Published online Jul 26, 2020. doi: 10.12998/wjcc.v8.i14.2917
Peer-review started: April 17, 2020
First decision: May 26, 2020
Revised: May 27, 2020
Accepted: June 12, 2020
Article in press: June 12, 2020
Published online: July 26, 2020
Processing time: 98 Days and 9.7 Hours
DNA damage is one of the critical contributors to the occurrence and development of some cancers. APEX1 and APEX2 are the most important molecules in the DNA damage, and APEX1 has been identified as a diagnostic and prognostic biomarker in liver hepatocellular carcinoma (LIHC). However, the expression of APEX2 and its functional mechanisms in LIHC are still unclear.
To examine the expression of APEX2 and the potential mechanism network in LIHC.
We conducted a pan-cancer analysis of the expression of APEX1 and APEX2 using the interactive TIMER tool. GEO datasets, including GSE14520, GSE22058, and GSE64041, were used to compare the APEX2 expression level in tumor tissues and adjacent non-tumor tissues. Then, we calculated the 5-year survival rate according to the web-based Kaplan-Meier analysis. We included the TCGA liver cancer database in GSEA analysis based on the high and low APEX2 expression, showing the potential mechanisms of APEX2 in LIHC. After that, we conducted Pearson correlation analysis using GEPIA2. Next, we performed quantitative polymerase chain reaction (qPCR) assay to examine the APEX2 levels in normal liver cell line LO2 and several liver cancer cell lines, including HepG2, Huh7, SMMC7721, and HCCLM3. APEX2 in HCCLM3 cells was knocked down using small interfering RNA. The role of APEX2 in cell viability was confirmed using CCK-8. Dual-luciferase reporter assay was performed to examine the promoter activity of CCNB1 and MYC.
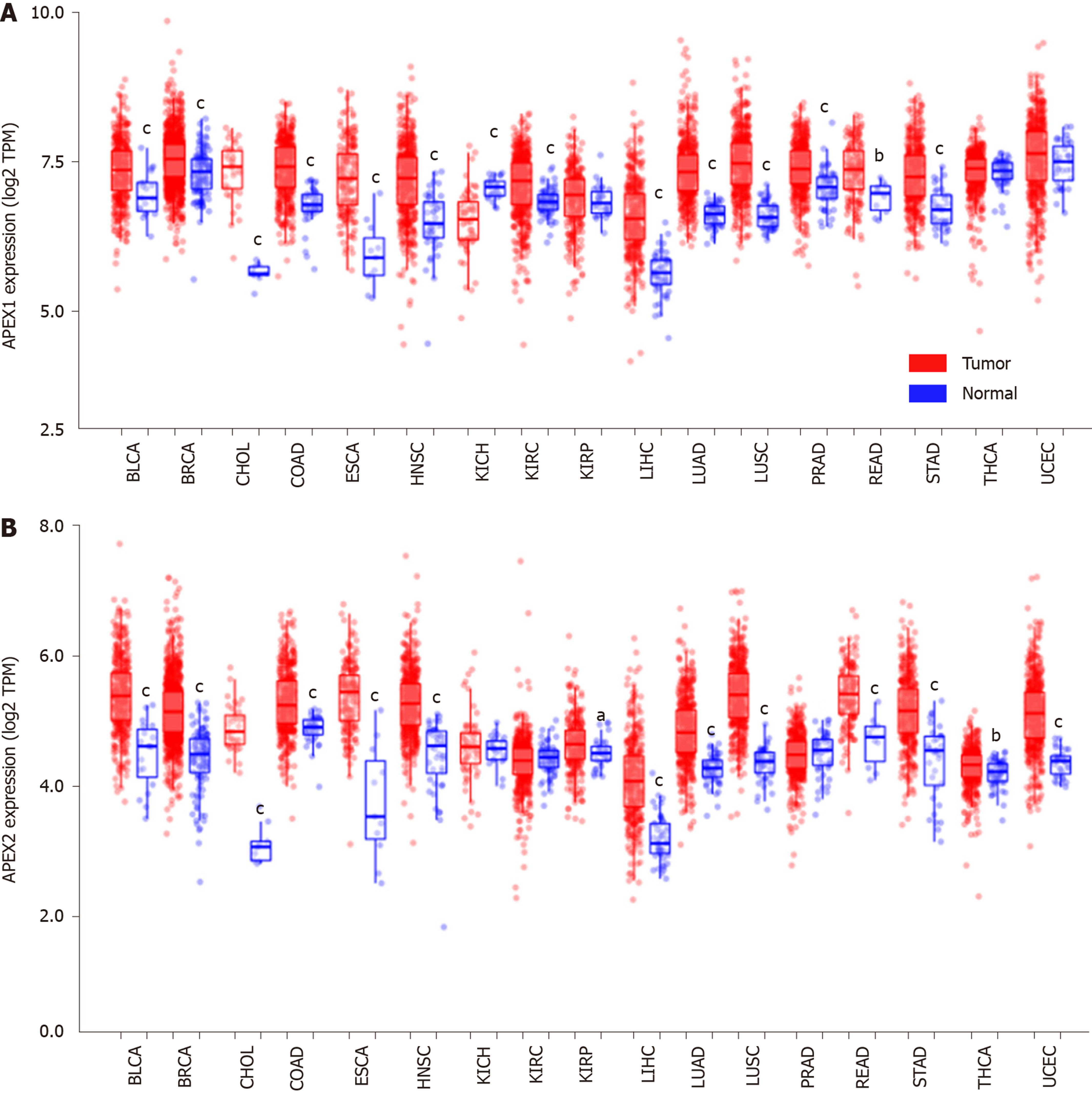
APEX1 and APEX2 are both highly expressed in the tumor tissues of BLCA, BRCA, CHOL, COAD, ESCA, HNSC, LIHC, LUAD, LUSC, READ, and STAD. APEX2 overexpression in LIHC was validated using GSE14520, GSE22058, and GSE64041 datasets. The survival analysis showed that LIHC patients with high expression of APEX2 had a lower overall survival rate, even in the AJCC T1 patients. High level of APEX2 could indicate a lower overall survival rate in patients with or without viral hepatitis. The GSEA analysis identified that kinetochore and spindle microtubules are the two main cellular components of APEX2 in GO Ontology. APEX2 was also positively associated with molecular function regulation of chromosome segregation and DNA replication. The results of KEGG analysis indicated that APEX2 expression was positively correlated with cell cycle pathway and pro-oncogenic MYC signaling. Pearson correlation analysis showed that APEX2 had a significant positive correlation with CCNB1 and MYC. APEX2 level was higher in liver cancer cell lines than in normal liver LO2 cells. Small interfering RNA could knock down the APEX2 expression in HCCLM3 cells. Knockdown of APEX2 resulted in a decrease in the viability of HCCLM3 cells as well as the expression and promoter activity of CCNB1 and MYC.
APEX2 is overexpressed in LIHC, and the higher APEX2 level is associated with a worse prognosis in overall survival. APEX2 is closely involved in the biological processes of chromosome segregation and DNA replication. APEX2 expression is positively correlated with the pro-oncogenic pathways. Knockdown of APEX2 could inhibit the cell viability and CCNB1 and MYC pathways, suggesting that APEX2 is an oncogene in LIHC, which could be a potential pharmaceutic target in the anti-tumor therapy.
Core tip: In this study, we identified that APEX2 is highly expressed in liver hepatocellular carcinoma (LIHC), and overexpression of APEX2 could act as a poor prognostic factor. The GO Ontology-based GSEA analysis suggested that APEX2 is an important regulator in the kinetochore and spindle microtubule. Besides, APEX2 is closely associated with the biological processes of chromosome segregation and DNA replication. The molecular mechanism analysis indicated that APEX2 is positively associated with cell cycle and MYC pathways. Knockdown of APEX2 can impede the viability of liver cancer cells, and inhibit CCNB1 and MYC pathways, suggesting that APEX2 is an oncogene in LIHC.
