significant difference between MMR protein deletion and NRAS mutation (P > 0.05).
In elderly CRC patients, the tumor is mainly located in the right colon, and the deletion rate of MMR protein is higher when the tumor diameter is greater than or equal to 5 cm; the deletion rate of MLH1 and PMS2 is more common; the mutation rate of KRAS gene is higher than that of the NRAS, BRAF and PIK3CA genes, the BRAF gene mutation has different degrees of correlation with clinicopathological characteristics; when the MMR protein is deleted, the BRAF and PIK3CA gene mutations are often present, and the KRAS gene mutation rate is low.
Core Tip: The mismatch repair (MMR) protein deletion is one of the causes of colorectal cancer (CRC). The RAS (KRAS/NRAS), BRAF, and PIK3CA genes are important gene targets in CRC treatment and are closely related to the prognosis and survival of patients. However, little is known regarding the relationship between the expression of MMR, RAS, BRAF, PIK3CA and the clinicopathological features in CRC patients. In this study, we analyzed four target genes to provide a further theoretical basis for clinicians in relation to the diagnosis, treatment and prognosis of CRC.
- Citation: Fan JZ, Wang GF, Cheng XB, Dong ZH, Chen X, Deng YJ, Song X. Relationship between mismatch repair protein, RAS, BRAF, PIK3CA gene expression and clinicopathological characteristics in elderly colorectal cancer patients. World J Clin Cases 2021; 9(11): 2458-2468
- URL: https://www.wjgnet.com/2307-8960/full/v9/i11/2458.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i11.2458
Colorectal cancer (CRC) is one of the most common malignant tumors in the digestive tract. Its incidence and mortality rates show an increasing trend worldwide. At present, the elderly are still the main group of patients with CRC[1]. Mismatch repair (MMR) protein deletion is one of the causes of CRC[2,3]. The RAS (KRAS/NRAS), BRAF, and PIK3CA genes are important gene targets in CRC treatment and are closely related to the prognosis and survival of patients[4-7]. This study analyzed pathological samples and clinical data from 327 elderly CRC patients to determine the relationship between MMR, RAS (KRAS/NRAS), BRAF, PIK3CA and clinicopathological characteristics, and the relationship between MMR and the four target genes to provide a further theoretical basis for clinicians in the diagnosis, treatment and prognosis of CRC.
Surgical resection specimens from 327 elderly patients with CRC were collected from April 2019 to January 2020 in the Department of Pathology, First Medical Center, People's Liberation Army General Hospital. The patients included 196 males and 131 females, aged 60-91 years with an average age of 70 years. There were 281 cases of adenocarcinoma, 43 cases of mucinous adenocarcinoma, and 3 cases of signet ring cell carcinoma. The tumor was located in the right colon in 78 cases, the left colon in 90 cases, and the rectum in 159 cases. There were 181 cases of ulcer type, 77 cases of protruding type, 45 cases of protruding type of ulcer, and 24 cases of infiltrating type and flat type. There were 112 cases with tumor diameter ≥ 5 cm, and 215 cases with tumor diameter < 5 cm. Four cases were well differentiated, 15 cases were well-moderately differentiated, 241 cases were moderately differentiated, 59 cases were moderately-poorly differentiated, and 8 cases were poorly differentiated. There were 130 cases with lymph node metastasis and 197 cases without lymph node metastasis.
