Published online Sep 6, 2023. doi: 10.12998/wjcc.v11.i25.5962
Peer-review started: May 13, 2023
First decision: July 3, 2023
Revised: July 16, 2023
Accepted: August 8, 2023
Article in press: August 8, 2023
Published online: September 6, 2023
Processing time: 111 Days and 4.5 Hours
Variants in the MYO7A gene commonly result in Usher syndrome, and in rare cases lead to autosomal dominant non-syndromic deafness (DFNA11). Currently, only nine variants have been reported to be responsible for DFNA11 and their clinical phenotypes are not identical. Here we present a novel variant causing DFNA11 identified in a three-generation Chinese family.
The proband was a 53-year-old Han male who presented with post-lingual bilateral symmetrical moderate sensorineural hearing loss. We learned from the patient’s medical history collection that multiple family members also had similar hearing loss, generally occurring around the age of 40. Subsequent investigation by high-throughput sequencing identified a novel MYO7A variant. To provide evidence supporting that this variant is responsible for the hearing loss in the studied family, we performed Sanger sequencing on 11 family members and found that the variant co-segregated with the deafness phenotype. In addition, the clinical manifestation of the 11 affected family members was found to be late-onset bilateral slowly progressive hearing loss, inherited in this family in an autosomal dominant manner. None of the affected family members had visual impairment or vestibular symptoms; therefore, we believe that this novel MYO7A variant is responsible for the rare DFNA11 in this family.
We report a novel variant leading to DFNA11 which further enriches the collection of MYO7A variants, and our review of the nine previous variants that have been identified to cause DFNA11 provides a reference for clinical genetic counseling.
Core Tip: Autosomal dominant non-syndromic hearing loss caused by the MYO7A variant (DFNA11) is rare and characterized by post-lingual sensorineural hearing loss with no or mild vestibular dysfunction. To date, only nine variants have been identified to be responsible for DFNA11. Here we present a novel variant (c.1531G>A) causing DFNA11 identified in a three-generation Chinese family. Progressive hearing loss is the only clinical manifestation in this family, and the onset age of affected members is later and more concentrated than that of other DFNA11 families. Our findings further enrich the collection of MYO7A mutations, and our review of the nine reported DFNA11 families can provide a reference for clinical genetic counseling.
- Citation: Xia CF, Yan R, Su WW, Liu YH. Autosomal dominant non-syndromic hearing loss caused by a novel mutation in MYO7A: A case report and review of the literature. World J Clin Cases 2023; 11(25): 5962-5969
- URL: https://www.wjgnet.com/2307-8960/full/v11/i25/5962.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i25.5962
Hearing loss is the most common clinical manifestation in otology, with both genetic and environmental etiologies. Genetic factors account for approximately 60% of cases[1,2]. To date, more than 150 genes have been linked to hearing loss (https://hereditaryhearingloss.org), including GJB2, GJB3, SLC26A4, and MYO7A. MYO7A is located at 11q13.5, has 49 exons encoding 2215 amino acids, and is mainly expressed in the inner ear, retina, testis, lungs, and kidneys[3]. Variants in MYO7A can cause both syndromic (Usher syndrome)[4-6] and non-syndromic deafness, with the latter including autosomal dominant (DFNA11) and autosomal recessive (DFNB2) inheritance patterns[7-9].
Usher syndrome is the most common consequence of MYO7A variants, in which patients experience congenital sensorineural hearing loss, progressive retinitis pigmentosa, and vestibular dysfunction. Furthermore, severe cases can lead to deaf-mutism, total blindness, intellectual disability, and other disorders[10,11]. In contrast, the clinical symptoms of DFNB2 caused by the MYO7A gene variant are milder than those of Usher syndrome and are mainly characterized by congenital sensorineural hearing loss and vestibular dysfunction. A study by Astuto et al[12] found late-onset mild visual impairment in members of a family with DFNB2. Hence, the authors determined that DFNB2 and Usher syndrome are different stages of the same disease. However, this conclusion remains controversial, and visual impairment is still commonly used as the primary criterion to distinguish syndromic from non-syndromic hearing loss caused by MYO7A. Moreover, DFNA11 is the rarest consequence of the MYO7A gene variant and mainly manifests as delayed post-lingual sensorineural hearing loss. Most patients with DFNA11 present with mild or no vestibular dysfunction, but all cases exhibit normal vision[13,14]. To date, no specific association between clinical manifestations and variant location has been found, and only nine variants have been identified as causing DFNA11. The rarity of DFNA11 combined with its occult and atypical clinical manifestations makes the diagnosis of DFNA11 more challenging.