- Citation: Zheng R, Zhu HL, Hu BR, Ruan XJ, Cai HJ. Identification of APEX2 as an oncogene in liver cancer. World J Clin Cases 2020; 8(14): 2917-2929
- URL: https://www.wjgnet.com/2307-8960/full/v8/i14/2917.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v8.i14.2917
DNA damage is one of the major risk factors for cancerization[1]. Apyrimidinic endodeoxyribonuclease has been identified as an essential molecule that is involved in the regulation of DNA damage[2]. Apyrimidinic endodeoxyribonuclease contains two members, namely, APEX1 and APEX2[3]. Having been widely reported in the pathogenesis of many cancers, especially in liver cancer[4-8], APEX1 was recently identified as a new diagnostic and prognostic biomarker for liver hepatocellular carcinoma (LIHC)[6,8]. However, the role of APEX2 in LIHC remains poorly understood.
LIHC is one of the most malignant tumors and the fifth leading cause of cancer-related death[9]. In recent years, many researchers have been focusing on the mechanism study of liver cancer and have developed several therapeutic drugs[10-12]. However, the prognosis of liver cancer patients is still unsatisfactory. The first choice of liver cancer therapy is hepatectomy, but it is only suitable for early-stage patients[13]. For advanced-stage liver cancer, surgery is still less effective. Another critical issue is the acquired drug resistance, with an incidence of about 20% according to clinical findings, with resistance to sorafenib being a typical example in liver cancer[14-16]. Thus, the identification of novel drug targets could be a practical strategy for liver cancer therapy.
In this study, we focused on the expression of APEX2 and its functional mechanisms in liver cancer. Through bioinformatic analysis, we found the overexpression of APEX2 in liver cancer, and showed the relationship between APEX2 expression and patients’ outcome. GSEA-based enrichment analysis and in vitro analysis confirmed APEX2 as an oncogene in liver cancer. The study of inhibitors against APEX2 could be practical in the treatment of liver cancer.
Dulbecco's Modified Eagle Medium (DMEM, Cat. 11965084), Roswell Park Memorial Institute (RPMI) 1640 Medium (Cat. 12633012), Opti-MEM Reduced Serum Medium (Cat. 51985091), fetal bovine serum (FBS, Cat. A3161001C), and penicillin-streptomycin (5000 U/mL, Cat. 15070063) were purchased from Thermo Fisher Technology (Pennsylvania, United States). RNA isolation kit (Cat. R0016), 1st strand cDNA synthesis kit (Cat. D7170M), SYBR Green qPCR Mix (Cat. D7265), agarose (Cat. ST004L), and dual-luciferase reporter activity detection kit (Cat. RG088S) were obtained from Beyotime (Shanghai, China).
We analyzed the APEX1 and APEX2 expression in the tumor and adjacent non-tumor tissues in the TCGA database with the TIMER web-interactive tool[17]. Only those cancers (BLCA: Bladder urothelial carcinoma; BRCA: Breast invasive carcinoma; CHOL: Cholangiocarcinoma; COAD: Colon adenocarcinoma; ESCA: Esophageal carcinoma; HNSC: Head and neck squamous cell carcinoma; KICH: Kidney Chromophobe; KIRC: Kidney renal clear cell carcinoma; KIRP: Kidney renal papillary cell carcinoma; LIHC: Liver hepatocellular carcinoma; LUAD: Lung adenocarcinoma; LUSC: Lung squamous cell carcinoma; PRAD: Prostate adenocarcinoma; READ: Rectum adenocarcinoma; STAD: Stomach adenocarcinoma; THCA: Thyroid carcinoma; UCEC: Uterine corpus endometrial carcinoma) containing enough samples of tumor and adjacent non-tumor tissues were included. The relative gene expression level is indicated as transcript per million (TPM), and the expression value was normalized by log transformation.
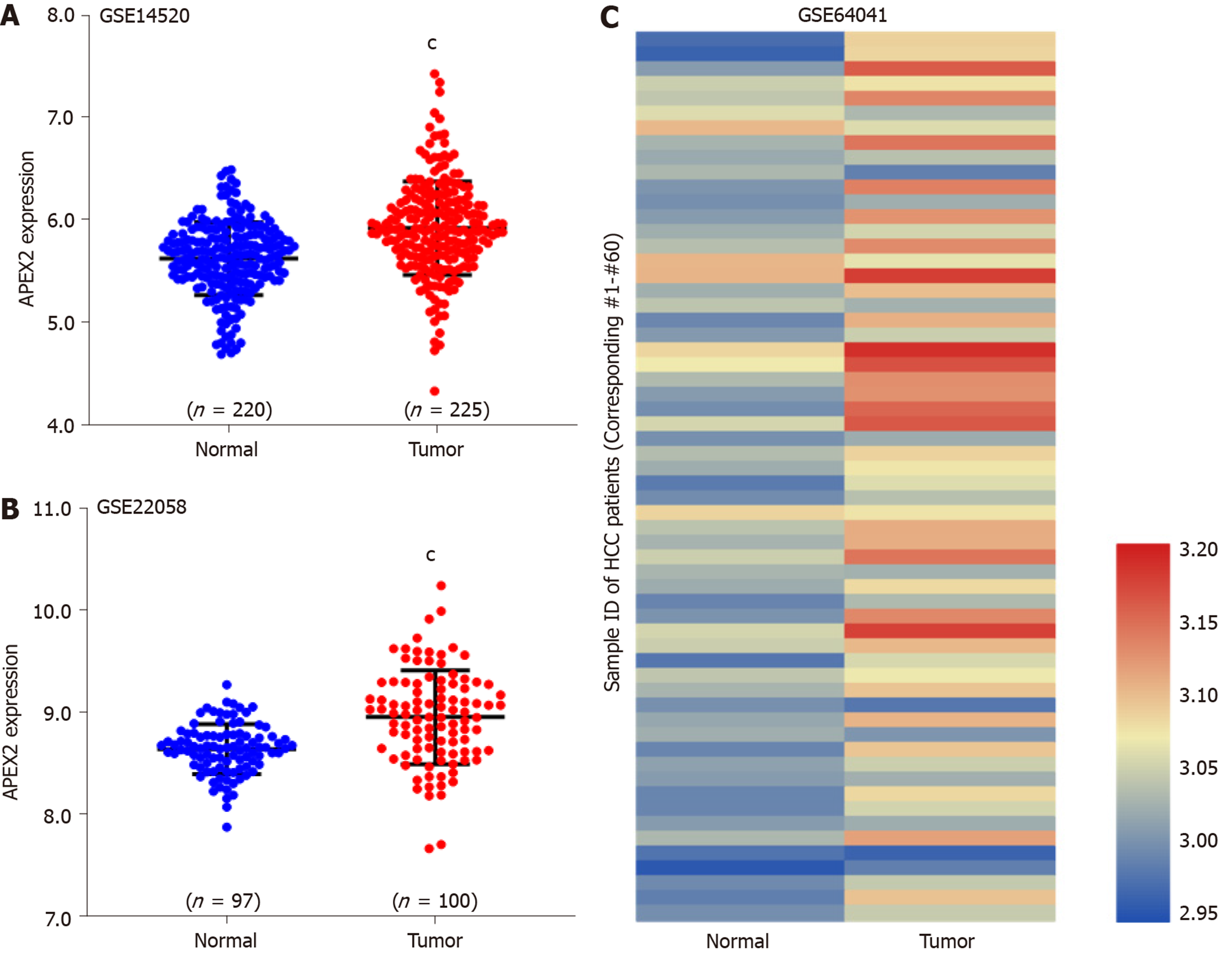
We analyzed GSE14520 (containing 220 adjacent non-tumor and 225 tumor tissues)[18], GSE22058 (containing 97 adjacent non-tumor and 100 tumor tissues)[19], and GSE64041 (containing 60 paired of adjacent non-tumor and tumor tissues)[20] datasets with GEO-2R tool. Then, we calculated the sample size by the distribution of valuable data. The Benjamini & Hochberg (False discovery rate, FDR) was applied to adjust the P value. The expression value was employed with log transformation.