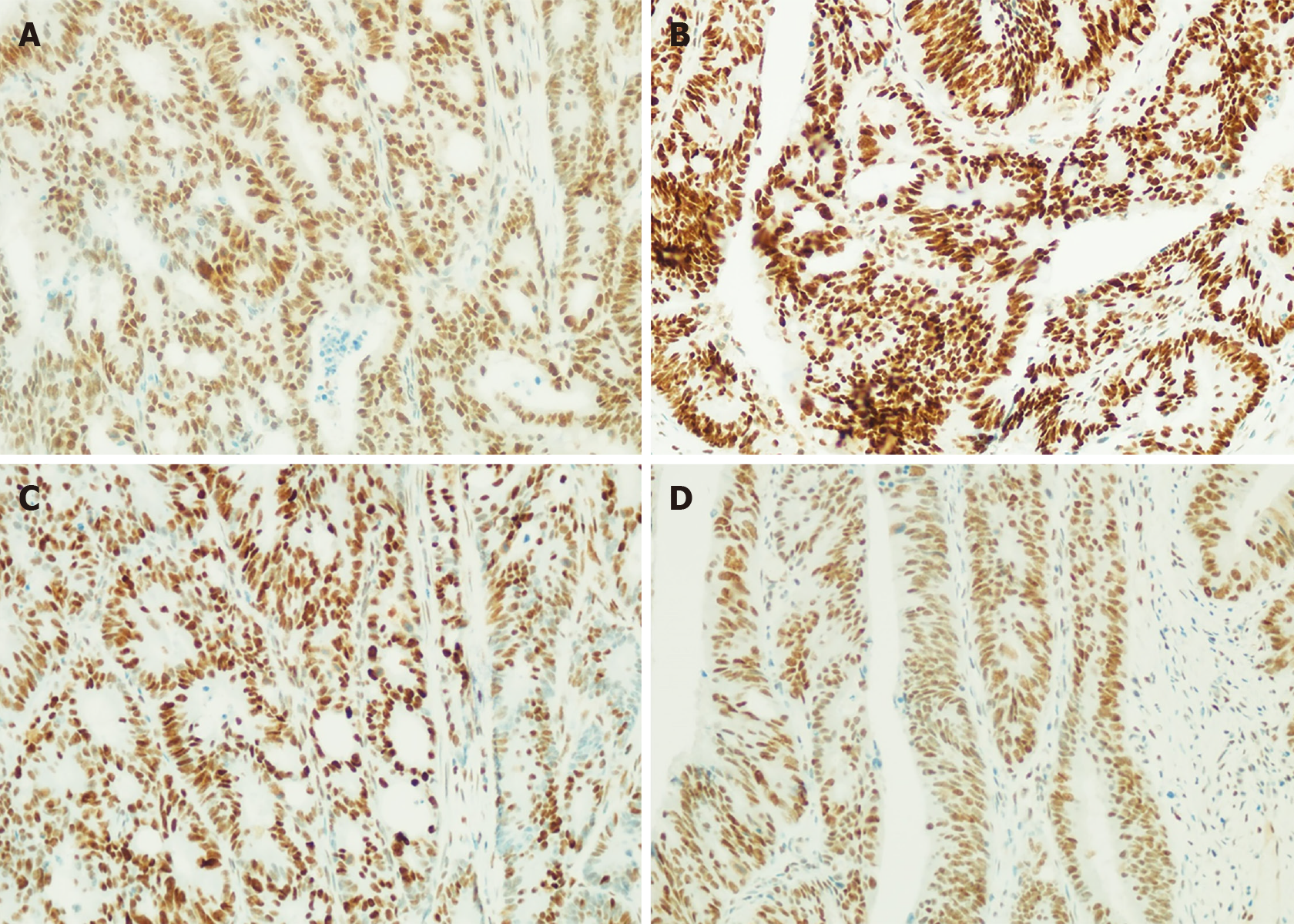
Cationic anti-stripping gel slides were used to cut the paraffin-embedded tissue sections after dehydration with neutral formalin to a thickness of 3-4 μm, which were then dried, placed in a 75°C baking table and baked for 20 min. After 20 min of xylene dewaxing, anhydrous ethanol X2, 95% ethanol X2, 85% ethanol X2 treatment, deionized water cleaning and high-pressure restoration of the antigen, the computer detected MSH2, MSH6, MLH1, and PMS2 MMR protein, antibody clone numbers were: RED2, EP49, ES05, EP51. The expression of MSH2, MSH6, MLH1, and PMS2 was observed under the microscope. When all four proteins were positive (> 30%), this was judged to be pMMR, and when at least one protein was missing, this was judged to be dMMR.
