Published online Sep 6, 2019. doi: 10.12998/wjcc.v7.i17.2463
Peer-review started: March 20, 2019
First decision: May 9, 2019
Revised: July 12, 2019
Accepted: July 27, 2019
Article in press: July 27, 2019
Published online: September 6, 2019
Processing time: 170 Days and 17.3 Hours
Crohn’s disease (CD) is a complex disorder resulting from the interaction of genetic, environmental, and microbial factors. The pathogenic process may potentially affect any segment of the gastrointestinal tract, but a selective location in the terminal ileum was reported in 50% of patients.
To characterize clinical sub-phenotypes (colonic and/or ileal) within the same disease, in order to identify new therapeutic targets.
14 consecutive patients undergoing surgery for ileal CD were recruited for this study. Peripheral blood samples from each patient were collected and the main polymorphisms of the gene Card15/Nod2 (R702W, G908R, and 1007fs) were analyzed in each sample. In addition, tissue samples were taken from both the tract affected by CD and from the apparently healthy and disease-free margins (internal controls). We used a multiplex gene assay in specimens obtained from patients with ileal localization of CD to evaluate the simultaneous expression of 24 genes involved in the pathogenesis of the disease. We also processed surgery gut samples with routine light microscopy (LM) and transmission electron microscopy (TEM) techniques to evaluate their structural and ultrastructural features.
We found a significant increase of Th17 (IL17A and IL17F, IL 23R and CCR6) and Th1 (IFN-γ) gene expression in inflamed mucosa compared to non-inflamed sites of 14 CD patients. DEFB4 and HAMP, two genes coding for antimicrobial peptides, were also strongly activated in inflamed ileal mucosa, suggesting the overwhelming stimulation of epithelial cells by commensal microbiota. IFN-γ and CCR6 were more expressed in inflamed mucosa of CD patients with ileal localization compared with patients with colonic localization suggesting a more aggressive inflammation process in this site. Morphological analysis of the epithelial lining of Lieberkün crypts disclosed enhanced release activity from goblet mucocytes, whereas the lamina propria contained numerous cells pertaining to various lines.
We observed that the expression of ileal genes related to Th1 and Th17 activity is strongly activated as well as the expression of genes involved in microbiota regulation.
Core tip: Multiplex Gene Assay in specimens obtained from patients with ileal localization of Crohn’s disease (CD) allowed the simultaneous analysis of messenger ribonucleic acid levels for 24 genes, known to be involved in the inflammation processes of CD pathogenesis. The result showed that the expression of genes related to Th1 and Th17 immune response is strongly activated as well as the expression of genes deputized to interact with the commensal microbiota, such as DEFB4 and HAMP, which code for antimicrobial peptides.
- Citation: Giudici F, Lombardelli L, Russo E, Cavalli T, Zambonin D, Logiodice F, Kullolli O, Giusti L, Bargellini T, Fazi M, Biancone L, Scaringi S, Clemente AM, Perissi E, Delfino G, Torcia MG, Ficari F, Tonelli F, Piccinni MP, Malentacchi C. Multiplex gene expression profile in inflamed mucosa of patients with Crohn’s disease ileal localization: A pilot study. World J Clin Cases 2019; 7(17): 2463-2476
- URL: https://www.wjgnet.com/2307-8960/full/v7/i17/2463.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v7.i17.2463
The pathogenesis of Crohn’s disease (CD), one of the major inflammatory bowel diseases (IBD) together with ulcerative colitis (UC), has been extensively investigated. It is generally accepted that both genetic and environmental factors contribute to the etiology of the disease. In CD patients, strong associations between genes involved in maintaining intestinal barrier function, epithelial anti- microbial defence, innate immune regulation, reactive oxygen species (ROS) generation, autophagy, and metabolic pathways have been identified[1,2].
Environmental risk factors involved in the progression of the disease include smoking, low-fiber and high-carbohydrate diet, gut microbiota (GM) alteration, and treatments with antibiotics or non-steroidal anti-inflammatory drugs[3].
CD is characterized by a transmural inflammation which can potentially affect any segment of the gastrointestinal tract[4]. However, recent studies reported a selective location in the terminal ileum in 50% of the patients and location in the colonic district in 20% of the patients. Ileum and colon district were involved in the remaining 30% of the patients. A different clinical course and surgical requirement was reported according to disease’s localization but currently the reasons underlying the differences in the clinical course have not been defined. In addition, the immunological pathways involved in colonic inflammation are different from those involved in ileal inflammation[5].
The mutual interplay between GM and the immune system is involved in the pathogenesis and prognosis of intestinal diseases[6] as the GM is a key modulator of intestinal inflammation[7]. In CD patients, a reduced GM diversity and lower bacterial load in inflamed vs non-inflamed tissues was observed[8]. In addition, several evidences report that the small bowel is responsible for the systemic tolerance towards microbes. A recent study revealed that the ileum harbors a distinctive niche of the GM that differs more from the colonic[9]. This different GM composition could be attributed to the activation of distinctive immunological pathways.
In the present study, we used the multiplex gene assay[10,11] to analyze surgical specimens of CD patients with prevalent ileal localization.
14 consecutive patients undergoing surgery for ileal CD aged 15 to 57 and hospitalized at the Surgery Unit of Azienda Ospedaliero-Universitaria Careggi, University of Florence, were recruited (Table 1). CD was diagnosed based on both histological and clinical/endoscopic criteria. Table 1 reports the clinical characteristics of the patients.
| Patient | Pt1 | Pt2 | Pt3 | Pt4 | Pt5 | Pt6 | Pt7 | Pt8 | Pt9 | Pt10 | Pt11 | Pt12 | Pt13 | Pt14 |
| Localization | L1 | L1 | L1 | L1 | L1 | L1 | L3 | L1 | L3 | L1 | L3-L4 | L1 | L3 | L1 |
| of CDa | ||||||||||||||
| Age of CD | 57 | 15 | 53 | 55 | 25 | 31 | 39 | 42 | 16 | 24 | 19 | 18 | 46 | 30 |
| onset (yr) | ||||||||||||||
| Surgery / relapse | 1st | 1st | Relapse | 1st | Relapse | Relapse | Relapse | 1st | Relapse | Relapse | Relapse | Relapse | Relapse | 1st |
| surgery | surgery | surgery | surgery | surgery | ||||||||||
| Disease behaviorb | B2 | B2, B3 | B2 | B3 | B2, B4 | B2 | B3 | B2 | B2 | B2 | B2 | B2 | B3 | B2 |
| Therapyc | C, F, I, B | F, C | C | No | F, C, I | C, F, I | I, B | F, C | C, I, F, B | C | F, C, B | F, C, I | C, F, B, I | F, C, I |
| Smoking statusd | No | Cur | No | No | 10月10日 | No | No | 20/30 | 10月10日 | Cur | 10月20日 | No | No | No |
| 20/15.5 | 10 | |||||||||||||
| Genotype | wt | R702 | wt | hzG881R | wt | wt | wt | wt | hzG881R | R702 | No seq | No seq | wt | wt |
Peripheral blood samples from each patient were collected in EDTA tubes and genomic DNA was extracted using QIA-AMP DNA Blood Maxi Kit (Qiagen GmbH, Hilden, Germany). The main polymorphisms of the gene Card15/Nod2 (R702W, G908R, and 1007fs) were analysed in each sample[12].
Tissue samples were taken both from the tract affected by CD and from the apparently healthy and disease-free margins (internal controls). The surgical specimens were opened longitudinally.