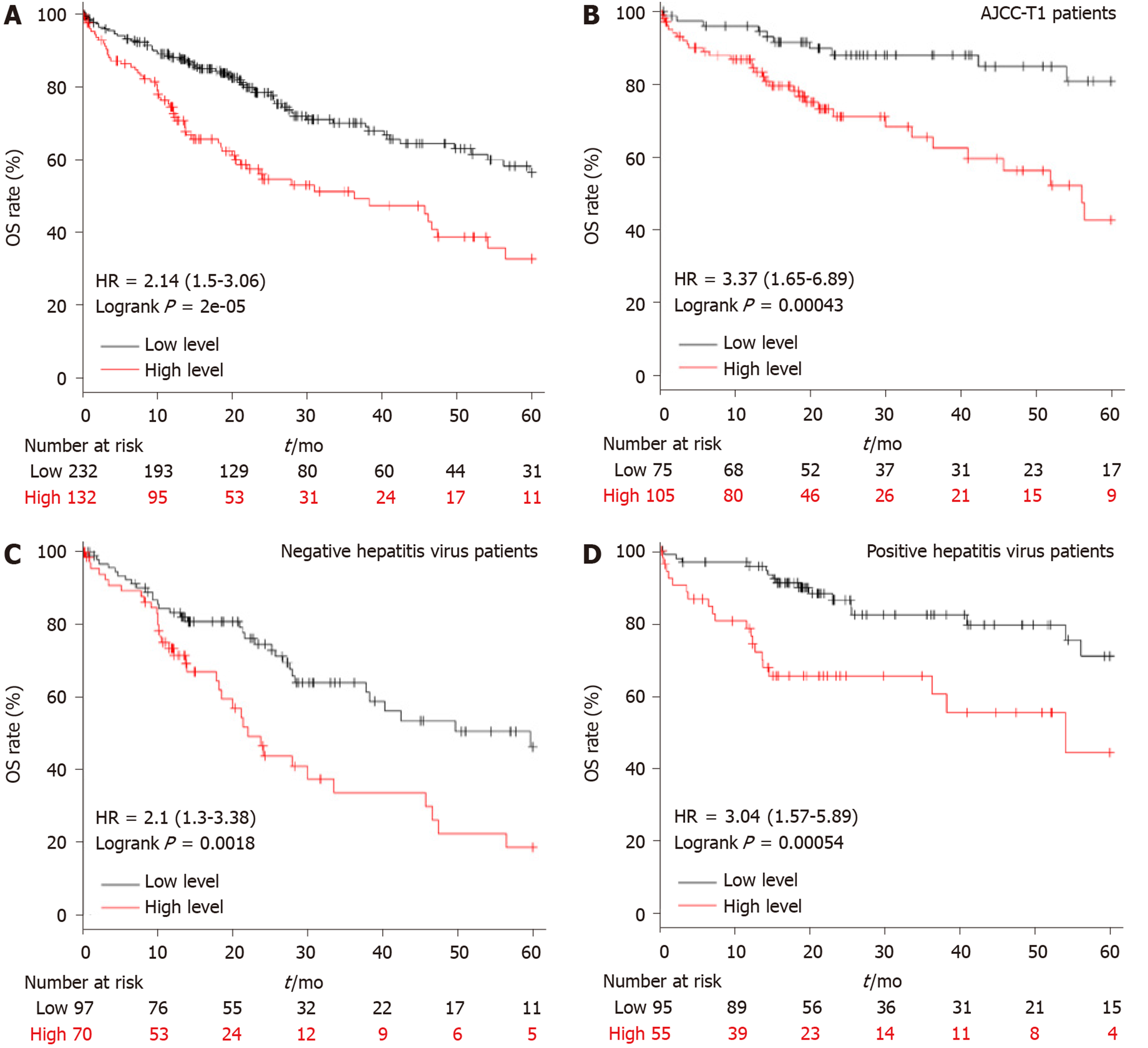
A total of 364 LIHC patients were included in the overall survival (OS) analysis, which was calculated from the date of the surgical resection to the 5-year follow-up event. The best cut-off was conducted in the Kaplan Meier plotter[21], and the patients were divided into high and low APEX2 transcription groups. The clinical features, such as the AJCC stage, hepatitis virus infection, were also included to evaluate the specific subgroup of patients.
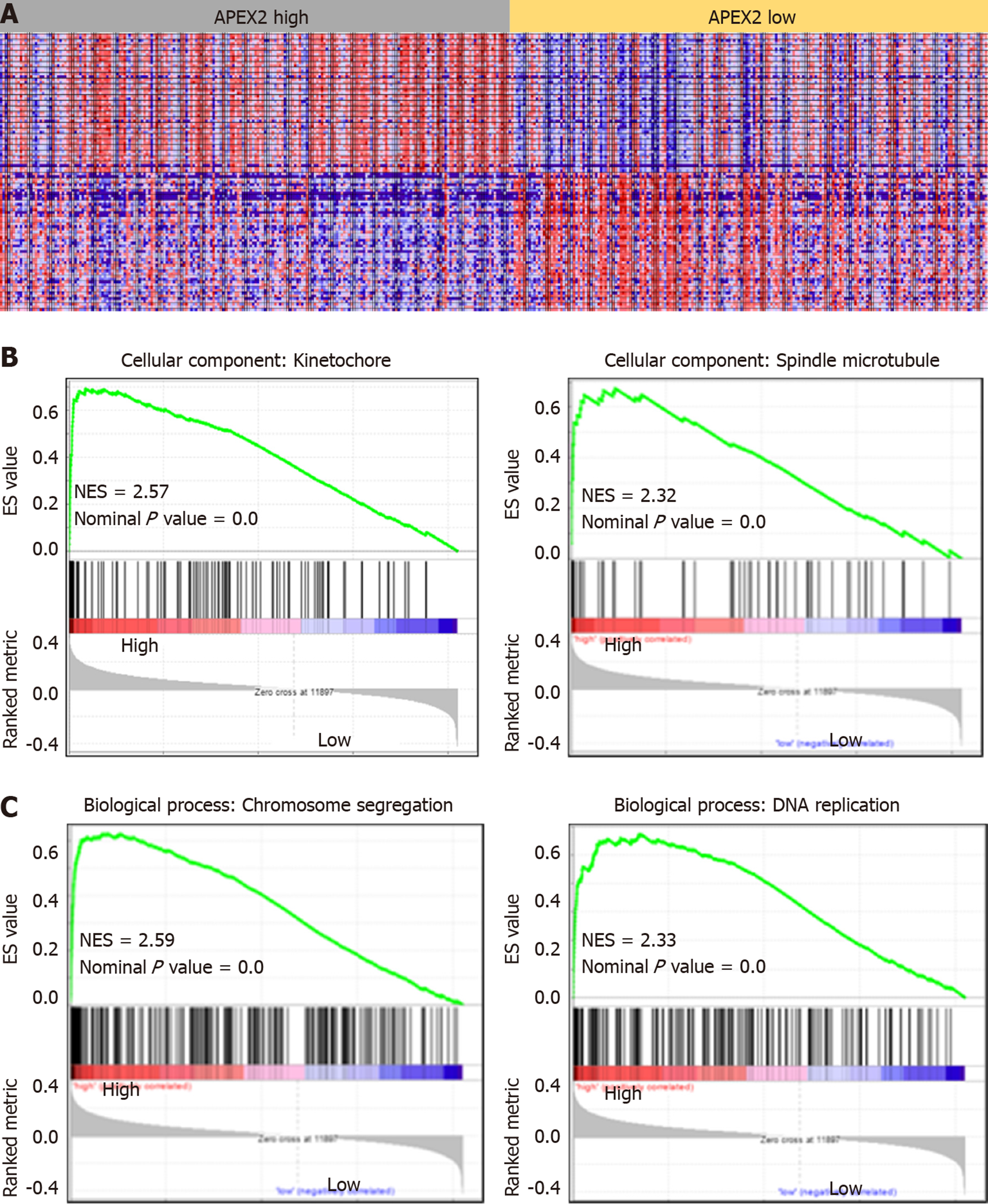
GSEA analysis was carried out as previously reported[22] to evaluate whether a significant set of genes are associated with the APEX2 difference in LIHC. The GO Ontology was selected as the gene sets database to confirm the cellular component and biological process. And the molecular function analysis was achieved by the KEGG v6.2 symbols. The symbol of oncogenic signatures was selected to confirm the potential mechanisms of APEX2 in the LIHC. We set the number of permutations as 1000, weighted the enrichment statistic, and showed the metric for ranking genes as Signal2Noise. The sets below 15 were excluded in the process of analysis.