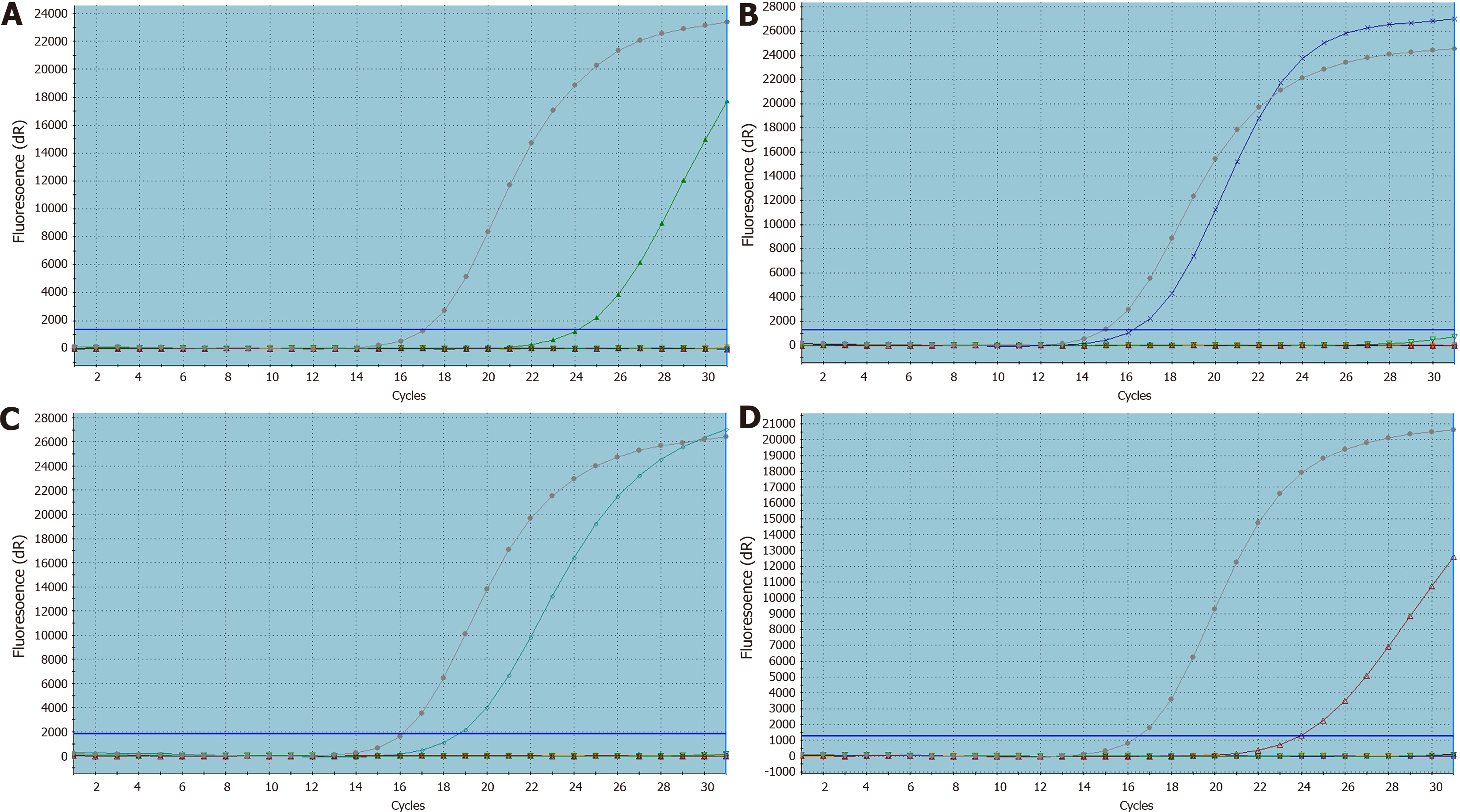
The paraffin embedded tissues were cut into 3-4 pieces 4 μm thick and placed in a clean Eppendorf test tube, and the sample DNA was extracted using the FFPE-DNA sample nucleic acid extraction kit (Xiamen Aide Biomedicine Company), and an ultraviolet spectrophotometer was used to determine the concentration and purity of the sample for quality control. The KRAS, NRAS, BRAF, PIK3CA four gene joint detection kits (Xiamen Aide Biopharmaceutical Company) were used to carry out the polymerase chain reaction (PCR), and after PCR was complete, the results were recorded. The PCR steps are shown in Table 1.
| Polymerase chain reaction steps | ||
| First stage | 95℃, 5 min | 1 cycle |
| Second stage | 95℃, 25 s | 15 cycles |
| 64℃, 20 s | ||
| 72℃, 20 s | ||
| Third stage | 93℃, 25 s | 31 cycles |
| 60℃, 35 s | ||
| 72℃, 20 s | ||
SPSS 19.0 statistical software was used for data analysis, and the χ2 test or Fisher's exact probability method were used to compare differences. A P value < 0.05 was considered statistically significant.