Here, we describe a novel variant in the MYO7A gene (c.1531G>A) that caused DFNA11 in a three-generation Chinese family. All affected members of this family presented with bilateral progressive sensorineural hearing loss beginning in adulthood. Additionally, we provide a summary of the clinical and genetic features observed in previously reported families with DFNA11.
The proband was a 53-year-old Han male who sought treatment at the outpatient clinic of Peking University First Hospital for progressive hearing loss in both ears, manifested for more than 10 years.
The patient presented with bilateral hearing loss that began around the age of 40 and gradually affected daily listening and communication. There were no vestibular disorders such as vertigo or unsteady walking.
The patient had no history of past illness.
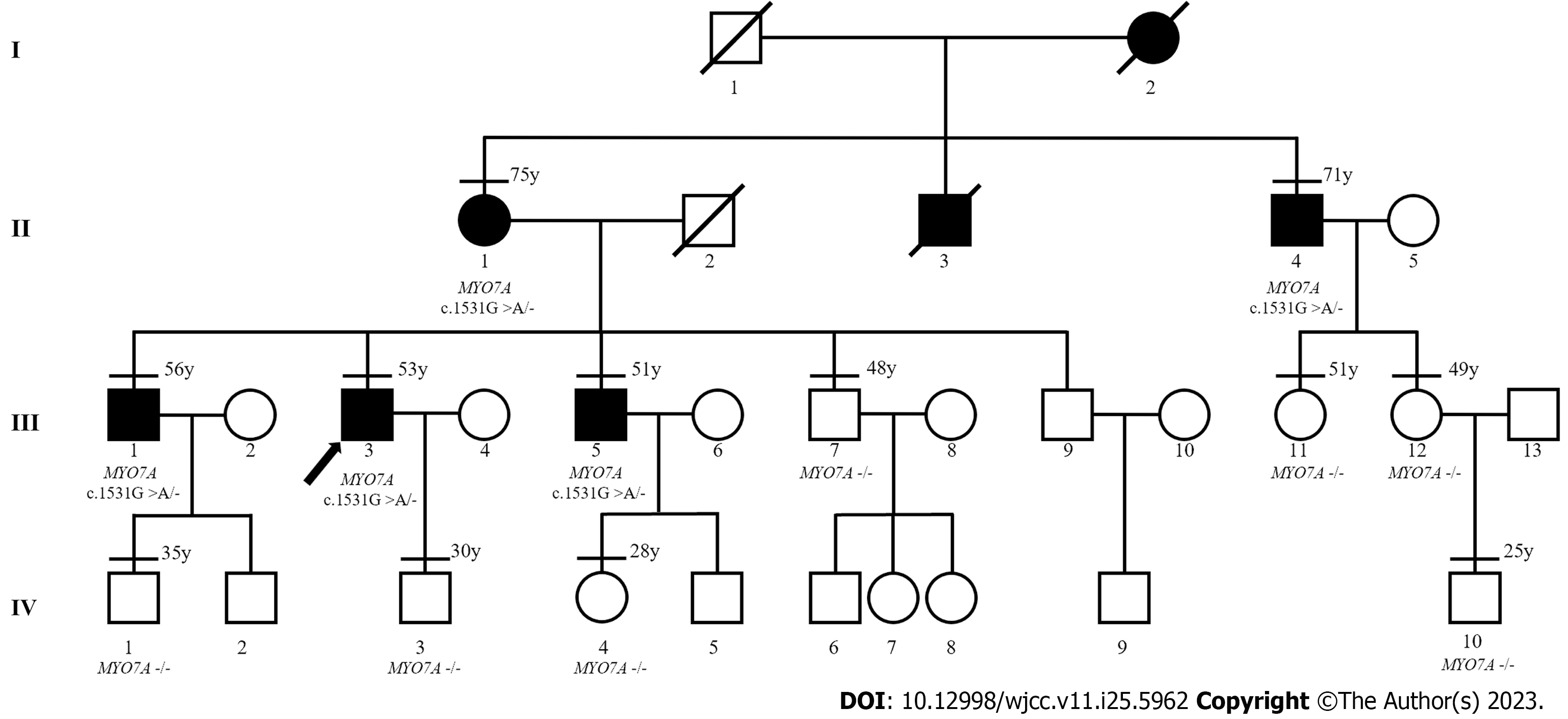
The patient had no history of noise exposure, otitis media, or ototoxic drug use. However, it was understood that several of the patient’s family members had similar symptoms, with slowly progressive bilateral hearing loss, also beginning around the age of 40. Therefore, we contacted 11 additional members of the patient’s family (including four members experiencing hearing loss) and obtained their consent to participate in our study (Figure 1). The first member of the third generation (III:1) had a history of noise exposure for about 20 years because of his work. The remaining 11 family members had no history of noise exposure, otitis media, or ototoxic drug use.