The normal liver cell line LO2 and liver cancer cell lines SMMC7721 and HCCLM3 were obtained from the Cell Bank of Chinese Academy of Science. The liver cancer cell lines HepG2 and Huh7 were acquired from American Type Culture Collection (ATCC). LO2 cells were cultured in RMPI-1640, and HepG2, SMMC7721, Huh7, and HCCLM3 cells were cultured in DMEM, supplemented with 10% FBS and 100 U/mL penicillin and streptomycin. Small interfering RNA (siRNA) transfection was conducted with Lipofectamine 2000 according to the manufacturer's instructions.
Total RNA was extracted using an RNA isolation kit, and the 1st strand cDNA was synthesized using a commercial kit according to the manufacturer’s protocol. Quantitative PCR was conducted using the SYBR Green kit. The specific primers were synthesized by Sangon Technology (Shanghai, China), with the sequences as follows: APEX2 (forward: 5’-CTGG AACA TCAA TGGG ATTC GG-3’; reverse: 5’-CCAG CTCG TCCA AAAT GCG-3’); GAPDH (forward: 5’-GGAG CGAG ATCC CTCC AAA AT-3’; reverse: 5’-GGCT GTTG TCAT ACTT CTCA TGG-3’). The amplicon sizes of APEX2 and GAPDH are 101 bp and 197 bp, respectively.
HCCLM3 cells were transfected with siRNA against APEX2 and scramble control. After transfection for 24 h, the cells were transfected with the pGL.3-Luc-CCNB1 or pGL.3-Luc-MYC reporter and pCMV-Rellina plasmids, respectively. After further culture for 48 h, the cells were collected and subjected to dual-luciferase reporter activity analysis using a commercial kit according to the manufacturer’s instructions.
The data analyses were conducted using Graphpad Prism version 8.0 (GraphPad Software Inc., La Jolla, CA, USA). The data are shown as the mean ± standard deviation (mean ± SD). The student t-test was applied to compare the statistical difference between groups. Correlation analysis was performed using the Pearson method. The Kaplan Meier method was used to analyze the overall survival, and the log-rank P method was applied to show the statistical difference. P < 0.05 was considered statistically significant.
Recently, APEX1 was identified as a new diagnostic and prognostic biomarker for LIHC, since APEX1 is overexpressed in the liver tumor. As another member of apyrimidinic endodeoxyribonuclease, we hypothesized that APEX2 also plays an important role in LIHC. The pan-cancer analysis showed that APEX1 and APEX2 were both overexpressed in BLCA, BRCA, CHOL, COAD, HNSC, LIHC, LUAD, LUSC, PRAD, READ, and STAD (Figure 1). APEX1 expression was decreased in KICH but increased in KIRC (Figure 1A), suggesting that APEX1 might also have a specific regulation pattern in different histological states. Unlike the specific APEX1 expression pattern in the kidney, APEX2 was only overexpressed in KIRP (Figure 1B). Based on the previous literature on the role of APEX1 in LIHC, and that APEX1 was identified as a poor prognostic factor, we evaluated the role of APEX2 in LIHC in this study.
TCGA-LIHC analysis showed the overexpression of APEX2 in LIHC tumor tissues. Next, several GEO datasets (including GSE14520, GSE22059, and GSE64041) containing a total of 377 adjacent non-tumor tissues and 385 tumor tissues, were used to validate the overexpression of APEX2 in LIHC. As expected, the APEX2 expression level was higher in the liver tumor tissues than in the adjacent non-tumor tissues (Figure 2). As shown in Figure 2C, the GSE64041 containing 60 paired adjacent non-tumor and tumor tissues, was included to present the APEX2 expression value as a heatmap (Figure 2C). It was clear that APEX2 was overexpressed in LIHC.
Similar to APEX1 in LIHC, APEX2 was also highly expressed in LIHC. Higher APEX1 expression was associated with a poorer prognosis in LIHC. Then we assessed the role of APEX2 in the prognosis prediction of LIHC. As shown in Figure 3A, the patients with high APEX2 level had a lower overall survival (OS) rate; the upper quartile survival of the low APEX2 group was 26.7 mo, which was two times more than that of the high APEX2 group (26.7 vs 11.6 mo). Furthermore, to fully determine the prognostic value of APEX2 in LIHC, we further included the tumor stage and viral hepatitis status in the OS analysis. A total of 154 LIHC patients with T1 stage (according to AJCC stage guideline) were subjected to OS analysis in the 5-year follow up. The results showed that the low APEX2 group had a higher OS rate of up to 80% in the 5-year follow up. However, the OS rate of the high APEX2 group was below 50% (Figure 3B). Also, OS rate was compared by APEX2 expression level in both the patients with hepatitis virus infection and those without. As shown in Figure 3C and D, the patients with high APEX2 expression showed a lower OS rate in patients with or without hepatitis virus infection. Collectively, these data suggested that APEX2 is also a poor prognostic marker in LIHC, even in the early-stage patients.
APEX2 is highly expressed in LIHC, and the overexpression of APEX2 is associated with a poor prognosis. Based on these data, we hypothesized that APEX2 is a critical molecule in the occurrence and development of LIHC. GSEA analysis based on the TCGA-LIHC tumor samples was performed in this study. As shown in Figure 4A, 195 LIHC patients were included in a high APEX2 group, and 205 patients were included in a low APEX2 group. The positive and negative correlated genes were represented. Furthermore, the results of cellular component module of GO Ontology suggested that APEX2 was closely associated with the kinetochore and spindle microtubule (Figure 4B), two main cellular components involved in cell proliferation[23]. The significant enrichment score suggested that APEX2 may be involved in the positive regulation of liver cancer cell proliferation. Further biological process module analysis showed that APEX2 was closely associated with chromosome segregation and DNA replication (Figure 4C), which are necessary for cell proliferation. This data was consistent with the cellular component module analysis. The significant enrichment analysis of APEX2 in the feedback regulation of cell proliferation, and the over-expression of APEX2, the lower OS rate, all showed that APEX2 might be an oncogene in LIHC.