Of the 327 elderly patients with CRC, the loss rate of MMR protein expression was 9.79% (32/327), and the loss rate of the four MMR proteins (MSH2, MSH6, MLH1, PMS2) was 1.83% (6/327), 3.06% (10/327), 7.65% (25/327), and 7.65% (25/327), respectively, and the loss rate of MLH1 and PMS2 was significantly higher than that of MSH2 and MSH6 (P < 0.001). MMR protein loss was not statistically different in terms of patient’s gender, pathological type, tumor morphology, degree of differentiation, lymph node metastasis, etc. (P > 0.05); however, a statistical difference was observed between MMR protein loss and both tumor diameter and tumor location (P = 0.048/P = 0.000). Patients with tumors ≥ 5 cm in diameter were more likely to have MMR protein loss or dMMR. The right colon was more likely to develop dMMR than the left colon and rectum (Table 2). Figure 1 shows the microscopic view of the immunohisto
| Clinical pathology data | n | MMR | χ2 | P value | |
| dMMR | pMMR | ||||
| Gender | 0.005 | 0.945 | |||
| Male | 196 | 19 | 177 | ||
| Female | 131 | 13 | 118 | ||
| Tumor size | 3.907 | 0.048 | |||
| ≥ 5 cm | 112 | 16 | 96 | ||
| < 5 cm | 215 | 16 | 199 | ||
| Pathological type | 0.456 | 0.796 | |||
| Adenocarcinoma | 281 | 27 | 254 | ||
| Mucinous adenocarcinoma | 43 | 5 | 38 | ||
| Other | 3 | 0 | 3 | ||
| Tumor morphology | 1.768 | 0.622 | |||
| Ulcer type | 181 | 19 | 162 | ||
| Raised type | 77 | 9 | 68 | ||
| Ulcer raised type | 45 | 3 | 42 | ||
| Other | 24 | 1 | 23 | ||
| Tumor site | 32.368 | 0.000 | |||
| Right colon | 78 | 19 | 59 | ||
| Left colon | 90 | 11 | 79 | ||
| Rectum | 159 | 2 | 157 | ||
| Differentiation | 8.181 | 0.085 | |||
| High | 4 | 1 | 3 | ||
| High-moderate | 15 | 1 | 14 | ||
| Moderate | 241 | 18 | 223 | ||
| Moderate-poor | 59 | 10 | 49 | ||
| Poor | 8 | 2 | 6 | ||
| Lymph node metastasis | 2.003 | 0.157 | |||
| Yes | 130 | 9 | 121 | ||
| No | 197 | 23 | 174 | ||
KRAS, NRAS, BRAF, PIK3CA gene mutation rates in elderly patients with CRC were 44.95% (147/327), 2.45% (8/327), 3.36% (11/327), and 2.75% (9/327), respectively. The mutation rate of KRAS gene was significantly higher than that of the other three genes (P < 0.001); the mutation rate of KRAS gene was closely related to tumor morphology, and the mutation rate of elevated CRC was significantly lower than that of ulcerative and other types (P = 0.002). Pathological features were not related (P > 0.05). There was no significant difference between NRAS gene mutation and various clinicopathological characteristics (P > 0.05), BRAF gene mutation showed significant differences in relation to pathological type, tumor location, degree of differentiation, and lymph node metastasis (P < 0.05). BRAF gene was more likely to occur in the right colon, in poorly differentiated mucinous adenocarcinoma with lymph node metastasis, which was not related to gender, tumor size, or tumor morphology (P > 0.05), and PIK3CA gene mutation showed no significant differences in relation to the above-mentioned clinicopathological characteristics (P > 0.05, Table 3). The PCR mutation curves of the four genes are shown in Figure 2.