No abnormality was found in the ear examination of the 12 family members.
The proband’s routine blood, liver function, renal function, and coagulation function tests showed no obvious abnormalities.
Five family members with deafness were examined by high-resolution computed tomography of the temporal bone, and no obvious abnormality was found.
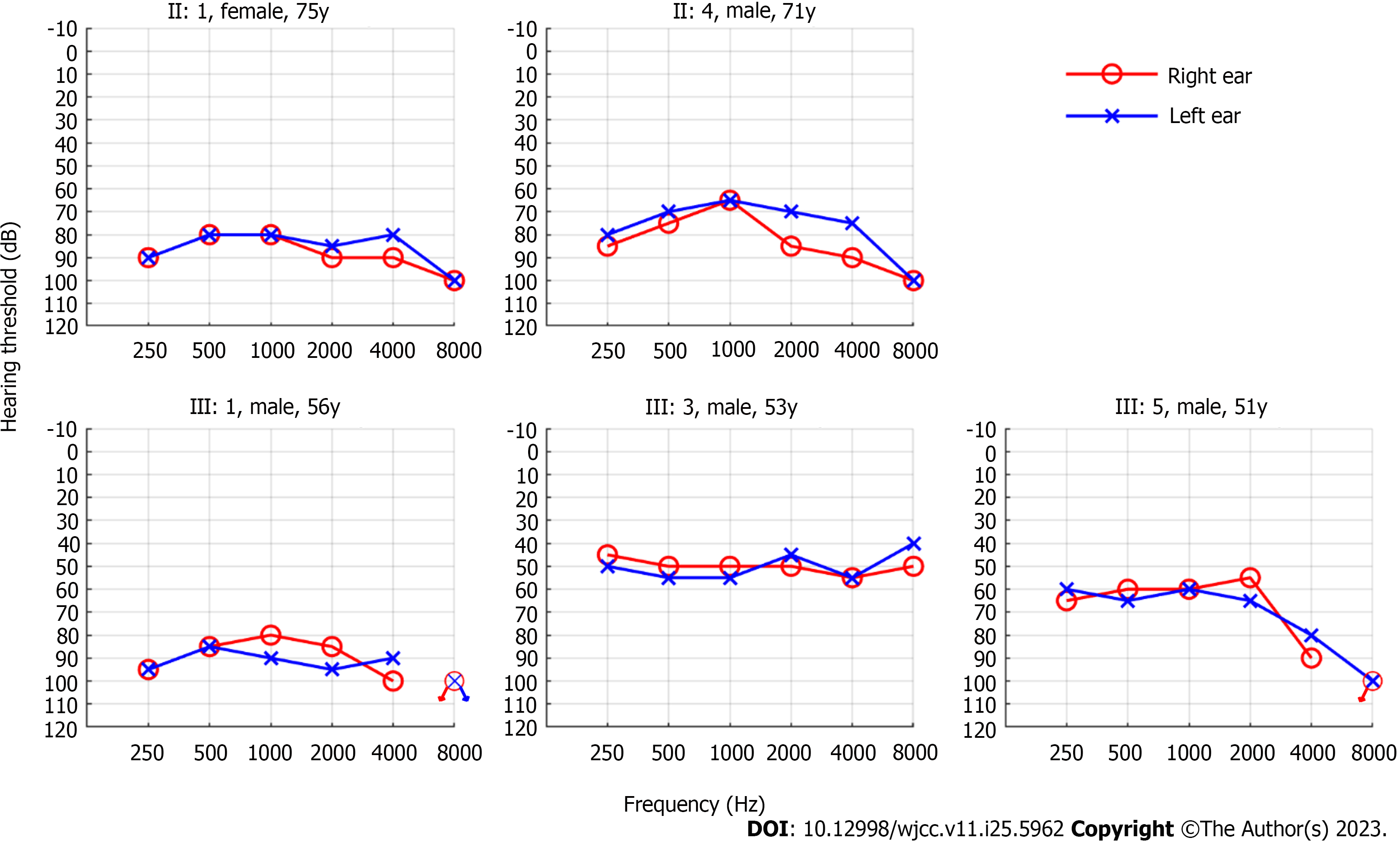
We found that all five affected members had bilateral symmetrical moderate to profound sensorineural hearing loss (Figure 2). The audiograms of the affected family members were flat or slightly sloping, and high-frequency hearing loss was particularly severe. And none of them had vestibular dysfunction or visual impairment.
