Published online Jul 16, 2023. doi: 10.12998/wjcc.v11.i20.4833
Peer-review started: April 1, 2023
First decision: May 8, 2023
Revised: May 13, 2023
Accepted: June 12, 2023
Article in press: June 12, 2023
Published online: July 16, 2023
Processing time: 102 Days and 6.8 Hours
Severe infection often results in bacteremia, which significantly increases mortality rate. Different therapeutic strategies are employed depending on whether the blood-borne infection is Gram-negative (G-) or Gram-positive (G+). However, there is no risk prediction model for assessing whether bacteremia patients are infected with G- or G+ pathogens.
To establish a clinical prediction model to distinguish G- from G+ infection.
A total of 130 patients with positive blood culture admitted to a single intensive care unit were recruited, and Th1 and Th2 cytokine concentrations, routine blood test results, procalcitonin and C-reactive protein concentrations, liver and kidney function test results and coagulation function were compared between G+ and G- groups. Least absolute shrinkage and selection operator (LASSO) regression analysis was employed to optimize the selection of predictive variables by running cyclic coordinate descent and K-fold cross-validation (K = 10). The predictive variables selected by LASSO regression analysis were then included in multivariate logistic regression analysis to establish a prediction model. A nomogram was also constructed based on the prediction model. Calibration chart, receiver operating characteristic curve and decision curve analysis were adopted for validating the prediction model.