All samples were stored in RNA later (Qiagen, Germany) before homogenization. Then each sample was weighed and the appropriate lysis solution was added to a final volume of 150 µL containing 50% Lysis Mixture (Thermo-Fisher, MA, United States) and 1 g/L proteinase. The mixture was agitated for 30 min at 65 °C to lyse the cells. The lysate was stored at -80 °C for later use. We used a microarray panel of 24 genes implicated in CD etiopathogenesis[10]. We evaluated the expression of these genes in both non-inflamed and inflamed ileal biopsies. Table 2 indicates the panel of the examined genes, the number of Mendelian Inheritance in Man (MIM) (used as a reference), accession number and their corresponding encoded product and function. To improve the analysis of the results, the selected genes were divided into four groups according to their biological role: (1) Transport across epithelia: ABCB1, SLC40A1, SLC22A4, SLC22A5, HAMP; (2) Immune response: CCR6, IL-17F, IL-17A, MICA, MYD88, STAT3, IL-23R, JAK2, IFNG, NOD2; (3) Antimicrobial activity: HAMP, CAMP, LRRK2.DEFB4; and (4) Physiological activities: STAT3, ESR1, LRRK2, TNFSF15, CARD14, DLG5 BMP2 ATG16L1.
| Symbol | Complete name | Group | Accession number | mim | Gene product function (s) | Ref. |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 | Low expression housekeeping gene | M26434 | 308000 | It plays a central role in the generation of purine nucleotides, chosen as a low expression housekeeping gene | [39] |
| ACTB | Actin betaprovided | High expression housekeeping gene | M28424 | 102630 | Is involved in the cell motility, structure, and integrity | [40] |
| SLC40A1 | Solute carrier family 40 (iron-regulated transporter), member1 | 1 | AF215636 | 604653 | Exports iron from duodenal epithelial cells | [41] |
| ABCB1 | ATP-binding cassette, sub-family B (MDR/TAP), member1 | 1 | M14758 | 171050 | Transports various molecules across extra- and intra-cellular membranes. It belongs to a protein sub-family involved in multidrug resistance | [42] |
| SLC22A5 | Solute carrier family 22 (organic cation/carnitine transporter), member 5 | 1 | AF057164 | 603377 | Transports several small organic cations in the liver, kidney, intestine. It is involved in elimination of drugs and environmental toxins | [43] |
| SLC22A4 | Solute carrier family 22 (organic cation/ergothioneine transporter), member 4 | 1 | AB007448 | 604190 | Polyspecific transporter of organic cations in the liver, kidney, intestine, and involved in the elimination of these molecules. | [44] |
| CCR6 | Chemokine (C-C motif) receptor6 | 2 | U68030 | 601835 | Induces B-lineage maturation and antigen-driven B-cell differentiation | [45] |
| IL17A | Interleukin 17A | 2 | U32659 | 603149 | Produced by Th17-type CD4+ cells. Regulates the activities of NF-kB and mitogen-activated protein kinases | [26] |
| IL17F | Interleukin 17F | 2 | AF384857 | 606496 | Produced by Th17-type CD4+ cells. Stimulates the production of other cytokines, including IL6,IL8.It also inhibits angiogenesis by endothelial | [46] |
| cells. | ||||||
| STAT3 | Signal transducer and activator of transcription 3 (acute-phase response factor) | 2-4 | BC014482 | 102582 | Activates transcription of cell growth and apoptosis’ genes asresponses to inflammation | [22,47] |
| MICA | MHC class I polypeptide-related sequence A | 2 | L14848 | 600169 | Acts as a stress-induced antigen broadly recognized by intestinal intra-epithelial gamma delta T cells. | [48,49] |
| MYD88 | Myeloid differentiation primary response gene (88) | 2 | U84408 | 602170 | Acts as an essential signal transducer in the interleukin-1 and Toll-like receptor signaling | [50] |
| pathways | ||||||
| IL23R | Interleukin 23 receptor | 2 | AF461422 | 607562 | Expressed on Th17 cells. Involvedin the IL23A signaling pathways with the receptor molecule IL12RB1/IL12Rbeta1. | [20] |
| JAK2 | Janus kinase 2 | 2 | 3717 | 147796 | Is involved in cytokine receptor signaling pathways and is required for responses to gamma interferon | [51] |
| IFNG | Interferon, gamma | 2 | 3458 | 147570 | It encodes a cytokine with antiviral,immunoregulatory and anti-tumor properties and activates macrophages | [52] |
| CAMP | Cathelicidin antimicrobial peptide | 3 | BC055089 | 600474 | It is an antimicrobial protein (defensin) | [53] |
| CARD15 | Nucleotide-binding oligomerization domain containing 2 | 2 | AF178930 | 605956 | Induces immune response to intracellular bacterial by recognizing the muramyl dipeptide (MDP) | [54] |
| DEFB4 | Defensin, beta 4A | 3 | AJ314835 | 602215 | Acts as an antibiotic peptide locally regulated by inflammation. | [55,56] |
| HAMP | Hepcidin antimicrobialpeptide | 1/2/3 | AF309489 | 606464 | It is involved in iron transport, antimicrobial, defence and inflammatory responses | [57,58] |
| LRRK2 | Leucine-rich repeat kinase 2 | 3/4 | AK026776 | 609007 | It is involved in autophagy and implicated in clearance of intracellular bacteria. | [59,60] |
| TNFSF15 | Tumor necrosis facto (ligand) superfamily, member 15 | 4 | AF039390 | 604052 | Induces apoptosis in endothelial cells | [61] |
| CARD14 | Caspase recruitment domain family, member 14 | 4 | AF322642 | 607211 | Regulates the molecular scaffolding process and activates NF-kappa B | [62,63] |
| ATG16L1 | ATG16 autophagy related 16-like 1 | 4 | AK000897 | 610767 | Induces autophagy processes involved in degradation of cell organelles | [64] |
| ESR1 | Estrogen receptor 1 | 4 | X03635 | 133430 | Involved in the metabolic pathway of the hormones and in several diseases including osteoporosis | [65] |
| BMP2 | Bone morphogenetic protein 2 | 4 | 650 | 112261 | Induces bone and cartilage formation | [66] |
| DLG5 | Discs, large homolog 5 | 4 | U61843 | 604090 | It encodes for scaffolding molecules involved in cell-cell contact and in the maintenance of epithelial cell integrity. Its products are also involved in the transmission of extracellular signals | [67] |
The messenger ribonucleic acid (mRNA) expression for CCR6, IL-17A, IL-17F, BMP2, TNFSF15, ABCB1, IL-23R, DEFB4, CARD14, STAT3, SLC40A1, JAK2, SLC22A5, ACTB, ATG16L1, CAMP, DLG5, ESR1,CARD15, MICA, MYD88, SLC22A4, IFN-γ, LRRK2, HAMP, ACTB (high expression housekeeping gene), HPTR1 (low expression housekeeping gene) was measured using the QuantiGene® Plex assay (Thermo-Fisher, MA, United States).