| Clinical pathology data | n | KRAS | χ2 | P | NRAS | χ2 | P | BRAF | χ2 | P | PIK3CA | χ2 | P value | ||||
| Wild | Mutant | Wild | Mutant | Wild | Mutant | Wild | Mutant | ||||||||||
| Gender | |||||||||||||||||
| Male | 196 | 108 | 88 | 0.001 | 0.98 | 192 | 4 | 0.046 | 0.829 | 189 | 7 | 0 | 1 | 191 | 5 | 0 | 1 |
| Female | 131 | 72 | 59 | 127 | 4 | 127 | 4 | 127 | 4 | ||||||||
| Tumor size | |||||||||||||||||
| ≥ 5 cm | 112 | 70 | 42 | 3.825 | 0.051 | 109 | 3 | 0 | 1 | 106 | 6 | 1.254 | 0.263 | 110 | 2 | 0.124 | 0.725 |
| < 5 cm | 215 | 110 | 105 | 210 | 5 | 210 | 5 | 218 | 7 | ||||||||
| Pathological type | |||||||||||||||||
| Adenocarcinoma | 281 | 157 | 124 | 0.908 | 0.635 | 273 | 8 | 1.338 | 0.512 | 273 | 8 | 8.712 | 0.013 | 274 | 7 | 0.734 | 0.693 |
| Mucinous adenocarcinoma | 43 | 22 | 21 | 43 | 0 | 41 | 2 | 41 | 2 | ||||||||
| Other | 3 | 1 | 2 | 3 | 0 | 2 | 1 | 3 | 0 | ||||||||
| Morphology | |||||||||||||||||
| Ulcer | 181 | 89 | 92 | 14.972 | 0.002 | 177 | 4 | 1.328 | 0.723 | 177 | 4 | 3.279 | 0.351 | 176 | 5 | 1.1 | 0.777 |
| Raised | 77 | 57 | 20 | 74 | 3 | 72 | 5 | 74 | 3 | ||||||||
| Ulcer-raised | 45 | 21 | 24 | 44 | 1 | 44 | 1 | 44 | 1 | ||||||||
| Other | 24 | 13 | 11 | 24 | 0 | 23 | 1 | 24 | 0 | ||||||||
| Site | |||||||||||||||||
| Right colon | 78 | 38 | 40 | 3.184 | 0.203 | 77 | 1 | 0.784 | 0.676 | 71 | 7 | 11.004 | 0.004 | 75 | 3 | 5.652 | 0.059 |
| Left colon | 90 | 56 | 34 | 88 | 2 | 90 | 0 | 85 | 5 | ||||||||
| Rectum | 159 | 86 | 73 | 154 | 5 | 155 | 4 | 158 | 1 | ||||||||
| Differentiation | |||||||||||||||||
| High | 4 | 2 | 2 | 1.791 | 0.774 | 4 | 0 | 5.926 | 0.205 | 3 | 1 | 18.408 | 0.001 | 4 | 0 | 4.738 | 0.315 |
| High-moderate | 15 | 8 | 7 | 15 | 0 | 15 | 0 | 15 | 0 | ||||||||
| Moderate | 241 | 134 | 107 | 237 | 4 | 234 | 7 | 236 | 5 | ||||||||
| Moderate-poor | 59 | 30 | 29 | 55 | 4 | 58 | 1 | 55 | 4 | ||||||||
| Poor | 8 | 6 | 2 | 8 | 0 | 6 | 2 | 8 | 0 | ||||||||
| Lymph | |||||||||||||||||
| Yes | 130 | 71 | 59 | 0.016 | 0.899 | 124 | 6 | 2.879 | 0.09 | 124 | 6 | 0.499 | 0.48 | 127 | 3 | 0.003 | 0.957 |
| No | 197 | 109 | 88 | 195 | 2 | 192 | 5 | 191 | 6 | ||||||||
A statistically significant difference was observed between MMR protein deletion and KRAS, BRAF, PIK3CA gene mutations in elderly patients with CRC. When MMR protein deletion or dMMR was present, the KRAS gene mutation rate was significantly lower than that when pMMR was present (P = 0.044). When dMMR occurred, the mutation rate of BRAF and PIK3CA genes was significantly higher than that when pMMR occurred (P = 0.000/P = 0.003), and no significant difference was with NRAS mutation (P > 0.05, Table 4).
| Gene name | n | MMR | χ2 | P value | |
| dMMR | pMMR | ||||
| KRAS | 4.06 | 0.044 | |||
| Wild | 180 | 23 | 157 | ||
| Mutant | 147 | 9 | 138 | ||
| NRAS | 0.116 | 0.733 | |||
| Wild | 319 | 32 | 287 | ||
| Mutant | 8 | 0 | 8 | ||
| BRAF | 12.489 | 0.000 | |||
| Wild | 316 | 27 | 289 | ||
| Mutant | 11 | 5 | 6 | ||
| PIK3CA | 8.879 | 0.003 | |||
| Wild | 318 | 28 | 290 | ||
| Mutant | 9 | 4 | 5 | ||
Many previous studies have shown that mutations or methylation inactivation of DNA MMR genes is not only the main cause of Lynch syndrome but also one of the causes of CRC[2,3]. The elderly are still the main patient group with CRC. The role of the MMR gene is mainly to repair base mismatches during DNA replication and recombination to ensure the stability of gene structure. When the MMR gene is mutated or methylated inactivated, it is prone to mutations of related oncogenes, leading to tumor formation. The most important protein families encoded by MMR genes are MSH2, MSH6, MLH1, and PMS2. They usually work in the form of the MLH1-PMS2 complex and MSH2-MSH6 complex. Previous studies have shown that the deletion rate of MMR protein is approximately 9.5%-34.3%, and the deletion rate of MLH1 is the most common[8,9]. In this study, the deletion rate of the MMR protein in elderly CRC patients was 9.79% (32/327), and the missing rate was slightly lower, which was consistent with previous studies on elderly patients with CRC. The missing rates of MSH2, MSH6, MLH1, and PMS2 were 1.83% (6/327), 3.06% (10/327), 7.65% (25/327), and 7.65% (25/327). The missing rates of MLH1 and PMS2 were significantly higher than those of MSH2 and MSH6 (P < 0.001), which is consistent with previous research results.