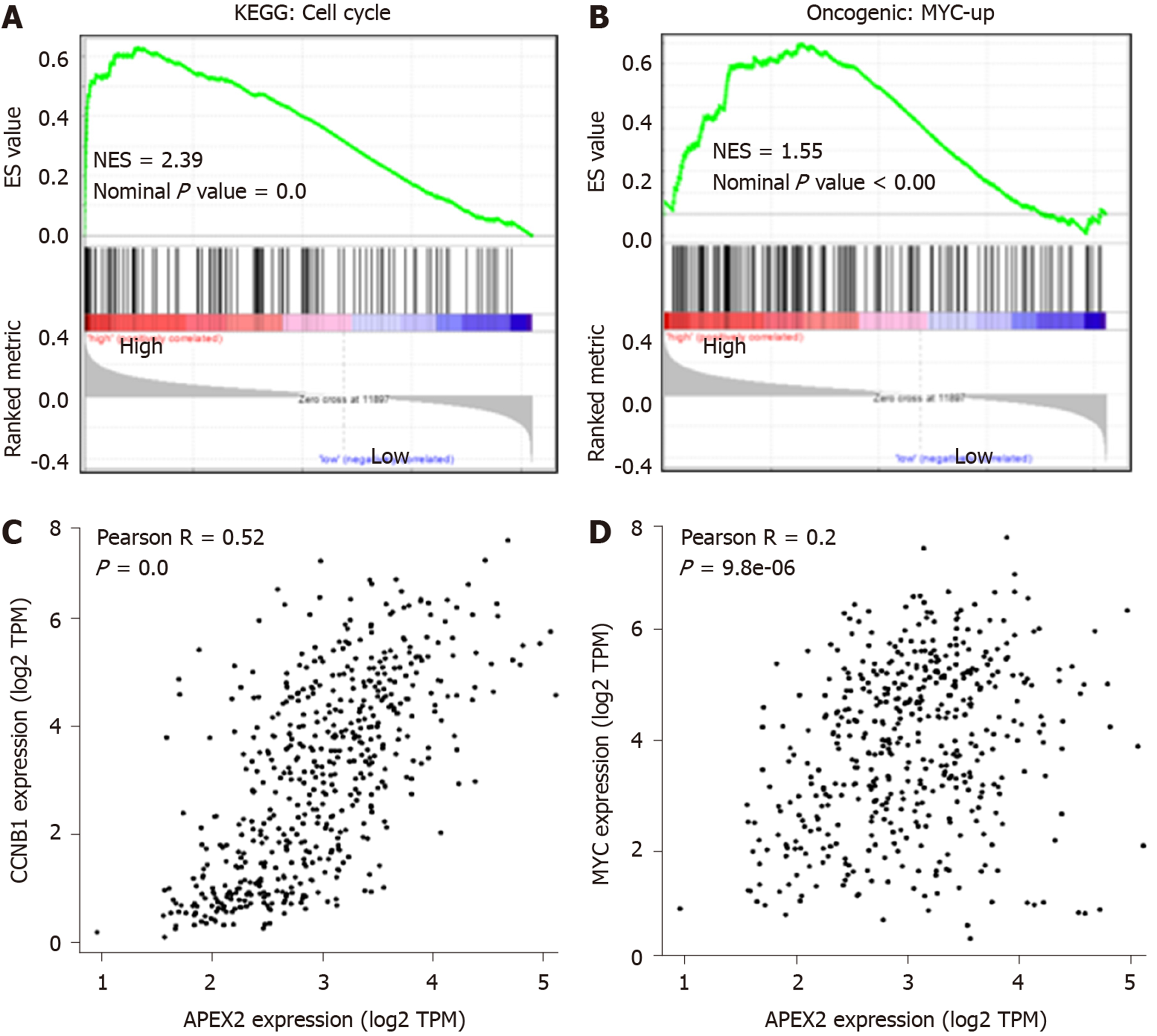
The GO enrichment analysis suggested the positive feedback regulation of APEX2 in DNA-related proliferation, and the detailed molecular mechanisms were further evaluated. As shown in Figure 5A and B, the cell cycle and MYC pathways were enriched with the APEX2 in LIHC. Furthermore, the correlation analysis showed that APEX2 was significantly positively correlated with CCNB1 and MYC (Figure 5C and D). The MYC upregulation is known as a pro-oncogene factor, which is widely activated in many cancers[24]. The regulation of the cell cycle is mainly dependent on the checkpoints. As an essential G2/M checkpoint, CCNB1 was also identified as a pro-oncogenic factor, whose activation could accelerate the transition from G2 to M phase[25]. The remarkable correlation between APEX2 and upregulated MYC and pro-oncogenic cell cycle checkpoint suggested that APEX2 might be an oncogene in the LIHC.
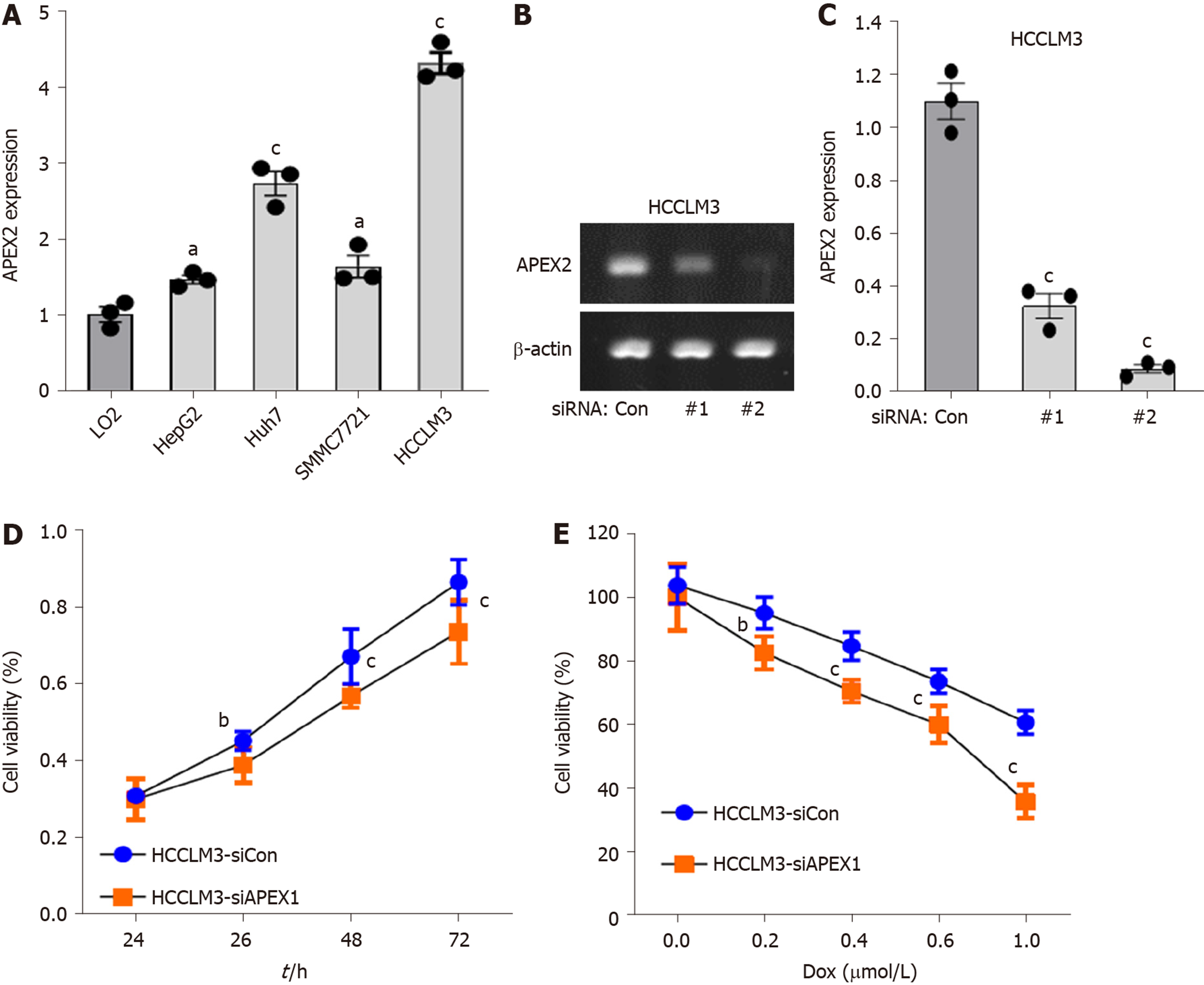
APEX2 overexpression is closely associated with the patients’ worse outcome, and the mechanism prediction demonstrated that APEX2 is an oncogene in LIHC. Next, we further evaluated the role of APEX2 in vitro. As shown in Figure 6A , the APEX2 expression was highly increased in several liver cancer cell lines, including HepG2, Huh7, SMMC7721 and HCCLM3. However, APEX2 expression in LO2, a normal liver cell line, was significantly lower than that in liver cancer cell lines. Interestingly, HCCLM3 cells showed the highest expression of APEX2. In terms of the phenotype of these cell lines, HCCLM3 is the most malignant[26], suggesting that APEX2 expression was closely associated with the malignancy of liver tumor.