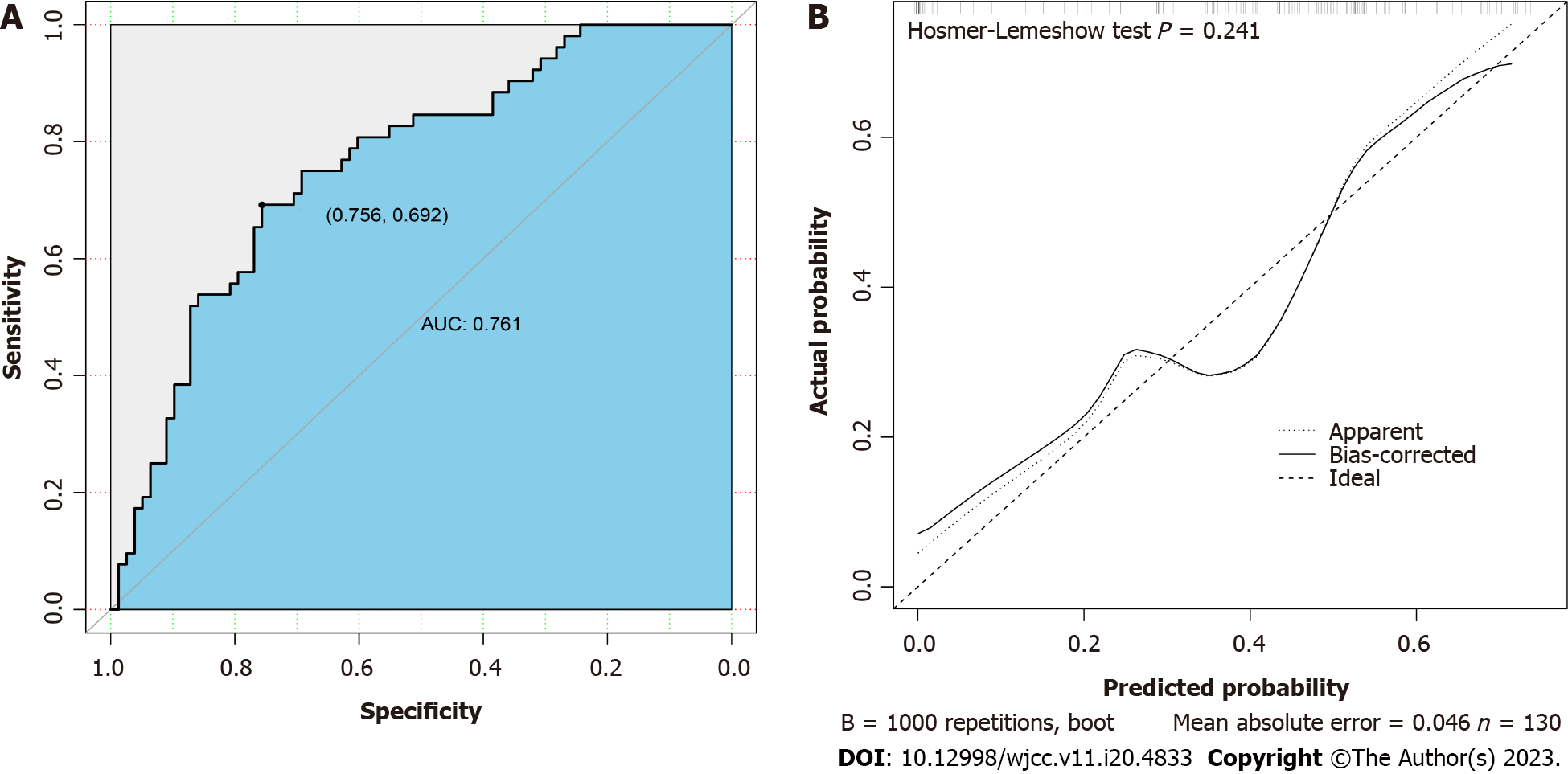
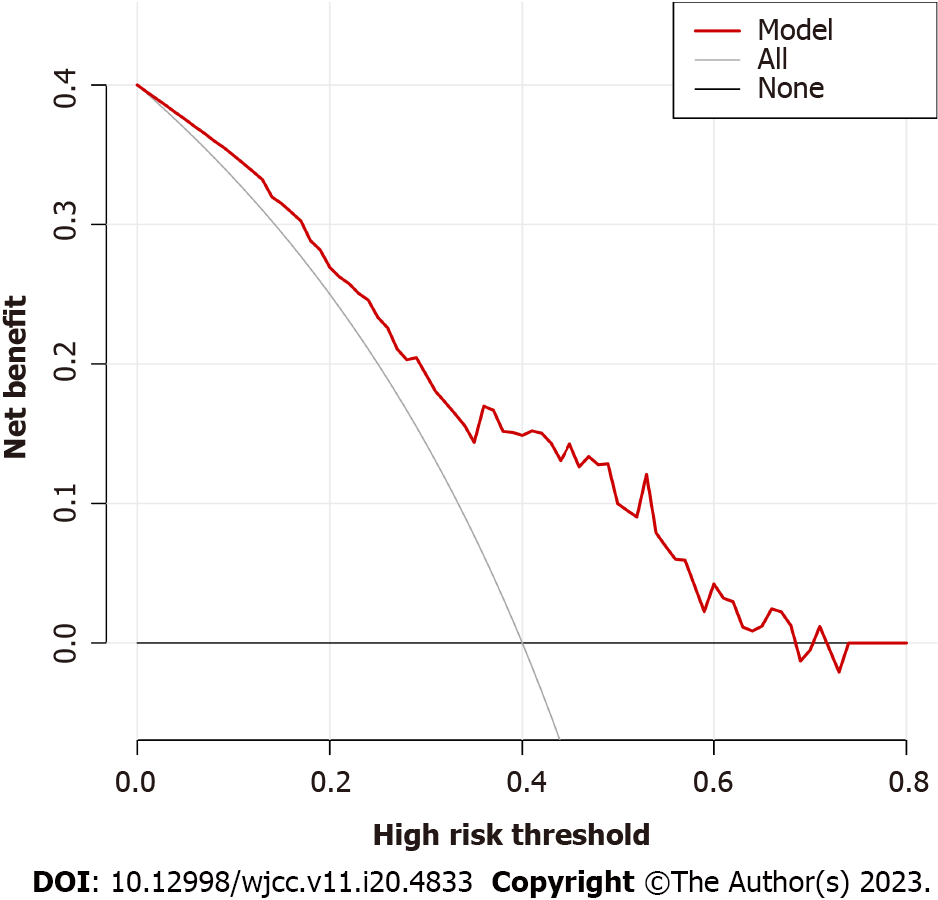
Age, plasma interleukin 6 (IL-6) concentration and plasma aspartate aminotransferase concentration were identified from 57 measured variables as potential factors distinguishing G+ from G- infection by LASSO regression analysis. Inclusion of these three variables in a multivariate logistic regression model identified age and IL-6 as significant predictors. In receiver operating characteristic curve analysis, age and IL-6 yielded an area under the curve of 0.761 and distinguished G+ from G- infection with specificity of 0.756 and sensitivity of 0.692. Serum IL-6 and IL-10 levels were upregulated by more than 10-fold from baseline in the G- bacteremia group but by less than ten-fold in the G+ bacteremia group. The calibration curve of the model and Hosmer-Lemeshow test indicated good model fit (P > 0.05). When the decision curve analysis curve indicated a risk threshold probability between 0% and 68%, a nomogram could be applied in clinical settings.
A simple prediction model distinguishing G- from G+ bacteremia can be constructed based on reciprocal association with age and IL-6 level.
Core Tip: This study was designed to assess whether the cytokine profile and other clinical variables can distinguish Gram-positive from Gram-negative bacteremia. A reliable predicted model could prove valuable for facilitating early identification of the causative pathogen and the rational use of antibiotics, thereby preventing progression into potentially fatal septic shock.