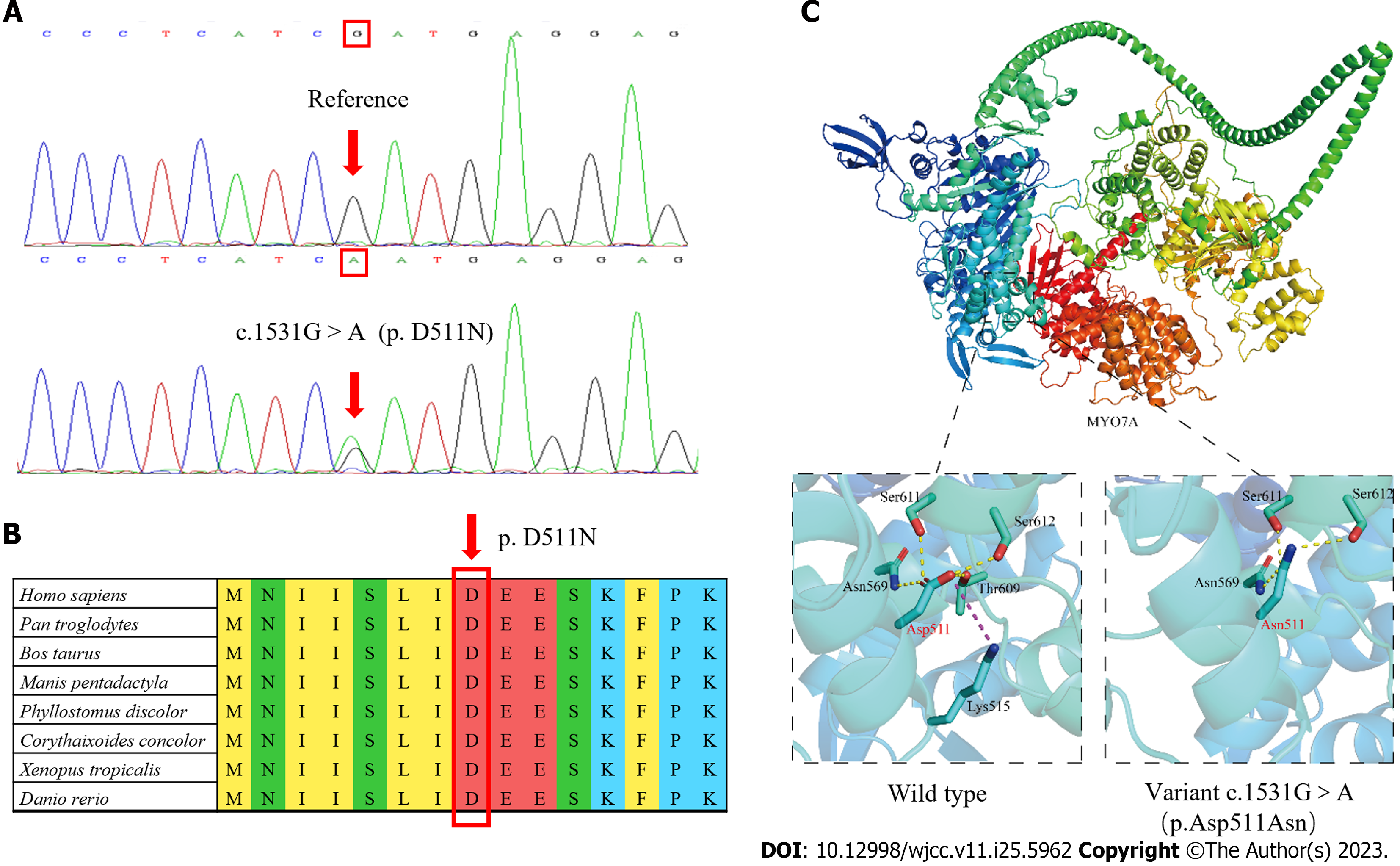
After written informed consent was obtained from all participants or their guardians, peripheral blood samples were collected for genetic analysis. High-throughput sequencing was used for the proband, including the exons of 415 genes, and partial deep intron regions of 147 genes reported by the Human Gene Mutation Database (https://www.hgmd.cf.ac.uk/) were also detected. The coverage density of the probe (GenCap deafness gene capture probe V4.0, https://www.mygeno.cn/) was increased in 29 common deafness pathogenic genes, among which GJB2, SLC26A4, and POU3F4 were covered in full length. Deafness-related genes in the mitochondrial circular DNA were also detected using GenCap Mitochondrial Loop Gene Capture Probe V1.0 (MyGenostics, Beijing, China). Then five candidate variants were identified in the proband: MYO7A (c.1531G>A), TRIOBP (c.3689C>T), WHRN (c.2090C>T), USH1C (1534G>A), and PDZD7 (c.1529G>A). These variants were annotated using ANNOVAR (v20200607)[15] and compared with the Exome Aggregation Consortium (ExAC, v0.3.1, Broad Institute, United States), 1000 Genomes (http://www.1000genomes.org/), and Genome Aggregation Database (gnomAD, http://www.gnomad-sg.org/) databases. The pathogenicity of the variants was predicted using Rare Exome Variant Ensemble Learner (REVEL, https://sites.google.com/site/revelgenomics/), Mutation Taster (https://www.mutationtaster.org/), and PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/) programs. At the same time, we obtained the spatial structure of the MYO7A protein with AlphaFold2 (https://alphafold.ebi.ac.uk), and then used PyMOL (v2.5.4, Schrodinge, United States) to map and analyze the effect of the mutation on the structure of MYO7A protein. Finally, Sanger sequencing of the remaining 11 family members showed that only MYO7A (c.1531G>A) co-segregated with the deafness phenotype. This variant resulted in the conversion of Asp to Asn at position 511 of the MYO7A protein (Figure 3A), an amino acid highly conserved among species (Figure 3B). This variant was not detected in the control group consisting of 200 Chinese Han individuals with normal hearing, or in the ExAC, 1000 Genomes, or gnomAD database. The pathogenicity of this variant was predicted to be “probably damaging” (0.994) by