Moreover, we examined the effect of APEX2 on the viability of liver cells. First, two siRNA sequences against APEX2 were transfected into HCCLM3 cells, a cell line highly expressing APEX2. The results showed that the second siRNA sequence could remarkably inhibit the APEX2 expression (Figure 6B and C). And this confirmed siRNA sequence was subjected to the subsequent analysis. Second, the knockdown of APEX2 in HCCLM3 cells resulted in distinctly impaired cell viability (Figure 6D). Third, after the knockdown of APEX2, the HCCLM3 cells become more sensitive to doxorubicin (DOX) (Figure 6E), which is a common drug in liver chemotherapy. Taken together, these results showed that APEX2 as an oncogene could affect the proliferation of liver cells.
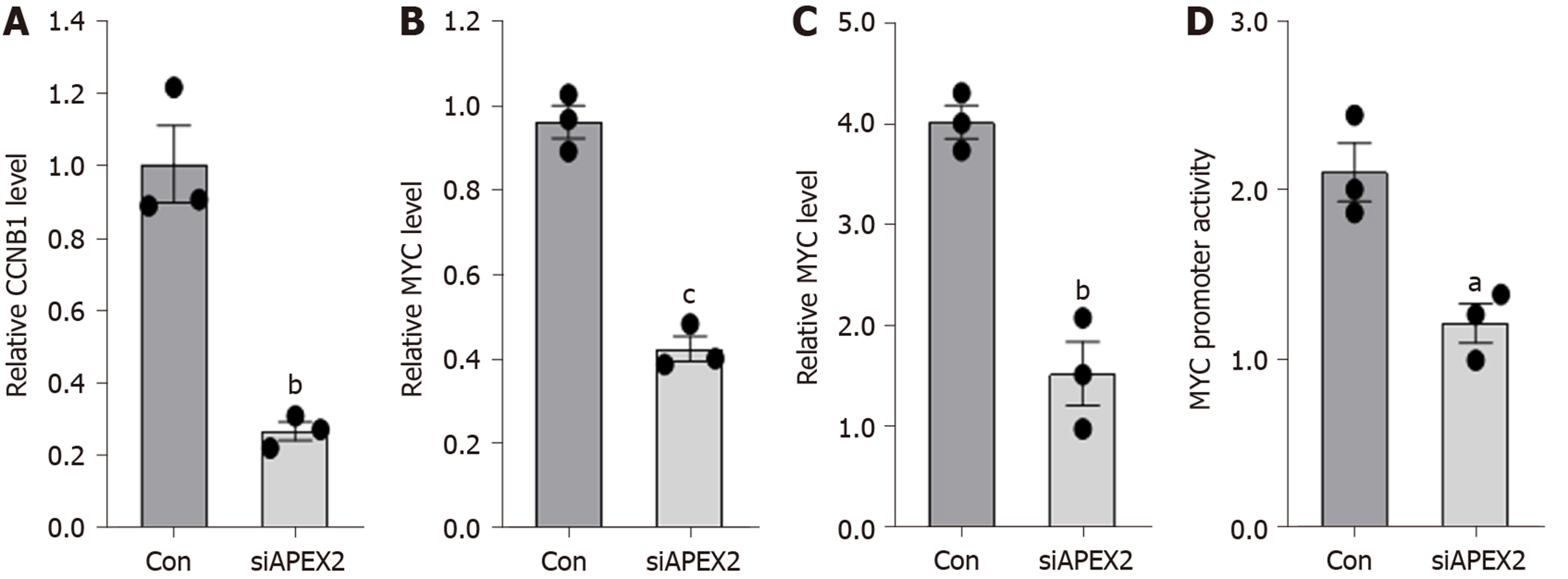
The GSEA analysis showed that APEX2 is closely associated with cell cycle checkpoints and pro-oncogenic MYC. Knockdown of APEX2 can inhibit the viability of liver cancer cells. Thus, we hypothesized that APEX2 might inhibit cell proliferation through CCNB1 and MYC-mediated pathways. First, we examined the mRNA levels of CCNB1 and MYC in HCCLM3 cells transfected with APEX2 siRNA or scramble control. As shown in Figure 7A and B, knockdown of APEX2 can decrease the CCNB1 and MYC expression. Since the regulation of promoter function is the most significant pattern that contributes to the dysregulation of gene expression, we also evaluated the effect of APEX2 on the promoter activity of CCNB1 and MYC. The results showed that the promoter activity of CCNB1 and MYC decreased significantly in the APEX2 siRNA group (Figure 7C and D), which was consistent with the enrichment analysis.
LIHC is a highly malignant tumor, which contributes to the 5th cancer-related death according to the cancer statistics[9]. Currently, the diagnosis of LIHC has improved a lot due to early screening. In clinical practice, some serum biomarkers have been applied in the early screening of LIHC, such as alpha-fetoprotein (AFP), glypican 3 (GPC3), and glutamic pyruvic transaminase (GPT)[27-29]. With the significant advancement of medical imaging such as computed tomography and magnetic resonance, radiologists could also check out the tumor occupation[30]. However, the prognosis of LIHC patients remains poor. The deficiency of powerful drugs and acquired drug resistance are the main reasons. Sorafenib is the only approved targeted drug for LIHC, and it is commonly reported that the drug resistance would be acquired in the process of therapy[31]. Although researchers have been trying their best to find underlying the mechanisms, there are much more unknown. In this sense, the identification of new therapeutic targets could provide a practical strategy.
Apyrimidinic endodeoxyribonuclease genes contain two members, APEX1 and APEX2. Some literature suggested that APEX1 and APEX2 are involved in DNA damage[2,32]. And DNA damage is a remarkable marker for canceration. Also, APEX1 was reported to be associated with some cancer types, including the cancers of colon, stomach, liver, kidney, and breast, as well as osteosarcoma[4-8,33]. Recent studies showed that APEX1 is an efficient biomarker for the diagnosis and prognosis of liver cancer[6,8,34]. However, the role of APEX2 in cancer is still poorly understood. Considering the importance of APEX1 in liver cancer, this study aimed to determine the role of APEX2 in liver cancer. First, we evaluated the expression of APEX1 and APEX2 in various cancer tissues to confirm the difference of APEX1 and APEX2. The common overexpression of APEX1 and APEX2 in cancer types suggested that they could be potential biomarkers for cancer screening (Figure 1). The overexpression of APEX2 in liver cancer was also validated based on other external data (Figure 2), which is consistent with the previous reports of APEX1[6]. To confirm the clinical value of APEX2 in liver cancer, this study also evaluated the relationship between APEX2 expression and survival time. As hypothesized, the overexpression of APEX2 in liver tumors suggested a lower overall survival rate, even in the early stage (Figure 3). This study also analyzed the potential regulatory mechanisms of APEX2 in liver cancer through bioinformatic methods. APEX2 is closely related to the regulation of chromosome segregation and DNA replication. The analysis of the signaling pathways also showed that APEX2 is involved in the regulation of cell cycle checkpoints and the pro-oncogenic MYC (Figures 5 and 6). Based on the bioinformatic analysis, the in vitro assays also validated the results (Figure 7).