- Citation: Zhang W, Chen T, Chen HJ, Chen N, Xing ZX, Fu XY. Risk prediction model for distinguishing Gram-positive from Gram-negative bacteremia based on age and cytokine levels: A retrospective study. World J Clin Cases 2023; 11(20): 4833-4842
- URL: https://www.wjgnet.com/2307-8960/full/v11/i20/4833.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i20.4833
Bacteremia is defined as the presence of bacteria in the blood circulation. If not effectively controlled, these bacteria and associated bacterial toxins (endotoxins and exotoxins) can induce sepsis, a condition characterized by severe tissue-damaging inflammation with high morbidity, mortality and treatment cost, mainly due to (septic) shock and multiple organ dysfunction syndrome[1-4]. Gram-negative (G-) bacteria such as Escherichia coli, Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae and Gram-positive (G+) bacteria such as Staphylococcus aureus, carbapenem-resistant Staphylococcus aureus and Enterococcus faecium are among the most common causative pathogens of septic infection[3,5]. The optimal treatment differs depending on whether the causative pathogen is G+ or G-. Therefore, distinguishing between these bacterial types during the early stages of infection is essential for preventing sepsis.
The profile of Th1 and Th2 cytokine release by activated neutrophils, lymphocytes and mononuclear phagocytes is also a critical determinant of whether bacteremia progresses to sepsis. Cytokines can be broadly divided into proinflammatory and anti-inflammatory, and the balance between these classes has a major influence on clinical course, including the incidence of sepsis[6]. Among patients with bacterial infection, interleukin 6 (IL-6), IL-10, tumor necrosis factor (TNF)-α and interferon (IFN)-γ are all elevated. In contrast, only IFN-γ levels may be elevated significantly (> 100 pg/mL) in patients with fungal infections or tuberculosis, although IL-6 level may also increase in severe cases. However, IFN-γ levels may increase only slightly (< 100 pg/mL), accompanied by elevations of IL-6 or IL-10[7,8].
This study was designed to assess if the cytokine profile and other clinical variables can distinguish G+ from G- bacteremia. A reliable predicted model could prove valuable for facilitating early identification of the causative pathogen and the rational use of antibiotics, thereby preventing progression into potentially fatal septic shock.
The data used to construct the predictive model were obtained from a retrospective review of sepsis patients admitted to the intensive care unit of our hospital from December 2018 to February 2020.
A total of 130 patients with sepsis were enrolled. Inclusion criteria were as follows: (1) Basic conditions for infection; (2) Positive for bacteremia according to blood bacterial culture; (3) Age ≥ 18 years and ≤ 80 years; (4) Initial quick Sequential Organ Failure Assessment score ≥ 2 (severe infection and organ dysfunction); and (5) Two or more of (a) body temperature > 38 C or < 36 C, (b) heart rate > 90/min, (c) respiratory frequency > 20/min or arterial partial pressure of carbon dioxide < 32 mmHg, (d) peripheral white blood cell count > 12 × 109/L or < 4 × 109/L, and (e) infected site confirmed by imaging examination. Exclusion criteria were: (1) Age < 18 years or > 80 years; and (2) Infection complicated by other serious physical diseases (malignant tumors or nervous system diseases, etc), chronic renal insufficiency or other infectious diseases.
Age, sex, family medical history, patient medical history, concentrations of plasma cytokines (IL-2, IL-4, IL-6, IL-10, TNF-α and IFN-γ), bacterial culture results (blood, sputum, secretions and effusions, etc), blood biochemistry results, C-reactive protein, procalcitonin (PCT) and fungal D-glucan concentrations, routine blood test results and chest or other imaging data were collected.
Venous blood samples was collected using EDTA anticoagulation tubes and sent for laboratory detection of plasma Th1/Th2 cytokine levels. Briefly, the plasma sample was centrifuged at 3000 rpm for 20 min at room temperature to separate plasma. A Th1/Th2 cytokine detection kit was purchased from Cellgene Biotech (Hangzhou, China). Plasma cytokine contents were determined using a BD FACS flow cytometer.
All statistical analyses were conducted using R language software (version 4.0.5). Patients were divided into G- and G+ groups according to blood culture results. Normally distributed continuous variables were expressed as mean ± SD and non-normally distributed continuous variables by median and interquartile range. Continuous variable normal distributions and homogeneity of variance were compared between groups by independent sample t-test, while non-normally distributed continuous variables were compared between groups by Mann-Whitney U-test. Counting data were compared by χ2 test.
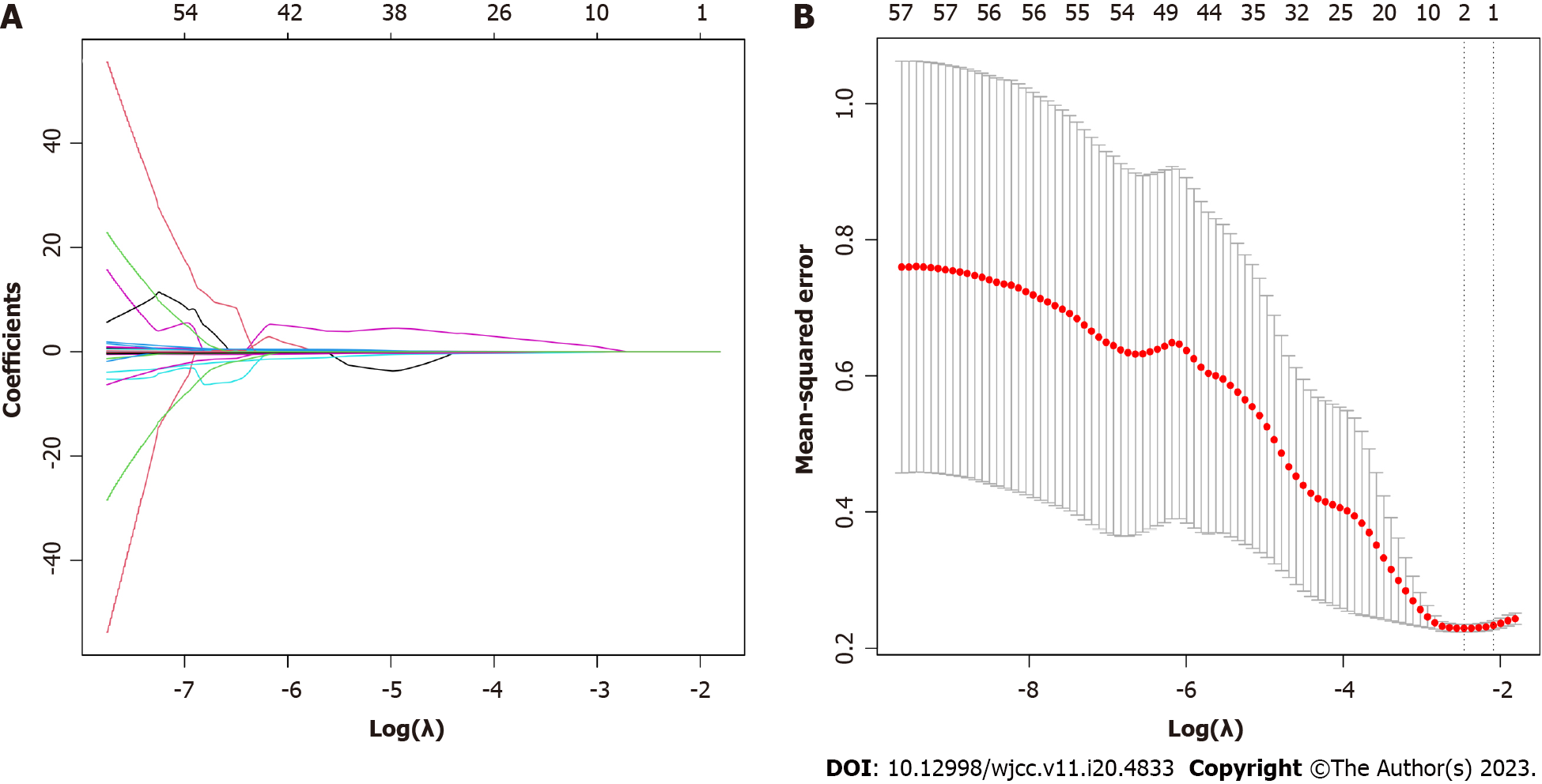
Least absolute shrinkage and selection operator (LASSO) regression analysis using the package is suitable for reducing high-dimensional data and identifying the optimal variables for further construction of multivariate prediction models. Variables with “non-zero” coefficients from the LASSO regression model were selected, and ten-fold cross validation was used to define an appropriate adjustment parameter (λ). The most significant variables screened by LASSO regression analysis were then included in a multivariate logistic regression model. Variables without significant correlation coefficients were excluded.
Model performance for distinguishing G+ from G- bacteremia was evaluated by receiver operating characteristic (ROC) curve analysis using the qROC package for R language. The calibration curve function of the rms package was utilized to evaluate the sepsis risk nomogram and the Hosmer-Lemeshow test to determine the model goodness of fit. Finally, the decision curve analysis (DCA) package ggDCA was used to determine the clinical utility of the model according to the net benefit under different threshold profiles. Finally, a visual nomogram was drawn using the rms package.
A total of 130 patients with sepsis were recruited (42 females and 88 males), including 52 cases caused by G+ bacteria and 78 cases caused by G- bacteria as determined by blood culture. All patients completed relevant examinations, yielding 57 variables, and each was compared between G- and G+ groups. Age, IL-2, IL-4, IL-6, IL-10, TNF-α, PCT, total bilirubin, direct bilirubin, aspartate aminotransferase (AST) and prealbumin differed significantly between groups (all P < 0.05). Both IL-6 and IL-10 were upregulated to higher levels in the G- group than the G+ group (more than ten-fold from baseline vs less than 10-fold from baseline (Table 1).
| Variable | G+, n = 52 | G-, n = 78 | P value |
| Male | 38 (35.2) | 50 (52.8) | 0.284 |
| Female | 14 (16.8) | 28 (25.8) | |
| Age in yr | 53.00 (36.25, 60.75) | 56.00 (46.50, 71.25) | 0.025 |
| APACHE II | 19.00 (16.00, 23.00) | 19.00 (17.00, 25.00) | 0.194 |
| SOFA | 5.00 (3.00, 6.00) | 5.00 (4.00, 7.00) | 0.243 |
| IL-2 in pg/mL | 0.12 (0.00, 0.63) | 0.36 (0.00, 1.15) | 0.017 |
| IL-4 in pg/mL | 0.15 (0.00, 0.71) | 0.55 (0.00, 1.10) | 0.057 |
| IL-6 in pg/mL | 31.68 (18.87, 82.93) | 297.41 (95.40, 1532.49) | < 0.001 |
| IL-10 in pg/mL | 9.64 (5.81, 18.67) | 32.67 (10.39, 18.07) | < 0.001 |
| TNF-α in pg/mL | 0.00 (0.00, 0.00) | 0.00 (0.00, 0.67) | 0.002 |
| IFN-γ in pg/mL | 0.16 (0.00, 0.78) | 0.23 (0.00, 1.38) | 0.123 |
| Fungal D in pg/mL | 35.81 (10.00, 219.23) | 93.02 (10.00, 259.73) | 0.257 |
| PCT in ng/mL | 1.42 (0.31, 12.56) | 3.79 (1.40, 18.68) | 0.04 |
| ALT in U/L | 43.00 (24.25, 125.25) | 35.30 (17.75, 74.00) | 0.067 |
| AST in U/L | 68.00 (39.25, 208.75) | 65.00 (27.50, 173.75) | 0.035 |
| TBIL in μmol/L | 15.650 (10.875, 24.750) | 19.650 (13.070, 29.600) | 0.034 |
| DBIL in μmol/L | 5.700 (3.425, 10.375) | 7.800 (4.100, 14.050) | 0.024 |
| IBIL in μmol/L | 8.750 (6.625, 13.725) | 11.550 (7.770, 14.630) | 0.094 |
| ALB in g/L | 28.050 (25.500, 32.525) | 28.250 (25.170, 32.920) | 0.968 |
| GLB in g/L | 22.850 (19.175, 26.800) | 21.950 (18.750, 26.725) | 0.714 |
| PA in mg/L | 114.00 (70.00, 148.25) | 91.50 (59.50, 120.50) | 0.047 |
| Urea in mmol/L | 8.350 (5.898, 14.528) | 8.630 (5.270, 13.970) | 0.887 |
| Cr in μmol/L | 77.00 (60.25, 147.25) | 82.50 (61.75, 134.25) | 0.960 |
| CysC in mg/L | 1.165 (0.863, 1.613) | 1.180 (0.900, 1.640) | 0.621 |
| GLU in mmol/L | 8.06 (6.11, 10.49) | 8.30 (6.55, 10.73) | 0.684 |
| CRP in mg/L | 154.55 (129.80, 176.40) | 167.40 (128.07, 185.93) | 0.155 |
| WBC as 109 | 13.17 (7.73, 19.22) | 14.15 (9.93, 17.77) | 0.378 |
| NEUT | 0.870 (0.820, 0.918) | 0.890 (0.850, 0.930) | 0.168 |
| LYMPH | 0.07 (0.04, 0.11) | 0.55 (0.03, 0.08) | 0.076 |
| HGB in g/L | 98.00 (79.00, 115.70) | 97.00 (86.75, 107.50) | 0.732 |
| HCT in L/L | 0.29 (0.24, 0.35) | 0.29 (0.26, 0.33) | 0.730 |
| PLT as 1012 | 134.00 (75.00, 225.75) | 117.00 (57.50, 212.75) | 0.626 |
| INR | 1.04 (0.93, 1.16) | 1.04 (0.95, 1.31) | 0.379 |
| PT | 96.90 (83.05, 114.38) | 97.00 (69.40, 113.00) | 0.382 |
| APTT | 36.10 (28.75, 44.20) | 36.50 (30.00, 48.65) | 0.460 |
All 57 measured variables were included in the LASSO regression model, and those with non-zero coefficients (age, AST and IL-6) were retained for further construction of a multivariate model (Figure 1).
The three variables identified by LASSO regression were then included as independent variables in a multivariate logistic regression model with type of bacterial infection (G- or G+) as the dependent variable (Table 2). However, the coefficient for AST was not significant and was excluded. Therefore, the final multivariate model included age and plasma IL-6 concentration.
| Variable | Prediction model | ||
| β | Odds ratio (95%CI) | P value | |
| Age | -0.026 | 0.951-0.999 | 0.040 |
| IL-6 in pg/mL | -0.002 | 0.997-0.999 | 0.009 |
| AST in U/L | 0.001 | 1.000-1.002 | 0.070 |
The area under the ROC curve was 0.761, and the prediction model demonstrated a specificity of 0.756 and sensitivity of 0.692 for distinguishing G- from G+ bacteremia (Figure 2A), with generally moderate performance. The calibration curve of the model is shown in Figure 2B. Here, the diagonal dashed line represents perfect prediction and the solid line represents the performance of the actual model. The difference (distance) is indicative of prediction performance. The Hosmer-Lemeshow test yielded a P > 0.05, indicating good model fit.
The DCA diagram indicated that when the risk threshold probability was between 0% and 68% (Figure 3), the model yielded higher accuracy for distinguishing G- from G+ bacteremia.
Finally, a nomogram was constructed for individual clinical use (Figure 4). A typical case was used to explain nomogram performance. If the critically ill patient was 60-years-old and plasma IL-6 level was 4500 pg/mL, the nomogram yielded a score of approximately 25 points (15 points plus 10 points), which is closer to the side of G- bacteremia indicating that G- bacteremia is more likely.
Sepsis patients deteriorate rapidly, and timely identification of the causative pathogen and treatment initiation, including rational antibiotic therapy, is essential[9,10]. The progression of bacterial infection into sepsis is strongly associated with the host cytokine profile. Multiple stressors, including infection, trauma and surgery, can disrupt the Th1-Th2 balance[11,12]. In patients with sepsis, macrophages release large amounts of IL-1β, TNF-α and prostaglandin E2 in response to stimulation by bacterial endotoxin or signals from damaged tissue. While IL-1β and TNF-α are generally proinflammatory, prostaglandin E2 inhibits IFN-γ release in response to endotoxin.
Concomitantly, the hypothalamus-pituitary-adrenal axis can secrete a large quantity of glucocorticoids under stress, which in turn induces IkB synthesis, inhibits NF-kB signaling and the release of inflammatory cytokines, enhances the differentiation of Th2 cells and promotes the release of multiple cytokines, such as IL-4, IL-10 and IL-6, that drive the transformation of Th1 cells into Th2 cells[13-15]. Consistent with Tang et al[7], cytokine levels were upregulated in both G- and G+ bacteremia patients. However, the increases in IL-6 and IL-10 relative to baseline were significantly greater in the G- group (P < 0.05), providing a metric for distinguishing G- from G+ bacteremia.
Among the 130 patients, age, IL-2, IL-4, IL-6, IL-10, TNF-α, PCT, total bilirubin, direct bilirubin, AST and prealbumin differed significantly between those infected with G+ bacteria and those infected with G- bacteria. These 11 variables were used to construct and validate a novel risk prediction model for G+vs G- blood-borne infection. This multivariate analysis indicated that age and IL-6 were the key distinguishing variables. Introducing other demographic and clinical parameters from biochemical and physical examinations into the risk nomogram may further enhance discrimination accuracy and allow the nomogram to be applied for larger clinical samples[16,17]. However, the sample size was small, and important predictive variables may have been excluded.
The multivariate model indicated that age and plasma IL-6 alone were interchangeable predictors of G+vs G- bacteremia, while the regression coefficient for AST was not significant. Further, the Hosmer-Lemeshow test yielded a P > 0.05 for age and IL-6, indicating goodness of fit. Specifically, older age was associated with higher serum IL-6 and greater incidence of G- bacteremia.
Nomograms are reliable and practical statistical algorithms that can rapidly predict the probability of clinical events by integrating multiple variables[18] and risk factors[19]. Further, these may be digitized for inclusion of additional factors for more accurate prediction and better clinical decision-making. A visual nomogram constructed in the R language considering these two variables predicted increased risk of G- bacterial infection (e.g., if the IL-6 level is 4500 pg/mL in a 60-year-old patient, total score is close to the G- bacteremia side on the nomogram).Thus, the older the patient and the higher the plasma IL-6 level, the greater the risk of G- bacterial infection, while a younger patient with lower IL-6 is more likely to have a G+ infection.
A reliable prediction model to distinguish G- from G+ bacteremia based on age and plasma IL-6 could facilitate more rapid diagnosis and initiation of rational treatment, possibly because elderly patients suffer from multiple primary diseases and low immune function, conditions that increase susceptibility to G- bacterial infection over G+ bacterial infection. Invasion by G- bacteria evokes a cytokine imbalance after cytokine storm, including elevated IL-6. Other cytokines are also elevated but may not have reached significance for model inclusion due to the small sample size.
Sepsis is a progressive disease, and it is essential to control infection and prevent deterioration through rational use of antibiotics and control of cytokine storm. For this, accurate risk prediction tools are necessary to identify the pathogenic species at an early stage and implement the most effective therapeutic measures. In the present study, we developed such a prediction tool to assist clinicians in prompt identification of bacteremia type.
In the present study, age and IL-6 concentration were found to distinguish G+ from G- bacteremia with high accuracy. Specifically, the probability of G- bacteremia is associated with older age and higher plasma IL-6 than G+ bacteremia. Plasma IL-6 and IL-10 levels were increased ten-fold from baseline in the G- group but by less than ten-fold in the G+ group. While this prediction model may have excluded other significant distinguishing factors due to the small sample size, it may still facilitate faster and more effective treatment.
Severe infection often results in bacteremia, which significantly increases mortality rates. Different therapeutic strategies are employed depending on whether the blood-borne infection is Gram-negative (G-) or Gram-positive (G+).
There is no risk prediction model for assessing whether bacteremia patients are infected with G- or G+ pathogens.
To establish a clinical prediction model to distinguish G- from G+ infection.
A total of 130 patients with positive blood culture admitted to a single intensive care unit were recruited, and Th1 and Th2 cytokine concentrations, routine blood test results, procalcitonin and C-reactive protein concentrations, liver and kidney function test results and coagulation function were compared between G+ and G- groups. Least absolute shrinkage and selection operator (LASSO) regression analysis was employed to optimize the selection of predictive variables by running cyclic coordinate descent and K-fold cross-validation (K = 10). The predictive variables selected by LASSO regression analysis were then included in multivariate logistic regression analysis to establish a prediction model. A nomogram was also constructed based on the prediction model. Calibration chart, receiver operating characteristic curve and decision curve analysis were adopted for validating the prediction model.
Age, plasma interleukin 6 (IL-6) concentration and plasma aspartate aminotransferase concentration were identified from 57 measured variables as potential factors distinguishing G+ from G- infection by LASSO regression analysis. Inclusion of these three variables in a multivariate logistic regression model identified age and IL-6 as significant predictors. In receiver operating characteristic analysis, age and IL-6 yielded an area under the curve of 0.761, and distinguished G+ from G- infection with a specificity of 0.756 and a sensitivity of 0.692. Serum IL-6 and IL-10 levels were upregulated by more than ten-fold from baseline in the G- bacteremia group but by less than ten-fold in the G+ bacteremia group. The calibration curve of the model and Hosmer-Lemeshow test indicated good model fit (P > 0.05). When the decision curve analysis curve indicated a risk threshold probability between 0% and 68%, a nomogram could be applied in clinical settings.
A simple prediction model distinguishing G- from G+ bacteremia can be constructed based on reciprocal association with age and IL-6 level.
Through the method of predicting pathogens, we can know that clinical preemptive treatment and relatively accurate use of antibiotics are beneficial to improve clinical outcomes. Through the method of predicting pathogens, we can know that clinical preemptive treatment and relatively accurate use of antibiotics are beneficial to improve clinical outcomes.
The authors would like to thank all participants and severe infection patients in this study. This study was supported by Zunyi Health Commission.
| 1. | Chen AX, Simpson SQ, Pallin DJ. Sepsis Guidelines. N Engl J Med. 2019;380:1369-1371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 2. | Cecconi M, Evans L, Levy M, Rhodes A. Sepsis and septic shock. Lancet. 2018;392:75-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 810] [Cited by in RCA: 1590] [Article Influence: 198.8] [Reference Citation Analysis (0)] |
| 3. | Meyhoff TS, Hjortrup PB, Wetterslev J, Sivapalan P, Laake JH, Cronhjort M, Jakob SM, Cecconi M, Nalos M, Ostermann M, Malbrain M, Pettilä V, Møller MH, Kjær MN, Lange T, Overgaard-Steensen C, Brand BA, Winther-Olesen M, White JO, Quist L, Westergaard B, Jonsson AB, Hjortsø CJS, Meier N, Jensen TS, Engstrøm J, Nebrich L, Andersen-Ranberg NC, Jensen JV, Joseph NA, Poulsen LM, Herløv LS, Sølling CG, Pedersen SK, Knudsen KK, Straarup TS, Vang ML, Bundgaard H, Rasmussen BS, Aagaard SR, Hildebrandt T, Russell L, Bestle MH, Schønemann-Lund M, Brøchner AC, Elvander CF, Hoffmann SKL, Rasmussen ML, Martin YK, Friberg FF, Seter H, Aslam TN, Ådnøy S, Seidel P, Strand K, Johnstad B, Joelsson-Alm E, Christensen J, Ahlstedt C, Pfortmueller CA, Siegemund M, Greco M, Raděj J, Kříž M, Gould DW, Rowan KM, Mouncey PR, Perner A; CLASSIC Trial Group. Restriction of Intravenous Fluid in ICU Patients with Septic Shock. N Engl J Med. 2022;386:2459-2470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 245] [Article Influence: 61.3] [Reference Citation Analysis (0)] |