PolyPhen-2, “damaging” (0.853) by REVEL, and “disease-causing” (1.000) by Mutation Taster. As shown in Figure 3C, the Asp at position 511 of wild-type MYO7A protein is located in the motor domain, has three hydrogen bonds with surrounding amino acids, and has electrostatic interaction with Lys at position 515. However, the p.Asp511Asn variant loses one hydrogen bond and the only electrostatic interaction.
The phenotype of deafness in this family was typical of late onset and progressive sensorineural hearing loss, which is consistent with that reported in other families with DFNA11. The MYO7A (c.1531G>A, p.D511N) variant co-segregated with the deafness phenotype in an autosomal dominant pattern in this family. The variant was not detected in 200 normal hearing controls, or in the ExAC, 1000 Genomes, or gnomAD database. Finally, the variant was predicted to be generally damaging by REVEL, Mutation Taster, and PolyPhen2. The p.511D is highly conserved among species, and PyMOL analysis suggests that the structural stability of the MYO7A protein was destroyed after Asp was replaced by Asn in the variant. Based on the above evidence and following the criteria of the American College of Medical Genetics and Genomics, we diagnosed the affected members of this family with autosomal dominant hearing loss caused by the MYO7A (c.1531G>A, p.D511N) variant.
We advised the five hearing-impaired members to protect their residual hearing by avoiding exposure to noise and ototoxic drugs. In addition, the family members were advised to wear hearing aids to improve hearing and daily communication.
Hearing aids greatly improved the hearing and quality of life for the affected family members, as we learned in our telephone follow-ups 3 mo later. Considering that all participants’ hearing loss will progress with age, they were encouraged to make regular follow-up visits and facilitate timely adjustment of hearing aid parameters.
We searched for articles in the PubMed database published up to December 2022 using the keywords “DFNA11”, “MYO7A”, “autosomal dominant inheritance”, and “hearing loss”. A total of 11 families with DFNA11, including nine MYO7A mutation sites, were identified by genetic analysis. The genotype, hearing status, age of onset, and other information of these families are shown in Table 1[8,13,14,16-22].
| Mutation | Exon | Structure | Patient number | Age of onset (yr) | Audiogram | Vestibular symptoms | Family origin | Ref. |
| c.2656-2664del/A886-K888Del | 22 | Coiled coil | 8/19 | 12-16 | Flat/sloping | Absent | Japan | [8] |
| c.652G>A/p.D218N | 7 | Motor domain | 11/29 | 20-47 | Flat/sloping | Absent | China | [13] |
| c.689C>T/p.A230V | 7 | Motor domain | 1 (sporadic case) | 4 | U-shaped | Absent | Japan | [16] |
| c.689C>T/p.A230V | 7 | Motor domain | 9/18 | Mean 6-7 | Flat/sloping | Bilateral areflexia | Italy | [17] |
| c.1373A>T/p.N458I | 13 | Motor domain | 11/26 | 4-43 | Ascending-flat-sloping | Vertigo and unsteady walking | Netherlands | [18] |
| c.2003G>A/p.R668H | 17 | Motor domain | 9/15 | 17-45 | Ascending-flat-sloping | - | China | [19] |
| c.2011G>A/p.G671S | 17 | Motor domain | 9/23 | 10-39 | Ascending-flat-sloping | Absent | China | [13] |
| c.2011G>A/p.G671S | 17 | Motor domain | 11/29 | 13-40 | Flat/sloping | Absent | China | [20] |
| c.2164G>C/p.G722R | 17 | Motor domain | 13/43 | 20-30 | Ascending-flat-sloping | Absent | United States | [21] |
| c.2557C>T/p.R853C | 21 | IQ 5 | 5/12 | 1 mo to puberty | - | Mild dysfunction | Germany | [22] |
| c.2558G>A/p.A853H | 21 | IQ 5 | 12/23 | 1-33 | Flat/sloping | Absent | Japan | [14] |
| c.1531G>A/p.D511N | 13 | Motor domain | 5/12 | 35-42 | Flat/sloping | Absent | China | This report |
Since the first family with DFNA11 was reported by Liu et al in 1997[8], nine variants from 11 families have been identified to be responsible for DFNA11. The clinical phenotypes of deafness in members of these families with DFNA11 varied; however, most presented with post-lingual progressive sensorineural hearing loss[16,20]. In the family of this study, the affected members presented bilateral symmetrical sensorineural hearing loss. The audiograms of the affected family members were flat or slightly sloping, and high-frequency hearing loss was particularly severe, which is consistent with the symptoms previously reported in families with DFNA11[13,14,20]. In the family of this study, the onset age of hearing loss among affected members was relatively late and concentrated, occurring at around 40 years old. This differs from most reported families with DFNA11, where the age span of onset is larger and hearing loss occurred before adulthood in many cases[14,17,22]. The hearing loss of the second-generation members was generally more severe than that of the third-generation members, which also horizontally reflects that hearing loss progresses with age. However, differences were observed between III:1, III:3, and III:5, who were in the same age group. Although the presence of genetic modifications cannot be excluded[23], environmental effects on the phenotype, such as chronic exposure to noise (i.e., III:1), should also be considered. Therefore, the preservation of residual hearing is one of the important therapeutic measures. Moreover, it is essential to select hearing aid devices according to the hearing threshold and actual needs of each patient, to minimize the impact of DFNA11 on their daily life.
In the inner ear, MYO7A is expressed in hair cells, utricles, and semicircular canals and is involved in the transmembrane transport of proteins and functional maintenance of hair cells[24,25]. In the eyes, MYO7A is mainly expressed in photoreceptors and pigment epithelial cells and its main function is to transport visual proteins together with connecting cilia[26,27]. Therefore, in most cases, variants in MYO7A cause dysfunction in the encoded protein, leading to both sensorineural hearing loss and visual impairment. In the eye, the dysfunctional protein may be compensated by other proteins in the retina[18]; nonetheless, its function in the inner ear is unique[25,28]. Therefore, patients with variants in MYO7A can present only hearing loss and mild or no vestibular dysfunction with no ocular symptoms[17,18,22], resulting in rare non-syndromic hearing loss. In the family of this study, the phenotype of hearing loss of the study participants is consistent with an autosomal dominant inheritance pattern, and clinical examination revealed no visual impairment or vestibular symptoms in the affected members. This is consistent with the symptoms reported in families with DFNA11. The only symptom of the studied family was slowly progressive hearing loss, which may also have contributed to the delay in seeking medical attention.
The protein encoded by MYO7A belongs to the myosin family, which is composed of three regions: the N-terminal head (motor domain), IQ5 neck, and C-terminal tail, the last of which begins with a single α-helix domain[29,30]. The motor domain contains the binding domains of adenosine triphosphate and actin, which are the core functional areas of the molecule[31]. The variant identified in this study leads to the loss of hydrogen bond and electrostatic interaction in the motor domain, which is predicted to cause a decrease in the stability of the local structure and subsequently affect the function of the MYO7A protein.