Furthermore, we performed the functional validation of APEX2. Knockdown of APEX2 in HCCLM3 cells could impair the proliferative index of liver cancer cells. However, this study aimed to explain the function of APEX2 in a macro-perspective manner. Further study on the mechanisms of APEX2 in the regulation of the cell cycle is needed. Based on the downregulation of CCNB1 by APEX2, the specific checkpoints need to be examined, such as CDK1, CCND1, and other molecules. Furthermore, the effect of APEX2 on the production of growth factors, such as the transcription factor E2F and the downstream target genes, needs more research. Another issue is the mutation of p53, a common phenomenon in liver cancer[35]. p53 mutation can lead to the phosphorylation of Rb and accelerate the transition of the cell cycle from G2 to M phase[36]. Thus, exploring the effect of APEX2 on the p53 activity is an essential work in the next study.
Overall, APEX2 is identified as an oncogene in liver cancer, the overexpression of APEX2 could be a poor prognosis factor, and knockdown of APEX2 can disrupt the proliferation activity; thus the inhibitor against APEX2 may be a promising therapeutic strategy forliver cancer.
Hepatocellular carcinoma (HCC) is a most malignant tumor; the deficiency of drugs is a critical factor for poor prognosis. Thus, the identification of novel drug targets is significant. APEX1 was reported as a tumor promoter in HCC. However, the expression and clinical significance of APEX2 in HCC are still unclear.
This study was to evaluate the role of APEX2 in HCC and identify the potential mechanisms associated with APEX2.
This study aimed to evaluate the levels of APEX2 in the HCC, the association between APEX2 and patients’ prognosis, and the potential regulatory mechanisms of APEX2.
We performed pan-cancer analysis about APEX1 and APEX2 in TIMER, and included the GEO datasets to confirm the APEX2 expression in liver tumor and adjacent normal tissues. Then, the clinical significance of APEX2 was confirmed with the Kaplan-Meier method. Next, we conducted the gene enrichment analysis of APEX2 with the GSEA method, and subjected the KEGG pathways to show the potential signaling pathways. The correlation between APEX2 and other genes was detected in the GEPIA2 tool. For in vitro studies, we performed qPCR to analyze the level of relative gene expression. Then, the knockdown assay was conducted using small interfering RNA. The cell viability was tested with CCK-8. The promoter activity was detected by dual-luciferase reporter assay.
The pan-cancer analysis showed that APEX1 and APEX2 were over-expressed in some cancers, especially in HCC. GSE14520, GSE64041 and GSE22058 datasets analysis confirmed that APEX2 was over-expressed in HCC. And the 5-year survival analysis showed that the patients with higher APEX2 expression had a lower survival rate, even in those with AJCC-T1 in the early stage. We also confirmed the poor prognosis prediction of APEX2 in the liver cancers with or without hepatitis virus infection. The gene enrichment analysis showed that APEX2 was enriched in the regulation of chromosome segregation and DNA replication. And the most significant pathways associated with APEX2 were cell cycle pathways and MYC target. The Pearson correlation analysis showed that APEX2 was positively correlated with CCNB1 and MYC. The over-expression of APEX2 in HCC was also confirmed in the liver cancer cell lines and normal LO2 liver cells. And knockdown of APEX2 could inhibit the cell viability of HCCLM3 cells, and decrease CCNB1 and MYC expression and promoter activity.
Our findings confirmed the over-expression of APEX2 in the HCC, which could serve as a poor prognostic marker. The mechanism regulation analysis identified that APEX2 was closely involved in the biological process of chromosome segregation and DNA replication, which was positively associated with pro-oncogenic pathways.
In this study, we identified APEX2 as an oncogenic gene in HCC, the clinical significance of which was also confirmed with the public tumor database. And in the next work, the association between APEX2 and clinical-pathological features should be included to further evaluate the clinical significance of APEX2. Another critical task is to study the regulation of APEX2 in the cell cycle, and the analysis of the signaling pathways involving APEX2 in HCC.
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A, A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Katuchova J, Ueda H S-Editor: Gong ZM L-Editor: A E-Editor: Liu JH
| 1. | Goldstein JB, Zhao L, Wang X, Ghelman Y, Overman MJ, Javle MM, Shroff RT, Varadhachary GR, Wolff RA, McAllister F, Futreal A, Fogelman DR. Germline DNA Sequencing Reveals Novel Mutations Predictive of Overall Survival in a Cohort of Patients with Pancreatic Cancer. Clin Cancer Res. 2020;26:1385-1394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 2. | Lin Y, Bai L, Cupello S, Hossain MA, Deem B, McLeod M, Raj J, Yan S. APE2 promotes DNA damage response pathway from a single-strand break. Nucleic Acids Res. 2018;46:2479-2494. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 3. | Dan Y, Ohta Y, Tsuchimoto D, Ohno M, Ide Y, Sami M, Kanda T, Sakumi K, Nakabeppu Y. Altered gene expression profiles and higher frequency of spontaneous DNA strand breaks in APEX2-null thymus. DNA Repair (Amst). 2008;7:1437-1454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 4. | Kim HB, Lee HJ, Kim GB, Lim HJ, Park JH, Park SG. Clinical Significance of Jagged-1 Activated by APEX1 as a Chemoresistance Factor in Advanced Gastric Cancer. Anticancer Res. 2020;40:1897-1904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 5. | Kim HB, Lim HJ, Lee HJ, Park JH, Park SG. Evaluation and Clinical Significance of Jagged-1-activated Notch Signaling by APEX1 in Colorectal Cancer. Anticancer Res. 2019;39:6097-6105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 6. | Kim JM, Yeo MK, Lim JS, Song IS, Chun K, Kim KH. APEX1 Expression as a Potential Diagnostic Biomarker of Clear Cell Renal Cell Carcinoma and Hepatobiliary Carcinomas. J Clin Med. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 7. | Malfatti MC, Gerratana L, Dalla E, Isola M, Damante G, Di Loreto C, Puglisi F, Tell G. APE1 and NPM1 protect cancer cells from platinum compounds cytotoxicity and their expression pattern has a prognostic value in TNBC. J Exp Clin Cancer Res. 2019;38:309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 8. | Cao L, Cheng H, Jiang Q, Li H, Wu Z. APEX1 is a novel diagnostic and prognostic biomarker for hepatocellular carcinoma. Aging (Albany NY). 2020;12:4573-4591. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 9. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12667] [Cited by in RCA: 15514] [Article Influence: 2585.7] [Reference Citation Analysis (6)] |
| 10. | Chan BQY, Cheng H, Liow SS, Dou Q, Wu YL, Loh XJ, Li Z. Poly(carbonate urethane)-Based Thermogels with Enhanced Drug Release Efficacy for Chemotherapeutic Applications. Polymers (Basel). 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 11. | Okushin K, Tsutsumi T, Ikeuchi K, Kado A, Enooku K, Fujinaga H, Moriya K, Yotsuyanagi H, Koike K. Helicobacter pylori infection and liver diseases: Epidemiology and insights into pathogenesis. World J Gastroenterol. 2018;24:3617-3625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 31] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 12. | Hernández-Aquino E, Muriel P. Beneficial effects of naringenin in liver diseases: Molecular mechanisms. World J Gastroenterol. 2018;24:1679-1707. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 194] [Cited by in RCA: 251] [Article Influence: 31.4] [Reference Citation Analysis (15)] |