A panel of oligonucleotide capture probes was covalently linked to carboxylated fluorescently encoded beads (Luminex, Bio-Rad, MA, United States). Each probe has a unique sequence of 15 bases. Each sample lysate diluted at 1:1 and 1:2 was mixed with the pooled capture beads in a round-bottom assay well and hybridized for 16 h at 54 °C (final volume in each well was 100 µL). The assay mixture was moved to a MultiScreen® Filter Plate (Millipore, Billerica, MA, United States) and unbound material was filter-washed from the wells by rinsing 3 times with wash buffer. The plate was hybridized with 100 µL/well of bDNA amplifier in Amplifier Diluent (Panomics, CA, United States) at 54 °C for 1 h. After the plate was filter- washed twice with wash buffer and incubated at 50 °C for 1 h with 100 µL/well of 5’-dT(Biotin)-conjugated label probe (Panomics, CA, United States) diluted in Label Probe Diluent (Panomics, CA, United States). After 2 washes, streptavidin-conjugated R-phycoerythrin diluted in SA-PE diluent (20 mmol/L Tris-HCl, 400 mmol/L lithium chloride, 1 mL/L Tween 20, 1 mL/L bovine serum albumin, and 5 mL/L Micr-O-protect) was added and the plate was shaken and incubated at room temperature for 30 min. We washed the beads to remove unbound SA-PE and then evaluated them with Bio-Plex® 200 system (Bio-Rad, MA, United States). The SA-PE fluorescence measured from each bead was proportional to the number of mRNA transcripts captured by the beads. Expression of target-specific RNA molecules was calculated as the mean values from triplicate cultures and normalized against Actin gene (high expression housekeeping gene).
A standard non-enzymatic method, using the QIA-AMP® DNA Blood Maxi Kit (Qiagen GmbH, Hilden, Germany) was used to extract Genomic DNA from peripheral blood leucocytes of all CD patients and healthy controls. In addition, DNA samples from 70 healthy Caucasian subjects (140 alleles) were analysed as controls. Three exon of the CARD15/NOD2 gene (Exon 4, Exon 8, Exon 11), were amplified by PCR using pairs of primers derived from the published sequence of the gene (available upon request). Each exon is associated with the three main single-nucleotide polymorphisms (SNPs) (R702W-C2104T; G908R-G2722C; 1007fs-3020insC). These three main variants, associated with susceptibility to CD, represented 32%, 18%, and 31%, respectively, of the total CD mutations[13-15].
The BigDye® Terminator Cycle Sequencing kit (Applied Biosystems, CA, United States) was used to perform direct sequencing of PCR amplified products (SNPs rs87950, rs127951, and rs137955) of the CARD15/NOD2 gene. The samples were analysed in an ABI Prism® 310 genetic analyzer (Applied Biosystems, CA, United States). The of the sequences were confirmed with the analysis of newly-amplified fragments and the sequencing of both DNA strands.
SSPS software vers. 10 (SPSS Inc., IL, United States) was used to perform the statistical analysis. All comparisons of genes mRNA expression in tissues (non-inflamed and inflamed areas) were performed by non-parametric assay (Mann-Whitney test, Wilcoxon test). Data are reported as mean and ranges unless otherwise stated. A P-value < 0.05 was accepted as statistically significant. Furthermore, to better characterize the different clinical CD phenotypes, we compared the results regarding the CARD15, CCR6, interferon gamma, and IL-17A genes to colonic CD patients previously examined for these same genes.
Once removed, tissue samples were rinsed in 0.1 M, pH 7.0 cacodylate buffer, the same used in prefixation and further steps of histological preparation. Samples were then placed in Karnovsky (1965)[16], aldehyde solution, and after 3 h prefixation (4°C), underwent prolonged washing in the buffer. Surgery specimens were reduced into approximately 20 mm3 fragments that were post-fixed (1 h 30 min, 4°C) with 1% OsO4 in cacodylate. These specimens were washed in the buffer, dehydrated in graded ethanol series, soaked in propylene oxide, and embedded in Epon 812. Flat blocks were obtained after polymerization, which were reduced into semi-thin sections (1.5 µm thick), using an 8800 ULTROTOME III LKB equipped with glass knives. Semi-thin sections were stained with borax buffered 1% toluidine blue, and observed with a LEITZ DMRB, in order to collect LM digital images (JPG) for structural analysis. Subsequent ultrastructural observations were carried out on ultrathin sections, obtained with an ULTROTOME NOVA LKB, using a DIATOME diamond knife. Ultrathin sections with gold yellow to silver gray interference colour were selected and collected on uncoated 200-300 mesh copper grids to be electron-dense stained with a hydroalcoholic saturated solution (25 mg/mL) of uranyl acetate, followed by alkaline lead citrate (2 mg/mL). These sections where finally observed (80 KV) with a PHILIPS 201 TEM (BIO, UNIFI), and analogic images were collected, which were later acquired and stored as digital TIFF files using a DIMAGE SCAN DUAL (MINOLTA).
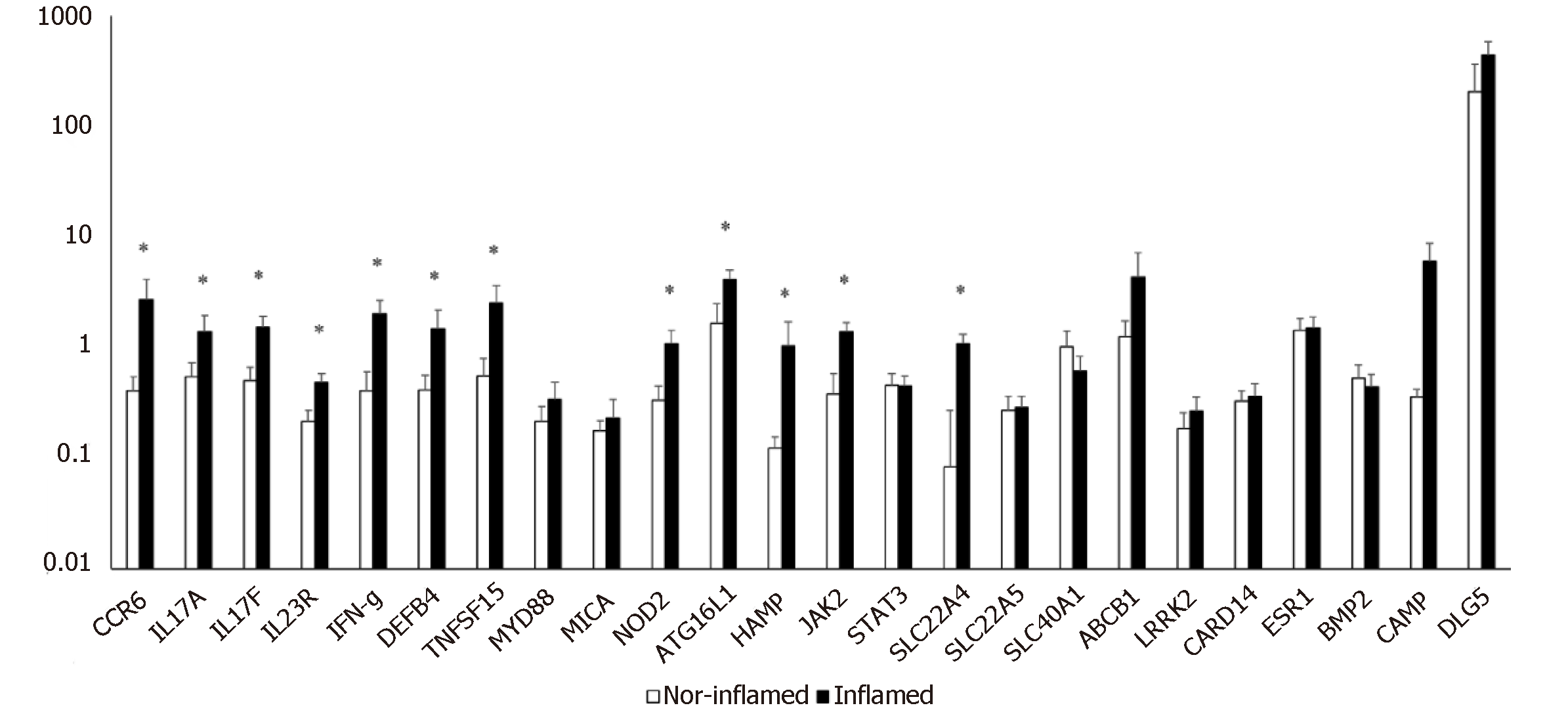
The simultaneous expression of 24 genes involved in the pathogenesis of CD was studied in surgical specimens from 14 CD patients with ileal localization of disease. The expression of genes in inflamed ileal mucosa was compared to that of non-inflamed ileal sites collected from the same patient. We observed a significant increase in mRNA levels of twelve genes compared to internal control (Figure 1).