| 4. | Berger RE, Rivers E, Levy MM. Management of Septic Shock. N Engl J Med. 2017;376:2282-2285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 5. | Venkatesh B, Finfer S, Cohen J, Rajbhandari D, Arabi Y, Bellomo R, Billot L, Correa M, Glass P, Harward M, Joyce C, Li Q, McArthur C, Perner A, Rhodes A, Thompson K, Webb S, Myburgh J; ADRENAL Trial Investigators and the Australian–New Zealand Intensive Care Society Clinical Trials Group. Adjunctive Glucocorticoid Therapy in Patients with Septic Shock. N Engl J Med. 2018;378:797-808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 521] [Cited by in RCA: 707] [Article Influence: 88.4] [Reference Citation Analysis (0)] |
| 6. | Giraldez MD, Carneros D, Garbers C, Rose-John S, Bustos M. New insights into IL-6 family cytokines in metabolism, hepatology and gastroenterology. Nat Rev Gastroenterol Hepatol. 2021;18:787-803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 111] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 7. | Meadows AT. Retinoblastoma: approaching international standards for recommending chemotherapy (Commentary on Chantada et al., page 692). Pediatr Blood Cancer. 2008;50:521-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 8. | Srisangthong P, Wongsa A, Kittiworawitkul P, Wattanathum A. Early IL-6 response in sepsis is correlated with mortality and severity score. Crit Care. 2013;17 Suppl 2:34. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 9. | Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, Hotchkiss RS, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, van der Poll T, Vincent JL, Angus DC. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA. 2016;315:801-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15803] [Cited by in RCA: 18859] [Article Influence: 1885.9] [Reference Citation Analysis (4)] |
| 10. | Seymour CW, Liu VX, Iwashyna TJ, Brunkhorst FM, Rea TD, Scherag A, Rubenfeld G, Kahn JM, Shankar-Hari M, Singer M, Deutschman CS, Escobar GJ, Angus DC. Assessment of Clinical Criteria for Sepsis: For the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA. 2016;315:762-774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3076] [Cited by in RCA: 2667] [Article Influence: 266.7] [Reference Citation Analysis (0)] |
| 11. | Yuan XH, Li YM, Shen YY, Yang J, Jin Y. Clinical and Th1/Th2 immune response features of hospitalized children with human rhinovirus infection. J Med Virol. 2020;92:26-33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 12. | Chen B, Li H, Xia W. Imiquimod regulating Th1 and Th2 cell-related chemokines to inhibit scar hyperplasia. Int Wound J. 2019;16:1281-1288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (1)] |
| 13. | Wang SY, Li TY, Zheng YQ. Effects of cardiopulmonary bypass on plasma pentraxin-3 level and the balance of Th1/Th2 function in children with congenital heart malformations. Zhonghua Yi Xue Za Zhi. 2009;89:1024-1027. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 14. | Bok SH, Seo JH, Bae CS, Kang B, Cho SS, Park DH. Allium hookeri root extract regulates asthmatic changes through immunological modulation of Th1/Th2related factors in an ovalbumininduced asthma mouse model. Mol Med Rep. 2019;20:3215-3223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 15. | Wang Z, Zhuo F, Chu P, Yang X, Zhao G. Germacrone alleviates collagen-induced arthritis via regulating Th1/Th2 balance and NF-κB activation. Biochem Biophys Res Commun. 2019;518:560-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 16. | Wei L, Champman S, Li X, Li S, Chen R, Bo N, Chater A, Horne R. Beliefs about medicines and non-adherence in patients with stroke, diabetes mellitus and rheumatoid arthritis: a cross-sectional study in China. BMJ Open. 2017;7:e017293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 123] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 17. | Mo R, Shi R, Hu Y, Hu F. Nomogram-Based Prediction of the Risk of Diabetic Retinopathy: A Retrospective Study. J Diabetes Res. 2020;2020:7261047. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 18. | Balachandran VP, Gonen M, Smith JJ, DeMatteo RP. Nomograms in oncology: more than meets the eye. Lancet Oncol. 2015;16:e173-e180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1119] [Cited by in RCA: 2584] [Article Influence: 234.9] [Reference Citation Analysis (0)] |
| 19. | Liang G, Chen X, Zha X, Zhang F. A Nomogram to Improve Predictability of Small-Incision Lenticule Extraction Surgery. Med Sci Monit. 2017;23:5168-5175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Infectious diseases
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Mogulkoc R, Turkey; Rodrigues AT, Brazil S-Editor: Yan JP L-Editor: Filipodia P-Editor: Yuan YY