| 13. | Bouvry C, Palard X, Edeline J, Ardisson V, Loyer P, Garin E, Lepareur N. Transarterial Radioembolization (TARE) Agents beyond 90Y-Microspheres. Biomed Res Int. 2018;2018:1435302. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 14. | Bai J, Liu Z, Liu J, Zhang S, Tian Y, Zhang Y, Ren L, Kong D. Mitochondrial metabolic study guided by proteomics analysis in hepatocellular carcinoma cells surviving long-term incubation with the highest dose of sorafenib. Aging (Albany NY). 2019;11:12452-12475. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 15. | Xia S, Pan Y, Liang Y, Xu J, Cai X. The microenvironmental and metabolic aspects of sorafenib resistance in hepatocellular carcinoma. EBioMedicine. 2020;51:102610. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 230] [Article Influence: 38.3] [Reference Citation Analysis (10)] |
| 16. | Xiang DM, Sun W, Zhou T, Zhang C, Cheng Z, Li SC, Jiang W, Wang R, Fu G, Cui X, Hou G, Jin GZ, Li H, Hou C, Liu H, Wang H, Ding J. Oncofetal HLF transactivates c-Jun to promote hepatocellular carcinoma development and sorafenib resistance. Gut. 2019;68:1858-1871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 17. | Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77:e108-e110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2728] [Cited by in RCA: 4292] [Article Influence: 476.9] [Reference Citation Analysis (0)] |
| 18. | Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX, Wang XW. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202-10212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 761] [Cited by in RCA: 809] [Article Influence: 50.6] [Reference Citation Analysis (0)] |
| 19. | Burchard J, Zhang C, Liu AM, Poon RT, Lee NP, Wong KF, Sham PC, Lam BY, Ferguson MD, Tokiwa G, Smith R, Leeson B, Beard R, Lamb JR, Lim L, Mao M, Dai H, Luk JM. microRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol Syst Biol. 2010;6:402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 167] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 20. | Makowska Z, Boldanova T, Adametz D, Quagliata L, Vogt JE, Dill MT, Matter MS, Roth V, Terracciano L, Heim MH. Gene expression analysis of biopsy samples reveals critical limitations of transcriptome-based molecular classifications of hepatocellular carcinoma. J Pathol Clin Res. 2016;2:80-92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 21. | Xu Z, Zhou Y, Cao Y, Dinh TL, Wan J, Zhao M. Identification of candidate biomarkers and analysis of prognostic values in ovarian cancer by integrated bioinformatics analysis. Med Oncol. 2016;33:130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 63] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 22. | Xiao Y, Zhu Z, Li J, Yao J, Jiang H, Ran R, Li X, Li Z. Expression and prognostic value of long non-coding RNA H19 in glioma via integrated bioinformatics analyses. Aging (Albany NY). 2020;12:3407-3430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 23. | Wimbish RT, DeLuca JG. Hec1/Ndc80 Tail Domain Function at the Kinetochore-Microtubule Interface. Front Cell Dev Biol. 2020;8:43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 24. | Liu H, Liu N, Zhao Y, Zhu X, Wang C, Liu Q, Gao C, Zhao X, Li J. Oncogenic USP22 supports gastric cancer growth and metastasis by activating c-Myc/NAMPT/SIRT1-dependent FOXO1 and YAP signaling. Aging (Albany NY). 2019;11:9643-9660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 25. | Huang L, Wang Y, Chen J, Wang Y, Zhao Y, Wang Y, Ma Y, Chen X, Liu W, Li Z, Zhao L, Shan B, Dong X, Li D, Shao S, Song Y, Zhan Q, Liu X. Long noncoding RNA PCAT1, a novel serum-based biomarker, enhances cell growth by sponging miR-326 in oesophageal squamous cell carcinoma. Cell Death Dis. 2019;10:513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 71] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 26. | Yang XM, Cao XY, He P, Li J, Feng MX, Zhang YL, Zhang XL, Wang YH, Yang Q, Zhu L, Nie HZ, Jiang SH, Tian GA, Zhang XX, Liu Q, Ji J, Zhu X, Xia Q, Zhang ZG. Overexpression of Rac GTPase Activating Protein 1 Contributes to Proliferation of Cancer Cells by Reducing Hippo Signaling to Promote Cytokinesis. Gastroenterology. 2018;155:1233-1249.e22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 84] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 27. | Lu Q, Li J, Cao H, Lv C, Wang X, Cao S. Comparison of diagnostic accuracy of Midkine and AFP for detecting hepatocellular carcinoma: a systematic review and meta-analysis. Biosci Rep. 2020;40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 28. | Zhang Q, Han Z, Tao J, Zhao M, Zhang W, Li P, Tang L, Gu Y. An innovative peptide with high affinity to GPC3 for hepatocellular carcinoma diagnosis. Biomater Sci. 2018;7:159-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 29. | Tahiri M, Drighil A, Jalal Y, Ghellab D, Hliwa W, Fouad H, Badre W, Bellabah A, Habbal R, Alaoui R. Chronic permanent hypoxemia predisposes to mild elevation of liver stiffness. World J Gastroenterol. 2014;20:10564-10569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 2] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 30. | Smajerova M, Petrasova H, Little J, Ovesna P, Andrasina T, Valek V, Nemcova E, Miklosova B. Contrast-enhanced ultrasonography in the evaluation of incidental focal liver lesions: A cost-effectiveness analysis. World J Gastroenterol. 2016;22:8605-8614. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 31] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (2)] |
| 31. | Cerrito L, Annicchiarico BE, Iezzi R, Gasbarrini A, Pompili M, Ponziani FR. Treatment of hepatocellular carcinoma in patients with portal vein tumor thrombosis: Beyond the known frontiers. World J Gastroenterol. 2019;25:4360-4382. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 82] [Cited by in RCA: 91] [Article Influence: 13.0] [Reference Citation Analysis (1)] |
| 32. | Lin Y, Raj J, Li J, Ha A, Hossain MA, Richardson C, Mukherjee P, Yan S. APE1 senses DNA single-strand breaks for repair and signaling. Nucleic Acids Res. 2020;48:1925-1940. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 33. | Zhao Z, Lin X, Tong Y, Li W. Silencing lncRNA ZFAS1 or elevated microRNA-135a represses proliferation, migration, invasion and resistance to apoptosis of osteosarcoma cells. Cancer Cell Int. 2019;19:326. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 34. | Tummanatsakun D, Proungvitaya T, Roytrakul S, Limpaiboon T, Wongkham S, Wongkham C, Silsirivanit A, Somintara O, Sangkhamanon S, Proungvitaya S. Serum Apurinic/Apyrimidinic Endodeoxyribonuclease 1 (APEX1) Level as a Potential Biomarker of Cholangiocarcinoma. Biomolecules. 2019;9:413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 35. | Rebouissou S, Nault JC. Advances in molecular classification and precision oncology in hepatocellular carcinoma. J Hepatol. 2020;72:215-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 408] [Article Influence: 68.0] [Reference Citation Analysis (2)] |
| 36. | Ni Z, Wang X, Zhang T, Li L, Li J. Comprehensive analysis of differential expression profiles reveals potential biomarkers associated with the cell cycle and regulated by p53 in human small cell lung cancer. Exp Ther Med. 2018;15:3273-3282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |