Published online Apr 26, 2023. doi: 10.12998/wjcc.v11.i12.2716
Peer-review started: November 1, 2022
First decision: January 30, 2023
Revised: February 12, 2023
Accepted: March 17, 2023
Article in press: March 17, 2023
Published online: April 26, 2023
Processing time: 171 Days and 22.5 Hours
Early identification of severe/critical coronavirus disease 2019 (COVID-19) is crucial for timely treatment and intervention. Chest computed tomography (CT) score has been shown to be a significant factor in the diagnosis and treatment of pneumonia, however, there is currently a lack of effective early warning systems for severe/critical COVID-19 based on dynamic CT evolution.
To develop a severe/critical COVID-19 prediction model using a combination of imaging scores, clinical features, and biomarker levels.
This study used an improved scoring system to extract and describe the chest CT characteristics of COVID-19 patients. The study also took into consideration the general clinical indicators such as dyspnea, oxygen saturation, alternative lengthening of telomeres (ALT), and androgen suppression treatment (AST), which are commonly associated with severe/critical COVID-19 cases. The study employed lasso regression to evaluate and rank the significance of different disease characteristics.
The results showed that blood oxygen saturation, ALT, IL-6/IL-10, combined score, ground glass opacity score, age, crazy paving mode score, qsofa, AST, and overall lung involvement score were key factors in predicting severe/critical COVID-19 cases. The study established a COVID-19 severe/critical early warning system using various machine learning algorithms, including XGBClassifier, Logistic Regression, MLPClassifier, RandomForestClassifier, and AdaBoost Classifier. The study concluded that the prediction model based on the improved CT score and machine learning algorithms is a feasible method for early detection of severe/cri
The findings of this study suggest that a prediction model based on improved CT scores and machine learning algorithms is effective in detecting the early warning signals of severe/critical COVID-19.
Core Tip: The computed tomography (CT) score is a relatively objective and clinically accessible semiquantitative assessment tool for patients with coronavirus disease 2019 (COVID-19). The CT scores of common, severe, and critically ill patients showed different trends, and there were differences between the groups of patients as the disease progressed. Patients who are recovering from the disease can be monitored via CT at reduced intervals to reduce their radiation exposure and financial burden. The 2 wk CT scores of the patients were important for predicting disease deterioration in hospitalized patients who have an average admission severity rating. The qSOFA score, aspartate aminotransferase, oxygenation, and dyspnea were important for the prediction of severe/critical COVID-19 disease.
- Citation: Li QY, An ZY, Pan ZH, Wang ZZ, Wang YR, Zhang XG, Shen N. Severe/critical COVID-19 early warning system based on machine learning algorithms using novel imaging scores. World J Clin Cases 2023; 11(12): 2716-2728
- URL: https://www.wjgnet.com/2307-8960/full/v11/i12/2716.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i12.2716
Since the outbreak of novel coronavirus pneumonia (COVID-19) in late December 2019, approximately 521 million more patients worldwide have been infected as of late May 2022, and over 6 million patients could have died from COVID-19. Some COVID-19 patients are not critical at the time of initial diagnosis, but their conditions may deteriorate, leading to severe illness. Currently, case reports in China show that most COVID-19 patients have mild disease, but 15% and 5% develop severe disease and become critically ill, respectively. The intensive care unit (ICU) mortality rate for critically ill patients is as high as 50%-60%. Hence, early identification of the warning signs of severe/critical COVID-19 cases and prompt intervention and treatment can help reduce mortality and improve cure rates[1,2]. At present, CT chest examination is an important part of the diagnosis and treatment of COVID-19 pneumonia and has been used to diagnose patients clinically. Its significance in indicating patient deterioration has been confirmed. However, there is still a lack of means to predict the early progression of severe/critical cases based on the dynamic evolution of CT, and how to determine the progression of CT lesions objectively and quantitatively has become an urgent clinical issue that needs to be addressed[3,4]. During the progression of COVID-19, the occurrence of an inflammatory storm plays a crucial role and the alteration of related inflammatory factors can directly damage the pulmonary capillary mucosa, promoting alveolar edema and inactivating surface active proteins, leading to diffuse alveolar damage and ventilation dysfunction. Inflammatory storms are also a major cause of acute respiratory distress syndrome and multiple organ dysfunction syndrome[5,6]. Numerous prognostic studies on COVID-19 have shown that the trends in related cytokines are crucial for early identification and treatment of critical cases[7,8]. Therefore, exploring the trends in biomarkers, such as cytokines, is important for further understanding the mechanisms of disease progression and regression in COVID-19 patients[9].
Identifying patients at the time of initial diagnosis and providing timely and aggressive interventions are currently the main challenges in treating COVID-19. There are several COVID-19 warning and scoring systems[9], such as the MuLBSTA scoring system, a mortality risk prediction model for COVID-19 that integrates four ML algorithms[10], the National Early Warning Score and Rapid Emergency Medicine Score for ER COVID-19 patients, and NEWS+age[11,12]. To optimize patient treatment and recovery using limited healthcare resources, early detection of prognostic biomarkers is crucial in distinguishing patients who may develop severe COVID-19 and assessing their associated mortality risk during a global pandemic. A model that combines multiple variables for early prediction of the prognosis in COVID-19 patients would help allocate healthcare resources effectively and reduce mortality. In this study, we aim to develop a severe/critical COVID-19 prediction model based on imaging scores, patient clinical features, and biomarker levels (Table 1).
| Variable | Classification | Overall (n = 153) | Common (n = 48) | Severe/critical (n = 105) | P value | |
| Basic information and vital signs | Age, median (IQR) | 69 (66, 74) | 69.0 (66.0, 74.0) | 69.0 (66.0, 76.0) | 0.79 | |
| Comorbidity | Sex, n (%) | Male | 78 (50.980) | 19 (39.583) | 59 (56.190) | 0.057 |
| Female | 75 (49.020) | 29 (60.417) | 46 (43.810) | |||
| Heart rate, median (IQR) | 90 (79, 102) | 87.0 (79.0, 98.0) | 91.0 (79.0, 104.0) | 0.329 | ||
| Temperature, median (IQR) | 36.7 (36.3, 37.1) | 36.7 (36.4, 37.0) | 36.7 (36.3, 37.2) | 0.576 | ||
| Body mass index, mean (SD) | 20.290 (9.929) | 20.789 (7.825) | 20.041 (10.819) | 0.715 | ||
| Respiratory rate, median (IQR) | 21 (20, 24) | 20.0 (20.0, 22.0) | 22.0 (20.0, 25.0) | P < 0.001 | ||
| Blood oxygen saturation, median (IQR) | 94 (90, 97) | 98.0 (96.0, 99.0) | 92.0 (85.0, 95.0) | P < 0.001 | ||
| Coronary heart disease, n (%) | No | 126 (82.353) | 38 (79.167) | 88 (83.810) | 0.485 | |
| Yes | 27 (17.647) | 10 (20.833) | 17 (16.190) | |||
| Pulmonary disease, n (%) | No | 124 (81.046) | 37 (77.083) | 87 (82.857) | 0.398 | |
| Yes | 29 (18.954) | 11 (22.917) | 18 (17.143) | |||
| Diabetes, n (%) | No | 99 (64.706) | 30 (62.500) | 69 (65.714) | 0.699 | |
| Yes | 54 (35.294) | 18 (37.500) | 36 (34.286) | |||
| Laboratory tests | Platelet, mean (SD) | 225.125 (86.792) | 215.563 (73.367) | 229.538 (91.997) | 0.359 | |
| Neutrophil, mean (SD) | 5.469 (3.729) | 3.821 (1.999) | 6.230 (4.080) | P < 0.001 | ||
| Lymphocyte, mean (SD) | 0.929 (0.418) | 1.028 (0.373) | 0.883 (0.429) | 0.047 | ||
| Hemoglobin, mean (SD) | 125.151 (17.785) | 117.958 (16.285) | 128.471 (17.464) | P < 0.001 | ||
| Alanine aminotransferase, median (IQR) | 24 (15, 40) | 16.0 (11.0, 24.0) | 27.0 (19.0, 44.0) | P < 0.001 | ||
| Aspartate aminotransferase, median (IQR) | 29 (21, 42) | 21.0 (17.0, 27.0) | 36.0 (26.0, 49.0) | P < 0.001 | ||
| Albumin, median (IQR) | 32.6 (30.1, 35.5) | 35.1 (31.5, 38.2) | 31.9 (29.6, 34.4) | P < 0.001 | ||
| Total bilirubin, median (IQR) | 10.0 (7.4, 14.2) | 9.5 (6.9, 12.5) | 10.7 (8.2, 14.6) | 0.066 | ||
| Direct bilirubin, median (IQR) | 4.4 (3.3, 6.2) | 3.7 (3.1, 5.1) | 4.9 (3.6, 7.0) | 0.005 | ||
| Lactate dehydrogenase, median (IQR) | 294 (239, 398) | 251.0 (224.0, 284.0) | 341.0 (266.0, 464.0) | P < 0.001 | ||
| Urea, median (IQR) | 4.8 (3.6, 6.6) | 4.0 (3.2, 5.0) | 5.2 (4.0, 7.8) | 0.002 | ||
| Creatinine, median (IQR) | 72 (59, 88) | 68.0 (59.0, 80.0) | 75.0 (59.0, 91.0) | 0.115 | ||
| Prothrombin time, median (IQR) | 14.2 (13.6, 14.9) | 14.1 (13.5, 14.5) | 14.3 (13.8, 15.1) | 0.019 | ||
| Activated partial thromboplastin time, median (IQR) | 40.0 (36.0, 44.5) | 38.9 (35.6, 42.9) | 40.6 (36.3, 45.4) | 0.035 | ||
| Fibrinogen, mean (SD) | 5.208 (1.495) | 4.640 (1.236) | 5.468 (1.531) | 0.001 | ||
| D-Dimer, mean (SD) | 3.394 (5.451) | 1.827 (3.304) | 4.125 (6.065) | 0.003 | ||
| C-Response Protein, mean (SD) | 61.199 (66.732) | 24.943 (30.023) | 77.583 (72.082) | P < 0.001 | ||
| calcitoninogen, mean (SD) | 0.483 (3.484) | 0.047 (0.053) | 0.678 (4.177) | 0.305 | ||
| Ferritin, mean (SD) | 956.507 (874.540) | 486.000 (328.994) | 1170.374 (957.550) | P < 0.001 | ||
| Cytokines | IL-1, mean (SD) | 3.639 (5.076) | 2.853 (1.351) | 3.996 (6.019) | 0.213 | |
| IL-2R, mean (SD) | 895.375 (630.955) | 621.844 (321.241) | 1019.707 (694.756) | P < 0.001 | ||
| IL-6, mean (SD) | 48.628 (88.255) | 13.359 (15.597) | 64.691 (101.986) | P < 0.001 | ||
| IL-10, mean (SD) | 6.464 (8.108) | 3.907 (5.734) | 7.626 (8.738) | 0.003 | ||
| IL-8, mean (SD) | 22.083 (22.814) | 13.900 (15.389) | 25.802 (24.600) | 0.004 | ||
| TNF, mean (SD) | 10.424 (8.969) | 9.158 (13.597) | 11.000 (5.649) | 0.256 | ||
| Scores | IL6/IL10, mean (SD) | 9.051 (15.654) | 3.848 (3.570) | 11.417 (18.241) | P < 0.001 | |
| CURB65, median (IQR) | 1 (1, 2) | 1.0 (1.0, 1.0) | 1.0 (1.0, 2.0) | 0.035 | ||
| qSOFA, median (IQR) | 1 (0, 1) | 0.0 (0.0, 1.0) | 1.0 (0.0, 1.0) | P < 0.001 | ||
A retrospective analysis was used to examine the medical records of adult COVID-19 patients who were admitted to the infectious disease wards of three medical centers in Beijing, Wuhan, and Nanchang. The retrospective cohort collected primary clinical data and CT imaging data, as well as collected the patients’ clinical data, including the patients' demographic data, medical history, laboratory tests after admission, inflammatory markers and cytokine levels, and CURB65 scores. All data were collected and obtained from the electronic medical record system, and if the electronic medical record system lacked relevant data records, these records were obtained by communicating with the attending physicians. This study was reviewed by the Ethics Committee of Peking University Third Hospital.
The case diagnostic criteria and clinical typing criteria included the following. All of the deceased patients were confirmed to have COVID-19 based on the pneumonia diagnosis and treatment protocol for novel coronavirus infection (trial version 7) that was issued by the National Health and Wellness Commission. 10 Confirmed cases were required to conform to the clinical manifestations in the diagnosis and treatment protocols, and pharyngeal swabs, sputum, and lower respiratory secretions were tested by real-time fluorescence RT–PCR using the Wuhan Centers for Disease Control and Prevention's 2019-nCoV nucleic acid test. The clinical typing criteria included the following: (1) Light: The clinical symptoms are mild, and no pneumonia manifestations are seen on imaging; (2) Common type: Fever and respiratory symptoms are present, and pneumonia is visible on imaging; (3) Severe disease: Any of the following are present: respiratory distress, a respiratory rate ≥ 30 breaths/min; a resting state oxygen saturation of ≤ 93%; and a PaO2/FiO2 ≤ 300 mmHg (1 mmHg = 0.133 kPa); and (4) Critical type: One of the following conditions is present: Respiratory failure requiring mechanical ventilation; shock, which may combined with other organ failure that requires ICU monitoring and treatment.
The CT images were independently interpreted by two emergency physicians with more than 10 years of experience, and the CT images were interpreted based on the Fleischner Society definition[13]. The training of the artificial intelligence was by respiratory and radiology specialists. If the scores were inconsistent, they were reassessed, and if an agreement could not be reached, the closer score was used to calculate the mean value. A new scoring system was developed based on the previous lung CT severity score 12, which is widely used in patients with interstitial lung disease, and this scoring system was modified by experts from the departments of respiratory medicine, radiology, critical care medicine, and emergency medicine: (1) The partitioning of the lung field was performed as follows. The lung field was divided into upper, middle, and lower parts, and a total of 6 regions were portioned on the left and right sides by the plane of the tracheal bulge and the plane of inferior pulmonary veins; (2) The target lesion types were defined as follows: (a) Ground glass opacity (GGO): Widespread, blurred increased density of the lung parenchyma with visible bronchial and vascular textures; (b) Pulmonary solidity: Uniformly increased density of the lung parenchyma, obscuring the bronchial and vascular shadows within it, with bronchial inflation signs; (c) Paving stone sign: Ground glass opacity combined with lobular septa; and (d) Pavement stone sign: The combination of lobular septum thickening with a ground glass shadow. The other types of lesions were not analyzed because they were relatively rare and were noncharacteristic[6]; and (3) Scoring: The overall extent of the involvement of the lesions, the extent of the ground glass shadows, and the extent of solid lesions in each region were scored separately: (a) 0: Normal lung tissue; (b) 1: The extent of the lesions is < 25%; (c) 2: The extent of lesions is 26%-50%; (d) 3: The extent of the lesions is 51%-75%; and (e) 4: The extent of the lesions is > 75%. The regional scores were accumulated (the total scores for the overall extent of the involvement, ground glass shadows, and solid lesions were 0-24 for each region). The total score of each was 0 to 24 points. The pavement stone sign was scored as 0 or 1 for the presence or absence of this sign, and the 6 area scores were also cumulative (total score 0 to 6)[13].
To understand the changes in the CT scores over time (time series), the median interval time between the recheck lung CTs was used as a cutoff point to segment the disease course time axis and to compare the differences in the lung CT scores among patients with common, severe, and critical disease at different periods within the course of the disease. The mean values of the 2 CT scores were taken within the same time period. To understand the spatial distribution characteristics (spatial sequence) of the intrapulmonary lesions, the lung fields were divided based on the aforementioned methods to compare the differences in the CT scores in the upper, middle and lower lung field regions (Table 2).
| Computed tomography scoring item | Overall (n = 110) | Common (n = 41) | Severe/Critical (n = 69) | P value | |
| Right upper area | Crazy-paving pattern score, median (IQR) | 0.000 (0.000, 0.000) | 0.000 (0.000, 0.000) | 0.000 (0.000, 0.000) | 0.1 |
| Consolidation score, median (IQR) | 0.000 (0.000, 0.000) | 0.000 (0.000, 0.000) | 0.000 (0.000, 1.000) | 0.027 | |
| GGO, median (IQR) | 1.000 (1.000, 2.000) | 1.000 (1.000, 1.000) | 2.000 (1.000, 3.000) | 0.003 | |
| Overall lung involment score, median (IQR) | 1.000 (1.000, 3.000) | 1.000 (1.000, 1.000) | 2.000 (1.000, 3.000) | 0.006 | |
| Left upper area | Crazy-paving pattern score, median (IQR) | 0.000 (0.000, 0.000) | 0.000 (0.000, 0.000) | 0.000 (0.000, 0.000) | 0.071 |
| Consolidation score, median (IQR) | 0.000 (0.000, 0.000) | 0.000 (0.000, 0.000) | 0.000 (0.000, 1.000) | 0.019 | |
| GGO, median (IQR) | 1.000 (1.000, 2.000) | 1.000 (0.000, 1.000) | 2.000 (1.000, 2.000) | P < 0.001 | |
| Overall lung involment score, median (IQR) | 1.000 (1.000, 2.000) | 1.000 (1.000, 1.000) | 2.000 (1.000, 3.000) | P < 0.001 | |
| Right medium area | Crazy-paving pattern score, median (IQR) | 0.000 (0.000, 0.000) | 0.000 (0.000, 0.000) | 0.000 (0.000, 1.000) | 0.085 |
| Consolidation score, median (IQR) | 0.000 (0.000, 1.000) | 0.000 (0.000, 0.000) | 0.000 (0.000, 1.000) | 0.007 | |