Previous studies found that lack of the MMR protein is closely related to the clinicopathological characteristics of CRC[10-13], and some research results show that lack of the MMR protein is related to lymph node metastasis and differentiation[13].
In this study, there were no statistically significant differences between lack of the MMR protein and the patient's sex, pathological type, tumor morphology, degree of differentiation, and lymph node metastasis (P > 0.05), which may have been related to the patient's age and sample data volume. At the same time, there were significant differences in the tumor diameter and tumor location (P = 0.048/P = 0.000). Patients with tumor diameters ≥ 5 cm were more likely to have MMR protein loss or dMMR, and the right colon was more likely to develop dMMR than the left colon and rectum. These results are consistent with previous research results.
Many previous studies have shown that KRAS, NRAS, BRAF, and PIK3CA gene mutations are closely related to the prognosis of CRC[6,7,14-16]. Of these mutations, the frequency of KRAS gene mutations is highest (approximately 30%-50%). When the KRAS gene is mutated, the RAS-RAF-MAPK signal transduction pathway can be activated, resulting in ineffective anti-EGFR inhibitor therapy[14]. In this study, the mutation frequency of KRAS was 44.95% (147/327), which was consistent with the results of previous studies, and we found that the mutation rate of KRAS (26.00%, 20/77) in late-stage CRC was significantly lower (P = 0.002). In previous studies, the mutation frequency of NRAS was below 5%[7,15], and the mutation rate of the BRAF gene was 2%-15%[17]. The PIK3CA gene belongs to the PI3K/AKT/mTOR signaling pathway and is also an EGFR signal. One of the pathways is related to cell proliferation. When PIK3CA is mutated, tumors are more aggressive and have a worse prognosis[6,18,19]. In this study, the mutation rates of the NRAS, BRAF and PIK3CA genes were 2.45% (8/327), 3.36% (11/327), and 2.75% (9/327), respectively, which are consistent with previous research results. In addition, we found that NRAS and PIK3CA were not related to the sex, tumor type, location, tumor size, morphology, and lymph node metastasis of CRC patients, while the BRAF gene is more common in the right colon, lymph node metastasis, mucinous carcinoma, and less differentiated tumors.
In this study, we analyzed the relationship between MMR protein deletion and the KRAS, NRAS, BRAF, and PIK3CA genes. We found that there was a significant correlation between deletion of the MMR protein in elderly CRC patients and KRAS, BRAF, and PIK3CA gene mutations and that there were no correlations between the MMR protein and NRAS mutations (P > 0.05). There was a significant negative correlation between MMR protein deletion and KRAS gene mutation. The mutation rate of the KRAS gene in dMMR was significantly lower than that in pMMR (P = 0.044). KRAS gene mutations can lead to ineffective anti-EGFR inhibitor therapy, which also confirms the results of previous studies on MMR and KRAS in CRC[20,21]. In addition, we also found that the mutation rate of the BRAF and PIK3CA genes was significantly higher than that of pMMR when dMMR occurred (P = 0.000/P = 0.003), which is consistent with the reports of Poulsen et al[22].
In summary, this study retrospectively analyzed several indicators of the MMR, KRAS, NRAS, BRAF, and PIK3CA genes in elderly CRC patients and found that they are related to multiple clinicopathological features, and there are also correlations between them. These findings provide additional support for the clinical diagnosis and treatment of CRC. The disadvantage of this study is that we do not have more data on the clinical treatment and prognosis of these patients and cannot explain the relationship between these indicators and the treatment or prognosis; these relationships need to be further investigated.