In a clinical setting, a diagnosis of this type of post-lingual hereditary hearing loss requires ruling out many lesions of the middle and inner ear, as well as nerve damage caused by noise, drugs, autoimmunity, or other factors. The key points of differential diagnosis are to inquire about the characteristics of hearing changes carefully, collect details on the patient’s personal and family medical history, and complete various clinical examinations. For rare variants, extensive clinical and genetic data collection from family members is a prerequisite for diagnosis. Based on the results presented above, we made the diagnosis and provided effective treatment recommendations and genetic counseling for all family members. However, owing to the limited experimental conditions, we did not conduct further verification of the pathogenic mechanism of this variant.
In this study, we report a new family with DFNA11 caused by a MYO7A variant, which provides a new screening site for hereditary deafness. At the same time, the late onset age of hearing loss in the studied family also provides new insights into the clinical phenotype of DFNA11.
The authors would like to thank the patients and their families.
| 1. | Lin X, Tang W, Ahmad S, Lu J, Colby CC, Zhu J, Yu Q. Applications of targeted gene capture and next-generation sequencing technologies in studies of human deafness and other genetic disabilities. Hear Res. 2012;288:67-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 83] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 2. | Rehm HL. Genetics and the genome project. Ear Hear. 2003;24:270-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 3. | Hasson T, Heintzelman MB, Santos-Sacchi J, Corey DP, Mooseker MS. Expression in cochlea and retina of myosin VIIa, the gene product defective in Usher syndrome type 1B. Proc Natl Acad Sci U S A. 1995;92:9815-9819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 315] [Cited by in RCA: 348] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 4. | Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston MD. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374:60-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 766] [Cited by in RCA: 779] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 5. | Weston MD, Kelley PM, Overbeck LD, Wagenaar M, Orten DJ, Hasson T, Chen ZY, Corey D, Mooseker M, Sumegi J, Cremers C, Moller C, Jacobson SG, Gorin MB, Kimberling WJ. Myosin VIIA mutation screening in 189 Usher syndrome type 1 patients. Am J Hum Genet. 1996;59:1074-1083. [PubMed] |
| 6. | Zina ZB, Masmoudi S, Ayadi H, Chaker F, Ghorbel AM, Drira M, Petit C. From DFNB2 to Usher syndrome: variable expressivity of the same disease. Am J Med Genet. 2001;101:181-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 7. | Liu XZ, Walsh J, Mburu P, Kendrick-Jones J, Cope MJ, Steel KP, Brown SD. Mutations in the myosin VIIA gene cause non-syndromic recessive deafness. Nat Genet. 1997;16:188-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 328] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 8. | Liu XZ, Walsh J, Tamagawa Y, Kitamura K, Nishizawa M, Steel KP, Brown SD. Autosomal dominant non-syndromic deafness caused by a mutation in the myosin VIIA gene. Nat Genet. 1997;17:268-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 216] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 9. | Tamagawa Y, Ishikawa K, Ishida T, Kitamura K, Makino S, Tsuru T, Ichimura K. Phenotype of DFNA11: a nonsyndromic hearing loss caused by a myosin VIIA mutation. Laryngoscope. 2002;112:292-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 10. | Fuster-García C, García-Bohórquez B, Rodríguez-Muñoz A, Aller E, Jaijo T, Millán JM, García-García G. Usher Syndrome: Genetics of a Human Ciliopathy. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 77] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 11. | Dammeyer J. Children with Usher syndrome: mental and behavioral disorders. Behav Brain Funct. 2012;8:16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 12. | Astuto LM, Kelley PM, Askew JW, Weston MD, Smith RJ, Alswaid AF, Al-Rakaf M, Kimberling WJ. Searching for evidence of DFNB2. Am J Med Genet. 2002;109:291-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 13. | Sun Y, Chen J, Sun H, Cheng J, Li J, Lu Y, Jin Z, Zhu Y, Ouyang X, Yan D, Dai P, Han D, Yang W, Wang R, Liu X, Yuan H. Novel missense mutations in MYO7A underlying postlingual high- or low-frequency non-syndromic hearing impairment in two large families from China. J Hum Genet. 2011;56:64-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 14. | Yamamoto N, Mutai H, Namba K, Goto F, Ogawa K, Matsunaga T. Clinical Profiles of DFNA11 at Diverse Stages of Development and Aging in a Large Family Identified by Linkage Analysis. Otol Neurotol. 2020;41:e663-e673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 15. | Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7976] [Cited by in RCA: 11038] [Article Influence: 689.9] [Reference Citation Analysis (0)] |