Figure 1 shows that genes related to innate immune response (NOD 2, ATG16L1, DEFB4), and to adaptive immune response (CCR6, IL17A, IL17F, IL23R, IFN-γ) were significantly increased in inflamed mucosa of CD patients compared with non-inflamed sites. Moreover, the levels of mRNA for genes involved in physiological functions of epithelial cells, such as JAK2, TNFSF15, and SLC22A4 were higher in inflamed mucosa compared to non-inflamed mucosa and the differences in expression reached statistical significance.
DNA samples obtained from peripheral blood were sequenced to investigate the presence of polymorphisms of CARD15/NOD2 gene. The results of this analysis showed that four patients (28.5%) included in this study are carriers of at least one of the polymorphisms investigated, suggesting that genetic factors might contribute to the dysregulated expression of CARD15/NOD2 gene[17-19].
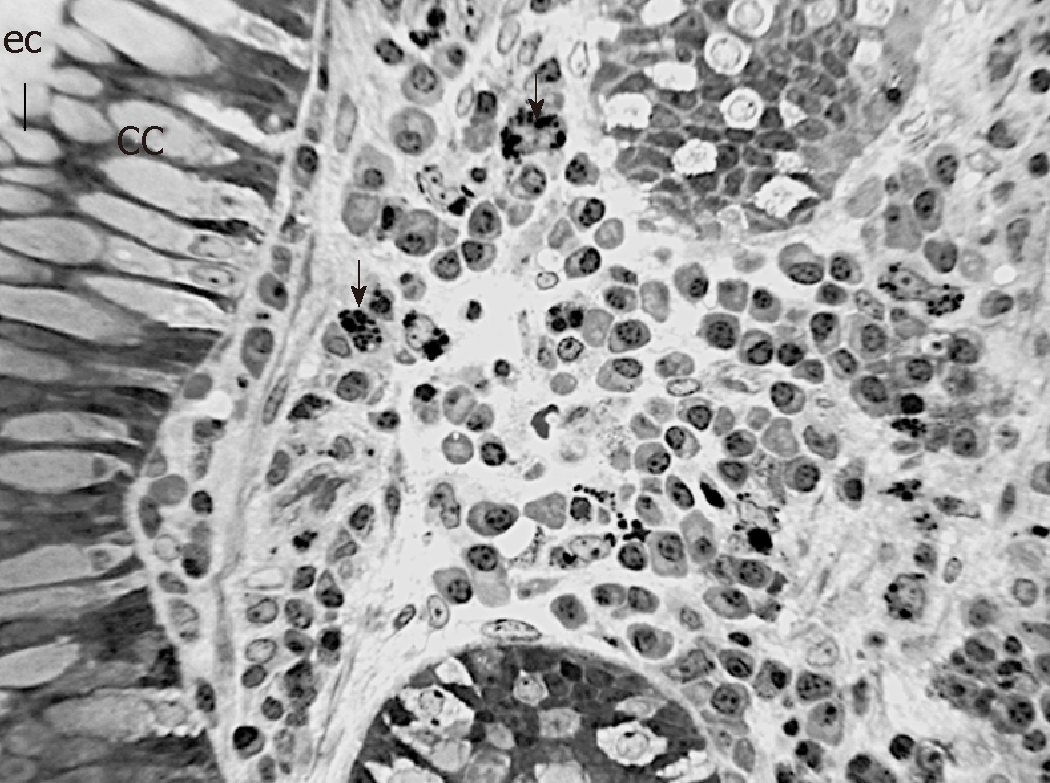
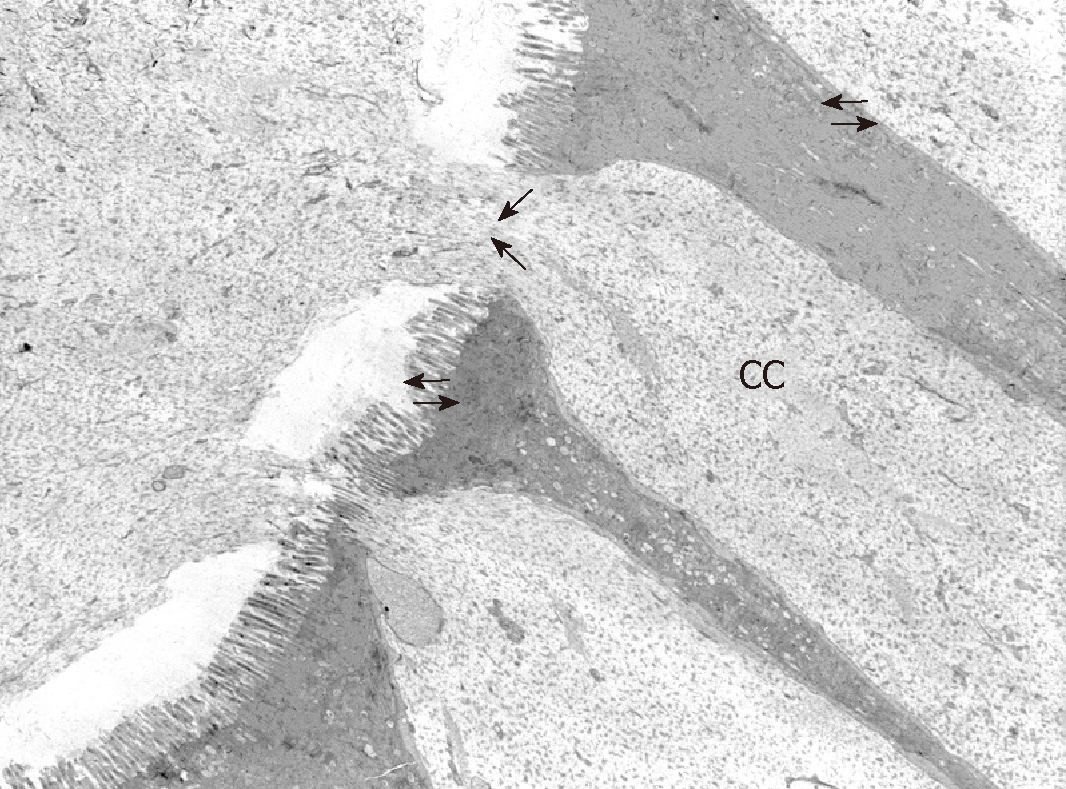
In order to observe the morphology of inflamed tissue samples, light microscopy (LM) and TEM micrographs were obtained. Both gut wall and Lieberkühn crypts retain usual features in both lining epithelium and lamina propria. Epithelial cells consist of constitutive enterocytes along with goblet mucocytes, whereas the underlying connective tissue contains large amounts of cells with wide morphological variety (Figure 2). Goblet cells are involved in impressive secretory processes, releasing a moderately opaque product into cryptal and gut lumen through gaps between enterocytes apices (Figure 3). As a consistent pattern, the lamina propria contains granulocytes and plasma cells.
Among the numerous genes that have been studied so far with respect to CD, strong and replicated associations have been identified with NOD2, IL23R, and ATG16L1 genes[20]. Environmental factors like smoking, low-fiber and, high-carbohydrate diet, altered GM, and medications such as non-steroidal anti-inflammatory drugs interact with genetic background and induce abnormal inflammation and dysregulation of the immune response. Clinical symptomatology relates to such dysregulation.
The clinical course of CD is conditioned by several parameters such as disease location, extra-intestinal manifestation, and age at onset[21]. Strictures and fistulas are more frequent in patients with ileal disease, whereas Crohn's colitis remains uncomplicated for many years. On the whole, almost 80% of patients with CD require intestinal surgery, with a permanent stoma required by almost 10%. The presence of selected mutations in the NOD2 gene (see, e.g., 605956.0001-605956.0003) (IBD1; 266600) has been associated with susceptibility to ileum-localized CD[22]; patients homozygous for the 1007fs mutation had an early disease onset with long-segment ileal stenoses and entero-enteral fistulas; they frequently needed surgical intervention and had a high risk of recurrence[23,24]. Beside NOD2 gene, huge genome-wide linkage-analyses and meta-analyses have described several CD susceptibility regions including IBD5 locus, DLG5, and autophagy- related 16-like 1 (ATG16L1) gene, JAK2, STAT3 interleukin-23 receptor (IL23R), SLC22A4 and SLC22A5 TNFSF15[14].
In this paper, we evaluated the expression of 24 genes that were associated to CD susceptibility[10]. mRNA was extracted from gut specimens obtained from patients with CD ileal localization of CD, undergoing surgery. We used a multiplex gene assay which directly quantifies the mRNA amounts without need of reverse transcription and gives a detailed picture of the inflammation process for each patient[11]. The same technique was used to quantify gene expression in colonic mucosa from surgical specimens or endoscopic bioptic fragments obtained by CD patients with predominant colonic (L2) location[10].
The analysis revealed a clear activation of immune-adaptive Th17 response in association with a Th1 response in inflamed mucosa of patients undergoing surgery suggesting a dysregulated and very aggressive immune-inflammatory response.
Here gene expression analysis of inflamed ileal mucosa revealed an increased expression of genes involved in adaptive immune response compared to non-inflamed tissue. In particular, we found a significant increase of IL17A and IL17F, IL 23R and CCR6 gene expression suggesting an activation of a Th17 adaptive response[25,26] similar to that found in gut mucosa of patients with colonic localization. According with this hypothesis, three additional genes involved in Th17 differentiation as JAK2, STAT3 and TNFSF15[27,28] were found to be overexpressed in inflamed ileal mucosa of CD patients compared to non- inflamed sites. Furthermore, as we expected, the expression of the antimicrobial peptides as defensin (DEFB4)[29] and Hepcidin 6 (HAMP)[30] were significantly increased in inflamed mucosa of CD patients compared with non-inflamed sites, suggesting the overwhelming stimulation of epithelial cells by commensal GM. Indeed, while the human β-defensin (HBD) 1 is constitutively expressed, other genes, like HBD2 (gene name DEFB4), show pathogen and/or inflammation dependent upregulation[31] while also being inducible by probiotic bacteria[32]. Conversely, HAMP transcription mediates the effects of host defence and inflammation. Shanmugam et al. provided persuasive evidence in support of an important role for the GM composition in influencing hepcidin expression during intestinal inflammation in mouse models of colitis[33].
As the position of the pathogenic tissue may condition not only the clinical course of the disease but also the probability to require surgery, we also compared with the same methodology (Quantigene 2.0) the expression of selected genes (IFN-γ, CCR6, IL17A, NOD2) involved in immune responses in inflamed mucosa with predominant ileal location with the one previously studied[10] in inflamed mucosa with colonic location. mRNA expression for IFN-γ γ and for the chemokine CCR6 appeared significantly higher in ileal site compared to colonic site (ileal CD = 2.7 ± 1.5; colonic CD = 0.2 ± 0.06; P = 0.01). The mRNA for IL-17 and NOD2 appeared to be expressed at higher levels in ileal site compared to colonic site, even if the difference is not statistically significant (P ≥ 0.05). The significant differences in the expression levels of IFN-γ gene (higher expression in specimens from patients with ileal localization compared to patients with colonic localization) may suggest an increased damage of the ileal mucosa due to the simultaneous presence of Th1 and Th17 effector cells and/or the shift of Th17 cells to Th1 effectors functionally more aggressive than Th17 unshifted cells[34,35].
Furthermore, according to a worse clinical course of patient with ileal localization of CD compared with patient with colonic localization[36], the increased expression of IL17 and NOD2 in mucosal fragments from patients with ileal CD compared to patients with colonic CD is in agreement with the NOD2 –dependent regulation of immunity in mouse intestinal tract[37]. We suppose that the above differences between the two gut tracts (ileal and colonic) may be due to the Paneth cells at the bottom of the crypts of Lieberkühn in the small intestine, which produce antimicrobial peptides and hinder commensal GM and pathogenic bacteria to penetrate gut mucosa. Initially described as innate immune cells producing antimicrobial products, Paneth cells have recently been suggested to constitute a cardinal component of the intestinal stem cell niche. In fact, Paneth cells contribute to controlling the luminal flora as well as repairing the intestinal barrier following an insult. Genomic alterations that impede the Paneth cell compartment functionality can potentially increase the propensity to develop CD[38].
As a consistent trait, cryptal globlet cells produce large amounts of mucus that performs the double role of barrier and holder of antimicrobial products. The microscopic anatomy analysis aims to provide some details that illustrate phenotypic features: the large cell variety in the lamina propria includes immune lines that represent a further defense tool. Although these morphological traits are not directly related to specific gene outputs, they illustrate the tissue responses to key gene deregulation.
As a pilot study, our study presents a low number of subjects investigated which may have influenced the statistical power of the results. To confirm these results, studies with a larger number of patients are needed. In addition, gene expression was evaluated with Multiplex Gene Assay only. This method directly quantifies the mRNA amounts without need of reverse transcription and gives a detailed picture of the metabolic processes for each patient but it should be validated by comparisons with additional techniques to evaluate gene expression.
One of the main purposes of our research is therefore to identify new molecules involved in metabolic pathways that could potentially represent new biological drugs to identify the appropriate therapy in relation to the clinical phenotype of the CD patient.
The interplay of environmental, genetic and microbial elements influences the etiopathogenesis of Crohn’s disease (CD). Differences in the clinical course of CD have recently been reported in patients with ileal or colonic localization of the inflammatory process.
Aim of this study was to define biochemical and histological differences in intestinal biopsies from patients with ileal or colonic localization of Crohn disease in order to identify new assays which can be useful for planning individual therapeutic strategies
Main objective of the current research was to investigate the expression of genes involved in immune-inflammatory pathways in gut mucosa from patients with ileal or colonic localization of CD and to correlate the results of gene expression with those obtained through a classical morphological analysis of surgical biopsies.
A Multiplex Gene Assay was used to assess the simultaneous expression of 24 genes related to immune-inflammatory process and to CD pathogenesis. Structural and ultrastructural features of gut samples were also evaluated through Light microscopy (LM) and Transmission Electron Microscopy (TEM) techniques.
We observed a strong activation of genes involved in TH-1- and TH-17 immune response in patients with ileal localization of CD compared to patients with colonic localization. In addition, the expression of genes for antimicrobial peptides as DEFB4 and HAMP was found highly stimulated in ileal mucosa from CD patients suggesting a possible interference with microbial commensals at this site.
Our results indicate that patients with ileal localization of CD have a stronger activation of TH-1 and TH-17 immune-inflammatory responses compared with patients with colonic localization of the disease thus defining a clear subclinical phenotype of CD.
These results may suggest that therapeutic strategies with biological drugs in CD patients can be differentiated depending on the location of the disease
We would like to thank Dr. Michele Tanturli for the statistical support, Dr. Giulia Ricciardi for additional manuscript revision, and all the patients who participated to this study,
| 1. | Boyapati R, Satsangi J, Ho GT. Pathogenesis of Crohn's disease. F1000Prime Rep. 2015;7:44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 68] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 2. | Ramos GP, Papadakis KA. Mechanisms of Disease: Inflammatory Bowel Diseases. Mayo Clin Proc. 2019;94:155-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 705] [Article Influence: 100.7] [Reference Citation Analysis (2)] |
| 3. | Abegunde AT, Muhammad BH, Bhatti O, Ali T. Environmental risk factors for inflammatory bowel diseases: Evidence based literature review. World J Gastroenterol. 2016;22:6296-6317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 125] [Cited by in RCA: 160] [Article Influence: 16.0] [Reference Citation Analysis (3)] |
| 4. | Li N, Shi RH. Updated review on immune factors in pathogenesis of Crohn's disease. World J Gastroenterol. 2018;24:15-22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 52] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (7)] |
| 5. | Weiser M, Simon JM, Kochar B, Tovar A, Israel JW, Robinson A, Gipson GR, Schaner MS, Herfarth HH, Sartor RB, McGovern DPB, Rahbar R, Sadiq TS, Koruda MJ, Furey TS, Sheikh SZ. Molecular classification of Crohn's disease reveals two clinically relevant subtypes. Gut. 2018;67:36-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 91] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 6. | Russo E, Taddei A, Ringressi MN, Ricci F, Amedei A. The interplay between the microbiome and the adaptive immune response in cancer development. Therap Adv Gastroenterol. 2016;9:594-605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 7. | Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1339] [Cited by in RCA: 1395] [Article Influence: 77.5] [Reference Citation Analysis (2)] |
| 8. | Sepehri S, Kotlowski R, Bernstein CN, Krause DO. Microbial diversity of inflamed and noninflamed gut biopsy tissues in inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:675-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 181] [Article Influence: 9.5] [Reference Citation Analysis (2)] |
| 9. | Villmones HC, Haug ES, Ulvestad E, Grude N, Stenstad T, Halland A, Kommedal Ø. Species Level Description of the Human Ileal Bacterial Microbiota. Sci Rep. 2018;8:4736. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 10. | Russo E, Lombardelli L, Giudici F, Cavalli T, Ficari F, Fazi M, Scaringi S, Biancone L, Logiodice F, Nesi M, Latiano A, Annese V, Torcia MG, Bechi P, Tonelli F, Piccinni MP, Malentacchi C. Crohn's Colitis: Development of a multiplex gene expression assay comparing mRNA levels of susceptibility genes. Clin Res Hepatol Gastroenterol. 2017;41:435-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 11. | Piccinni MP, Lombardelli L, Logiodice F, Tesi D, Kullolli O, Biagiotti R, Giudizi M, Romagnani S, Maggi E, Ficarra G. Potential pathogenetic role of Th17, Th0, and Th2 cells in erosive and reticular oral lichen planus. Oral Dis. 2014;20:212-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 67] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 12. | Palmieri O, Bossa F, Valvano MR, Corritore G, Latiano T, Martino G, D'Incà R, Cucchiara S, Pastore M, D'Altilia M, Scimeca D, Biscaglia G, Andriulli A, Latiano A. Crohn's Disease Localization Displays Different Predisposing Genetic Variants. PLoS One. 2017;12:e0168821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Barreiro-de Acosta M, Peña AS. Clinical applications of NOD2/CARD15 mutations in Crohn's disease. Acta Gastroenterol Latinoam. 2007;37:49-54. [PubMed] |
| 14. | Huang H, Fang M, Jostins L, Umićević Mirkov M, Boucher G, Anderson CA, Andersen V, Cleynen I, Cortes A, Crins F, D'Amato M, Deffontaine V, Dmitrieva J, Docampo E, Elansary M, Farh KK, Franke A, Gori AS, Goyette P, Halfvarson J, Haritunians T, Knight J, Lawrance IC, Lees CW, Louis E, Mariman R, Meuwissen T, Mni M, Momozawa Y, Parkes M, Spain SL, Théâtre E, Trynka G, Satsangi J, van Sommeren S, Vermeire S, Xavier RJ; International Inflammatory Bowel Disease Genetics Consortium, Weersma RK, Duerr RH, Mathew CG, Rioux JD, McGovern DPB, Cho JH, Georges M, Daly MJ, Barrett JC. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 2017;547:173-178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 349] [Cited by in RCA: 449] [Article Influence: 49.9] [Reference Citation Analysis (0)] |
| 15. | Keh C, Shatari T, Yamamoto T, Menon A, Clark MA, Keighley MR. Jejunal Crohn's disease is associated with a higher postoperative recurrence rate than ileocaecal Crohn's disease. Colorectal Dis. 2005;7:366-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 16. | Karnovsky MJ. A formaldehyde-glutaraldehyde fixative of high osmolarity for use in electron microscopy. J Cell Biol. 1965;27:137A. |
| 17. | Seiderer J, Schnitzler F, Brand S, Staudinger T, Pfennig S, Herrmann K, Hofbauer K, Dambacher J, Tillack C, Sackmann M, Göke B, Lohse P, Ochsenkühn T. Homozygosity for the CARD15 frameshift mutation 1007fs is predictive of early onset of Crohn's disease with ileal stenosis, entero-enteral fistulas, and frequent need for surgical intervention with high risk of re-stenosis. Scand J Gastroenterol. 2006;41:1421-1432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 56] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 18. | Sidiq T, Yoshihama S, Downs I, Kobayashi KS. Nod2: A Critical Regulator of Ileal Microbiota and Crohn's Disease. Front Immunol. 2016;7:367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 90] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 19. | Cucchiara S, Latiano A, Palmieri O, Staiano AM, D'Incà R, Guariso G, Vieni G, Rutigliano V, Borrelli O, Valvano MR, Annese V. Role of CARD15, DLG5 and OCTN genes polymorphisms in children with inflammatory bowel diseases. World J Gastroenterol. 2007;13:1221-1229. [PubMed] |
| 20. | Naser SA, Arce M, Khaja A, Fernandez M, Naser N, Elwasila S, Thanigachalam S. Role of ATG16L, NOD2 and IL23R in Crohn's disease pathogenesis. World J Gastroenterol. 2012;18:412-424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 62] [Cited by in RCA: 82] [Article Influence: 5.9] [Reference Citation Analysis (1)] |
| 21. | Lazarev M, Huang C, Bitton A, Cho JH, Duerr RH, McGovern DP, Proctor DD, Regueiro M, Rioux JD, Schumm PP, Taylor KD, Silverberg MS, Steinhart AH, Hutfless S, Brant SR. Relationship between proximal Crohn's disease location and disease behavior and surgery: a cross-sectional study of the IBD Genetics Consortium. Am J Gastroenterol. 2013;108:106-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 162] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 22. | Noble CL, Abbas AR, Lees CW, Cornelius J, Toy K, Modrusan Z, Clark HF, Arnott ID, Penman ID, Satsangi J, Diehl L. Characterization of intestinal gene expression profiles in Crohn's disease by genome-wide microarray analysis. Inflamm Bowel Dis. 2010;16:1717-1728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 76] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 23. | Quezada SM, Steinberger EK, Cross RK. Association of age at diagnosis and Crohn's disease phenotype. Age Ageing. 2013;42:102-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 24. | Sehgal R, Berg A, Hegarty JP, Kelly AA, Lin Z, Poritz LS, Koltun WA. NOD2/CARD15 mutations correlate with severe pouchitis after ileal pouch-anal anastomosis. Dis Colon Rectum. 2010;53:1487-1494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 47] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 25. | Cosmi L, Liotta F, Maggi E, Romagnani S, Annunziato F. Th17 and non-classic Th1 cells in chronic inflammatory disorders: two sides of the same coin. Int Arch Allergy Immunol. 2014;164:171-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 26. | Pelletier M, Maggi L, Micheletti A, Lazzeri E, Tamassia N, Costantini C, Cosmi L, Lunardi C, Annunziato F, Romagnani S, Cassatella MA. Evidence for a cross-talk between human neutrophils and Th17 cells. Blood. 2010;115:335-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 527] [Cited by in RCA: 605] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 27. | Floss DM, Klöcker T, Schröder J, Lamertz L, Mrotzek S, Strobl B, Hermanns H, Scheller J. Defining the functional binding sites of interleukin 12 receptor β1 and interleukin 23 receptor to Janus kinases. Mol Biol Cell. 2016;27:2301-2316. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 28. | Van Limbergen J, Wilson DC, Satsangi J. The genetics of Crohn's disease. Annu Rev Genomics Hum Genet. 2009;10:89-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 194] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 29. | Wehkamp J, Koslowski M, Wang G, Stange EF. Barrier dysfunction due to distinct defensin deficiencies in small intestinal and colonic Crohn's disease. Mucosal Immunol. 2008;1 Suppl 1:S67-S74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 107] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 30. | Arnold J, Sangwaiya A, Bhatkal B, Geoghegan F, Busbridge M. Hepcidin and inflammatory bowel disease: dual role in host defence and iron homoeostasis. Eur J Gastroenterol Hepatol. 2009;21:425-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 31. | Zilbauer M, Dorrell N, Boughan PK, Harris A, Wren BW, Klein NJ, Bajaj-Elliott M. Intestinal innate immunity to Campylobacter jejuni results in induction of bactericidal human beta-defensins 2 and 3. Infect Immun. 2005;73:7281-7289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 67] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 32. | Schlee M, Harder J, Köten B, Stange EF, Wehkamp J, Fellermann K. Probiotic lactobacilli and VSL#3 induce enterocyte beta-defensin 2. Clin Exp Immunol. 2008;151:528-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 262] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 33. | Shanmugam NK, Trebicka E, Fu LL, Shi HN, Cherayil BJ. Intestinal inflammation modulates expression of the iron-regulating hormone hepcidin depending on erythropoietic activity and the commensal microbiota. J Immunol. 2014;193:1398-1407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 34. | Annunziato F, Romagnani S. The transient nature of the Th17 phenotype. Eur J Immunol. 2010;40:3312-3316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 49] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 35. | Jeschke JC, Mayne CG, Ziegelbauer J, DeCiantis CL, Singh S, Kumar SN, Suchi M, Iwakura Y, Drobyski WR, Salzman NH, Williams CB. A model of TH17-associated ileal hyperplasia that requires both IL-17A and IFNγ to generate self-tolerance and prevent colitis. Mucosal Immunol. 2018;11:1127-1137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 36. | Hovde Ø, Moum BA. Epidemiology and clinical course of Crohn's disease: results from observational studies. World J Gastroenterol. 2012;18:1723-1731. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 85] [Cited by in RCA: 83] [Article Influence: 5.9] [Reference Citation Analysis (2)] |
| 37. | Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nuñez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1334] [Cited by in RCA: 1336] [Article Influence: 63.6] [Reference Citation Analysis (2)] |
| 38. | Roda G, Sartini A, Zambon E, Calafiore A, Marocchi M, Caponi A, Belluzzi A, Roda E. Intestinal epithelial cells in inflammatory bowel diseases. World J Gastroenterol. 2010;16:4264-4271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 87] [Cited by in RCA: 102] [Article Influence: 6.4] [Reference Citation Analysis (4)] |
| 39. | Fedrigo O, Warner LR, Pfefferle AD, Babbitt CC, Cruz-Gordillo P, Wray GA. A pipeline to determine RT-QPCR control genes for evolutionary studies: application to primate gene expression across multiple tissues. PLoS One. 2010;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 40. | Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12858] [Cited by in RCA: 14838] [Article Influence: 618.3] [Reference Citation Analysis (0)] |
| 41. | Benyamin B, Esko T, Ried JS, Radhakrishnan A, Vermeulen SH, Traglia M, Gögele M, Anderson D, Broer L, Podmore C, Luan J, Kutalik Z, Sanna S, van der Meer P, Tanaka T, Wang F, Westra HJ, Franke L, Mihailov E, Milani L, Hälldin J, Winkelmann J, Meitinger T, Thiery J, Peters A, Waldenberger M, Rendon A, Jolley J, Sambrook J, Kiemeney LA, Sweep FC, Sala CF, Schwienbacher C, Pichler I, Hui J, Demirkan A, Isaacs A, Amin N, Steri M, Waeber G, Verweij N, Powell JE, Nyholt DR, Heath AC, Madden PAF, Visscher PM, Wright MJ, Montgomery GW, Martin NG, Hernandez D, Bandinelli S, van der Harst P, Uda M, Vollenweider P, Scott RA, Langenberg C, Wareham NJ, van Duijn C, Beilby J, Pramstaller PP, Hicks AA, Ouwehand WH, Oexle K, Gieger C, Metspalu A, Camaschella C, Toniolo D, Swinkels DW, Whitfield JB. Corrigendum: Novel loci affecting iron homeostasis and their effects in individuals at risk for hemochromatosis. Nat Commun. 2015;6:6542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 42. | Brant SR, Panhuysen CI, Nicolae D, Reddy DM, Bonen DK, Karaliukas R, Zhang L, Swanson E, Datta LW, Moran T, Ravenhill G, Duerr RH, Achkar JP, Karban AS, Cho JH. MDR1 Ala893 polymorphism is associated with inflammatory bowel disease. Am J Hum Genet. 2003;73:1282-1292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 172] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 43. | Peltekova VD, Wintle RF, Rubin LA, Amos CI, Huang Q, Gu X, Newman B, Van Oene M, Cescon D, Greenberg G, Griffiths AM, St George-Hyslop PH, Siminovitch KA. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 2004;36:471-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 572] [Cited by in RCA: 566] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 44. | Girardin M, Dionne S, Goyette P, Rioux J, Bitton A, Elimrani I, Charlebois P, Qureshi I, Levy E, Seidman EG. Expression and functional analysis of intestinal organic cation/L-carnitine transporter (OCTN) in Crohn's disease. J Crohns Colitis. 2012;6:189-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 45. | Fransen K, van Sommeren S, Westra HJ, Veenstra M, Lamberts LE, Modderman R, Dijkstra G, Fu J, Wijmenga C, Franke L, Weersma RK, van Diemen CC. Correlation of genetic risk and messenger RNA expression in a Th17/IL23 pathway analysis in inflammatory bowel disease. Inflamm Bowel Dis. 2014;20:777-782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 46. | Ueno A, Ghosh A, Hung D, Li J, Jijon H. Th17 plasticity and its changes associated with inflammatory bowel disease. World J Gastroenterol. 2015;21:12283-12295. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 63] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 47. | Nguyen PM, Putoczki TL, Ernst M. STAT3-Activating Cytokines: A Therapeutic Opportunity for Inflammatory Bowel Disease? J Interferon Cytokine Res. 2015;35:340-350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 85] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 48. | Allez M, Tieng V, Nakazawa A, Treton X, Pacault V, Dulphy N, Caillat-Zucman S, Paul P, Gornet JM, Douay C, Ravet S, Tamouza R, Charron D, Lémann M, Mayer L, Toubert A. CD4+NKG2D+ T cells in Crohn's disease mediate inflammatory and cytotoxic responses through MICA interactions. Gastroenterology. 2007;132:2346-2358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 170] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 49. | Muro M, López-Hernández R, Mrowiec A. Immunogenetic biomarkers in inflammatory bowel diseases: role of the IBD3 region. World J Gastroenterol. 2014;20:15037-15048. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 50. | Santarlasci V, Cosmi L, Maggi L, Liotta F, Annunziato F. IL-1 and T Helper Immune Responses. Front Immunol. 2013;4:182. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 115] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 51. | Danese S, Grisham M, Hodge J, Telliez JB. JAK inhibition using tofacitinib for inflammatory bowel disease treatment: a hub for multiple inflammatory cytokines. Am J Physiol Gastrointest Liver Physiol. 2016;310:G155-G162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 137] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 52. | Tuller T, Atar S, Ruppin E, Gurevich M, Achiron A. Common and specific signatures of gene expression and protein-protein interactions in autoimmune diseases. Genes Immun. 2013;14:67-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 77] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 53. | Koczulla AR, Bals R. Antimicrobial peptides: current status and therapeutic potential. Drugs. 2003;63:389-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 220] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 54. | Hugot JP. CARD15/NOD2 mutations in Crohn's disease. Ann N Y Acad Sci. 2006;1072:9-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 54] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 55. | Wehkamp J, Stange EF, Fellermann K. Defensin-immunology in inflammatory bowel disease. Gastroenterol Clin Biol. 2009;33 Suppl 3:S137-S144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 56. | Gersemann M, Becker S, Kübler I, Koslowski M, Wang G, Herrlinger KR, Griger J, Fritz P, Fellermann K, Schwab M, Wehkamp J, Stange EF. Differences in goblet cell differentiation between Crohn's disease and ulcerative colitis. Differentiation. 2009;77:84-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 230] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 57. | Verga Falzacappa MV, Muckenthaler MU. Hepcidin: iron-hormone and anti-microbial peptide. Gene. 2005;364:37-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 72] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 58. | Mleczko-Sanecka K, Casanovas G, Ragab A, Breitkopf K, Müller A, Boutros M, Dooley S, Hentze MW, Muckenthaler MU. SMAD7 controls iron metabolism as a potent inhibitor of hepcidin expression. Blood. 2010;115:2657-2665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 88] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 59. | Gardet A, Benita Y, Li C, Sands BE, Ballester I, Stevens C, Korzenik JR, Rioux JD, Daly MJ, Xavier RJ, Podolsky DK. LRRK2 is involved in the IFN-gamma response and host response to pathogens. J Immunol. 2010;185:5577-5585. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 347] [Cited by in RCA: 335] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 60. | Schapansky J, Nardozzi JD, Felizia F, LaVoie MJ. Membrane recruitment of endogenous LRRK2 precedes its potent regulation of autophagy. Hum Mol Genet. 2014;23:4201-4214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 196] [Cited by in RCA: 185] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 61. | Bamias G, Martin C, Marini M, Hoang S, Mishina M, Ross WG, Sachedina MA, Friel CM, Mize J, Bickston SJ, Pizarro TT, Wei P, Cominelli F. Expression, localization, and functional activity of TL1A, a novel Th1-polarizing cytokine in inflammatory bowel disease. J Immunol. 2003;171:4868-4874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 243] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 62. | Tsoi LC, Spain SL, Knight J, Ellinghaus E, Stuart PE, Capon F, Ding J, Li Y, Tejasvi T, Gudjonsson JE, Kang HM, Allen MH, McManus R, Novelli G, Samuelsson L, Schalkwijk J, Ståhle M, Burden AD, Smith CH, Cork MJ, Estivill X, Bowcock AM, Krueger GG, Weger W, Worthington J, Tazi-Ahnini R, Nestle FO, Hayday A, Hoffmann P, Winkelmann J, Wijmenga C, Langford C, Edkins S, Andrews R, Blackburn H, Strange A, Band G, Pearson RD, Vukcevic D, Spencer CC, Deloukas P, Mrowietz U, Schreiber S, Weidinger S, Koks S, Kingo K, Esko T, Metspalu A, Lim HW, Voorhees JJ, Weichenthal M, Wichmann HE, Chandran V, Rosen CF, Rahman P, Gladman DD, Griffiths CE, Reis A, Kere J; Collaborative Association Study of Psoriasis (CASP); Genetic Analysis of Psoriasis Consortium; Psoriasis Association Genetics Extension; Wellcome Trust Case Control Consortium 2, Nair RP, Franke A, Barker JN, Abecasis GR, Elder JT, Trembath RC. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341-1348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 777] [Cited by in RCA: 793] [Article Influence: 56.6] [Reference Citation Analysis (0)] |
| 63. | Bertin J, Wang L, Guo Y, Jacobson MD, Poyet JL, Srinivasula SM, Merriam S, DiStefano PS, Alnemri ES. CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-kappa B. J Biol Chem. 2001;276:11877-11882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 282] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 64. | Parkes M. Evidence from genetics for a role of autophagy and innate immunity in IBD pathogenesis. Dig Dis. 2012;30:330-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 65. | Aguirre JI, Plotkin LI, Gortazar AR, Millan MM, O'Brien CA, Manolagas SC, Bellido T. A novel ligand-independent function of the estrogen receptor is essential for osteocyte and osteoblast mechanotransduction. J Biol Chem. 2007;282:25501-25508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 108] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 66. | Cejalvo T, Sacedón R, Hernández-López C, Diez B, Gutierrez-Frías C, Valencia J, Zapata AG, Varas A, Vicente A. Bone morphogenetic protein-2/4 signalling pathway components are expressed in the human thymus and inhibit early T-cell development. Immunology. 2007;121:94-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 67. | Stoll M, Corneliussen B, Costello CM, Waetzig GH, Mellgard B, Koch WA, Rosenstiel P, Albrecht M, Croucher PJ, Seegert D, Nikolaus S, Hampe J, Lengauer T, Pierrou S, Foelsch UR, Mathew CG, Lagerstrom-Fermer M, Schreiber S. Genetic variation in DLG5 is associated with inflammatory bowel disease. Nat Genet. 2004;36:476-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 314] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
Manuscript source: Invited manuscript
Specialty type: Medicine, Research and Experimental
Country of origin: Italy
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: El-Hussuna A, Mattar MC, Christodoulou DK, Day AS S-Editor: Dou Y L-Editor: AE-Editor: Zhou BX