Mismatch repair (MMR) protein deletion is one of the causes of colorectal cancer (CRC). The RAS (KRAS/NRAS), BRAF, and PIK3CA genes are important gene targets in CRC treatment and are closely related to the prognosis and survival of patients.
This study provides a further theoretical basis for clinicians in the diagnosis, treatment and prognosis of CRC.
This study aimed to explore the relationship between MMR, RAS (KRAS/NRAS), BRAF, PIK3CA and clinicopathological characteristics, and the relationship between MMR and the four target genes.
The MMR protein was detected by immunohistochemistry, and real-time polymerase chain reaction was performed to detect KRAS, NRAS, BRAF, PIK3CA genes.
There were no significant differences between MMR protein deletion and sex, pathological type, tumor morphology, differentiation degree or lymph node metastasis, but there was a significant difference between MMR protein deletion and tumor diameter and tumor location. The KRAS gene mutation was closely related to tumor morphology, but not to other clinicopathological features, and there were no significant differences between NRAS gene mutation and clinicopathological features, MMR protein deletion and NRAS mutation.
In elderly CRC patients, the deletion rate of MLH1 and PMS2 is more common; the mutation rate of KRAS gene is higher than that of the NRAS, BRAF and PIK3CA genes.
The relationship between these indicators and the treatment or prognosis requires further investigation.
| 1. | Park H, Parys S, Tan J, Entriken F, Hodder R. Post-operative outcomes in the elderly following colorectal cancer surgery. ANZ J Surg. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 2. | Carethers JM, Jung BH. Genetics and Genetic Biomarkers in Sporadic Colorectal Cancer. Gastroenterology 2015; 149: 1177-1190. e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 356] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 3. | Ghanipour L, Jirström K, Sundström M, Glimelius B, Birgisson H. Associations of defect mismatch repair genes with prognosis and heredity in sporadic colorectal cancer. Eur J Surg Oncol. 2017;43:311-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 4. | Benson AB, Venook AP, Al-Hawary MM, Cederquist L, Chen YJ, Ciombor KK, Cohen S, Cooper HS, Deming D, Engstrom PF, Garrido-Laguna I, Grem JL, Grothey A, Hochster HS, Hoffe S, Hunt S, Kamel A, Kirilcuk N, Krishnamurthi S, Messersmith WA, Meyerhardt J, Miller ED, Mulcahy MF, Murphy JD, Nurkin S, Saltz L, Sharma S, Shibata D, Skibber JM, Sofocleous CT, Stoffel EM, Stotsky-Himelfarb E, Willett CG, Wuthrick E, Gregory KM, Freedman-Cass DA. NCCN Guidelines Insights: Colon Cancer, Version 2.2018. J Natl Compr Canc Netw. 2018;16:359-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 635] [Cited by in RCA: 723] [Article Influence: 90.4] [Reference Citation Analysis (3)] |
| 5. | Rosty C, Young JP, Walsh MD, Clendenning M, Walters RJ, Pearson S, Pavluk E, Nagler B, Pakenas D, Jass JR, Jenkins MA, Win AK, Southey MC, Parry S, Hopper JL, Giles GG, Williamson E, English DR, Buchanan DD. Colorectal carcinomas with KRAS mutation are associated with distinctive morphological and molecular features. Mod Pathol. 2013;26:825-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 131] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 6. | Deming DA, Leystra AA, Nettekoven L, Sievers C, Miller D, Middlebrooks M, Clipson L, Albrecht D, Bacher J, Washington MK, Weichert J, Halberg RB. PIK3CA and APC mutations are synergistic in the development of intestinal cancers. Oncogene. 2014;33:2245-2254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 7. | Palomba G, Doneddu V, Cossu A, Paliogiannis P, Manca A, Casula M, Colombino M, Lanzillo A, Defraia E, Pazzola A, Sanna G, Putzu C, Ortu S, Scartozzi M, Ionta MT, Baldino G, Sarobba G, Capelli F, Sedda T, Virdis L, Barca M, Gramignano G, Budroni M, Tanda F, Palmieri G. Prognostic impact of KRAS, NRAS, BRAF, and PIK3CA mutations in primary colorectal carcinomas: a population-based study. J Transl Med. 2016;14:292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 8. | Kim MJ, Chang GJ, Lim HK, Song MK, Park SC, Sohn DK, Chang HJ, Kim DY, Park JW, Jeong SY, Oh JH. Oncological Impact of Lateral Lymph Node Dissection After Preoperative Chemoradiotherapy in Patients with Rectal Cancer. Ann Surg Oncol. 