| 16. | Kaneko Y, Nakano A, Arimoto Y, Nara K, Mutai H, Matsunaga T. The first sporadic case of DFNA11 identified by next-generation sequencing. Int J Pediatr Otorhinolaryngol. 2017;100:183-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 17. | Di Leva F, D'Adamo P, Cubellis MV, D'Eustacchio A, Errichiello M, Saulino C, Auletta G, Giannini P, Donaudy F, Ciccodicola A, Gasparini P, Franzè A, Marciano E. Identification of a novel mutation in the myosin VIIA motor domain in a family with autosomal dominant hearing loss (DFNA11). Audiol Neurootol. 2006;11:157-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 18. | Luijendijk MW, Van Wijk E, Bischoff AM, Krieger E, Huygen PL, Pennings RJ, Brunner HG, Cremers CW, Cremers FP, Kremer H. Identification and molecular modelling of a mutation in the motor head domain of myosin VIIA in a family with autosomal dominant hearing impairment (DFNA11). Hum Genet. 2004;115:149-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 42] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 19. | Sang Q, Yan X, Wang H, Feng R, Fei X, Ma D, Xing Q, Li Q, Zhao X, Jin L, He L, Li H, Wang L. Identification and functional study of a new missense mutation in the motor head domain of myosin VIIA in a family with autosomal dominant hearing impairment (DFNA11). PLoS One. 2013;8:e55178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 20. | Li L, Yuan H, Wang H, Guan J, Lan L, Wang D, Zong L, Liu Q, Han B, Huang D, Wang Q. Identification of a MYO7A mutation in a large Chinese DFNA11 family and genotype-phenotype review for DFNA11. Acta Otolaryngol. 2018;138:463-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 21. | Kallman JC, Phillips JO, Bramhall NF, Kelly JP, Street VA. In search of the DFNA11 myosin VIIA low- and mid-frequency auditory genetic modifier. Otol Neurotol. 2008;29:860-867. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 22. | Bolz H, Bolz SS, Schade G, Kothe C, Mohrmann G, Hess M, Gal A. Impaired calmodulin binding of myosin-7A causes autosomal dominant hearing loss (DFNA11). Hum Mutat. 2004;24:274-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 23. | Street VA, Li J, Robbins CA, Kallman JC. A DNA variant within the MYO7A promoter regulates YY1 transcription factor binding and gene expression serving as a potential dominant DFNA11 auditory genetic modifier. J Biol Chem. 2011;286:15278-15286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 24. | Kachar B, Battaglia A, Fex J. Compartmentalized vesicular traffic around the hair cell cuticular plate. Hear Res. 1997;107:102-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 70] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 25. | Li S, Mecca A, Kim J, Caprara GA, Wagner EL, Du TT, Petrov L, Xu W, Cui R, Rebustini IT, Kachar B, Peng AW, Shin JB. Myosin-VIIa is expressed in multiple isoforms and essential for tensioning the hair cell mechanotransduction complex. Nat Commun. 2020;11:2066. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 26. | Williams DS. Usher syndrome: animal models, retinal function of Usher proteins, and prospects for gene therapy. Vision Res. 2008;48:433-441. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 134] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 27. | Liu X, Udovichenko IP, Brown SD, Steel KP, Williams DS. Myosin VIIa participates in opsin transport through the photoreceptor cilium. J Neurosci. 1999;19:6267-6274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 212] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 28. | Self T, Mahony M, Fleming J, Walsh J, Brown SD, Steel KP. Shaker-1 mutations reveal roles for myosin VIIA in both development and function of cochlear hair cells. Development. 1998;125:557-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 193] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 29. | Li J, Chen Y, Deng Y, Unarta IC, Lu Q, Huang X, Zhang M. Ca(2+)-Induced Rigidity Change of the Myosin VIIa IQ Motif-Single α Helix Lever Arm Extension. Structure. 2017;25:579-591.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 30. | Chen ZY, Hasson T, Kelley PM, Schwender BJ, Schwartz MF, Ramakrishnan M, Kimberling WJ, Mooseker MS, Corey DP. Molecular cloning and domain structure of human myosin-VIIa, the gene product defective in Usher syndrome 1B. Genomics. 1996;36:440-448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 104] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 31. | Joo SY, Na G, Kim JA, Yoo JE, Kim DH, Kim SJ, Jang SH, Yu S, Kim HY, Choi JY, Gee HY, Jung J. Clinical Heterogeneity Associated with MYO7A Variants Relies on Affected Domains. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Mirsalehi M, Iran; Naz S, Pakistan S-Editor: Wang JJ L-Editor: Wang TQ P-Editor: Cai YX