2020;27:3525-3533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 9. | Lindor NM, Burgart LJ, Leontovich O, Goldberg RM, Cunningham JM, Sargent DJ, Walsh-Vockley C, Petersen GM, Walsh MD, Leggett BA, Young JP, Barker MA, Jass JR, Hopper J, Gallinger S, Bapat B, Redston M, Thibodeau SN. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20:1043-1048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 349] [Cited by in RCA: 476] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 10. | Kim ST, Klempner SJ, Park SH, Park JO, Park YS, Lim HY, Kang WK, Kim KM, Lee J. Correlating programmed death ligand 1 (PD-L1) expression, mismatch repair deficiency, and outcomes across tumor types: implications for immunotherapy. Oncotarget. 2017;8:77415-77423. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 69] [Article Influence: 7.7] [Reference Citation Analysis (1)] |
| 11. | Kasi PM. Circulating tumor DNA and plasma microsatellite instability during PD-1 blockade. J Gastrointest Oncol. 2020;11:826-828. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 12. | Zhou G, Noordam L, Sprengers D, Doukas M, Boor PPC, van Beek AA, Erkens R, Mancham S, Grünhagen D, Menon AG, Lange JF, Burger PJWA, Brandt A, Galjart B, Verhoef C, Kwekkeboom J, Bruno MJ. Blockade of LAG3 enhances responses of tumor-infiltrating T cells in mismatch repair-proficient liver metastases of colorectal cancer. Oncoimmunology. 2018;7:e1448332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 13. | Andersen HS, Bertelsen CA, Henriksen R, Campos AH, Kristensen B, Ingeholm P, Gögenur I. The pathological phenotype of colon cancer with microsatellite instability. Dan Med J. 2016;63. [PubMed] |
| 14. | Punt CJ, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol. 2017;14:235-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 311] [Cited by in RCA: 472] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 15. | Wang Y, Liu H, Hou Y, Zhou X, Liang L, Zhang Z, Shi H, Xu S, Hu P, Zheng Z, Liu R, Tang T, Ye F, Liang Z, Bu H. Performance validation of an amplicon-based targeted next-generation sequencing assay and mutation profiling of 648 Chinese colorectal cancer patients. Virchows Arch. 2018;472:959-968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 16. | Yao S, Wang X, Li C, Zhao T, Jin H, Fang W. Kaempferol inhibits cell proliferation and glycolysis in esophagus squamous cell carcinoma via targeting EGFR signaling pathway. Tumour Biol. 2016;37:10247-10256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 17. | Taieb J, Le Malicot K, Shi Q, Penault-Llorca F, Bouché O, Tabernero J, Mini E, Goldberg RM, Folprecht G, Luc Van Laethem J, Sargent DJ, Alberts SR, Emile JF, Laurent Puig P, Sinicrope FA. Prognostic Value of BRAF and KRAS Mutations in MSI and MSS Stage III Colon Cancer. J Natl Cancer Inst. 2017;109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 211] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 18. | Besson A, Robbins SM, Yong VW. PTEN/MMAC1/TEP1 in signal transduction and tumorigenesis. Eur J Biochem. 1999;263:605-611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 101] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 19. | Therkildsen C, Bergmann TK, Henrichsen-Schnack T, Ladelund S, Nilbert M. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: A systematic review and meta-analysis. Acta Oncol. 2014;53:852-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 307] [Cited by in RCA: 311] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 20. | Webber EM, Kauffman TL, O'Connor E, Goddard KA. Systematic review of the predictive effect of MSI status in colorectal cancer patients undergoing 5FU-based chemotherapy. BMC Cancer. 2015;15:156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 82] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 21. | Armstrong SA, Malley R, Weinberg BA. Molecular Profiling in Metastatic Colorectal Cancer. Oncology (Williston Park). 2020;34:352-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 22. | Poulsen TS, de Oliveira DVNP, Espersen MLM, Klarskov LL, Skovrider-Ruminski W, Hogdall E. Frequency and coexistence of KRAS, NRAS, BRAF and PIK3CA mutations and occurrence of MMR deficiency in Danish colorectal cancer patients. APMIS. 2021;129:61-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/Licenses/by-nc/4.0/
Manuscript source: Unsolicited manuscript
Specialty type: Pathology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Higuchi K S-Editor: Zhang H L-Editor: Webster JR P-Editor: Li X