| GGO, median (IQR) | 2.000 (1.000, 3.000) | 1.000 (1.000, 2.000) | 2.000 (1.000, 3.000) | 0.002 | |
| Overall lung involment score, median (IQR) | 2.000 (1.000, 3.000) | 1.000 (1.000, 2.000) | 2.000 (2.000, 3.000) | P < 0.001 | |
| Left medium area | Crazy-paving pattern score, median (IQR) | 0.000 (0.000, 0.000) | 0.000 (0.000, 0.000) | 0.000 (0.000, 1.000) | 0.279 |
| Consolidation score, median (IQR) | 0.000 (0.000, 1.000) | 0.000 (0.000, 0.000) | 0.000 (0.000, 2.000) | 0.006 | |
| GGO, median (IQR) | 2.000 (1.000, 3.000) | 1.000 (1.000, 1.000) | 2.000 (1.000, 3.000) | 0.001 | |
| Overall lung involment score, median (IQR) | 2.000 (1.000, 3.000) | 1.000 (1.000, 2.000) | 2.000 (1.000, 3.000) | P < 0.001 | |
| Right lower area | Crazy-paving pattern score, median (IQR) | 0.000 (0.000, 1.000) | 0.000 (0.000, 0.000) | 0.000 (0.000, 1.000) | 0.122 |
| Consolidation score, median (IQR) | 0.000 (0.000, 1.000) | 0.000 (0.000, 0.000) | 0.000 (0.000, 2.000) | 0.002 | |
| GGO, median (IQR) | 1.000 (1.000, 3.000) | 1.000 (1.000, 2.000) | 2.000 (1.000, 3.000) | 0.004 | |
| Overall lung involment score, median (IQR) | 2.000 (1.000, 3.000) | 1.000 (1.000, 2.000) | 2.000 (1.000, 3.000) | P < 0.001 | |
| Left lower area | Crazy-paving pattern score, median (IQR) | 0.000 (0.000, 0.000) | 0.000 (0.000, 0.000) | 0.000 (0.000, 0.000) | 0.357 |
| Consolidation score, median (IQR) | 0.000 (0.000, 1.000) | 0.000 (0.000, 0.000) | 0.000 (0.000, 1.000) | 0.007 | |
| GGO, median (IQR) | 1.000 (1.000, 3.000) | 1.000 (1.000, 1.000) | 2.000 (1.000, 3.000) | P < 0.001 | |
| Overall lung involment score, median (IQR) | 2.000 (1.000, 3.000) | 1.000 (1.000, 1.000) | 2.000 (1.000, 3.000) | P < 0.001 |
Cronbach′s alpha index was used to evaluate the reliability of the scores of the two reviewers. The measurement data are expressed as the median and interquartile range, and the count data are expressed as the frequency (percentage). The Mann–Whitney U test was used to evaluate the continuous variables, and the chi-square test or Fisher exact test was used to evaluate the rank variables. The Wilcoxon test or the Friedman test was used to compare two/multiple samples of interest. To evaluate the predictive validity of the CT scores, receiver operating characteristic (ROC) curves were traced, and the area under the curve (AUC) was calculated. The statistical analysis was performed using SPSS 22.0 software. MedCalc 19.1 software was used for the comparison of the AUCs of the ROC curves. All of the tests were two-sided; the significance level was set at P < 0.05.
A cohort of 153 COVID-19 patients was included in the study, with 105 patients in severe or critical condition and 48 with common disease. The median age of the patients was 89 years and 50.98% were male. The results of the chi-square test, Mann–Whitney U test, and t test showed that there was no significant difference between the two groups in terms of basic information and vital signs, such as age, sex, heart rate, blood pressure, maximum body temperature, and body mass index. However, the respiratory rate and oxygenation index were significantly different. No significant difference was found between the two groups in terms of underlying diseases, such as coronary artery disease, pulmonary disease, and diabetes mellitus. Clinical symptoms were not significantly different between the two groups, except for dyspnea. Significant differences were found between the two groups in terms of absolute neutrophil values, hemoglobin, liver function tests, albumin, urea, blood coagulation tests, CRP, ferritin, glucose, and T3. The qSOFA score and CURB-65 score were also significantly different between the groups. Significant differences were also found in the characteristics of lung CT scores for patients with different types of COVID-19. Of the 110 patients who underwent CT scans, 114 (94.6%) had GGOs, 68 (61.9%) had solid lesions, and 43 (39.1%) had paving stone signs. Bilateral involvement was seen in 103 (93.6%) of the patients. The CT scans were performed at intervals of 3 to 25 d, with a median interval of 7 d. Disease progression was segmented by week, with the CT scores of common and severe patients peaking during the 3rd to 4th week, while the CT scores of critically ill patients progressed more rapidly, peaking during the 2nd to 3rd week. During the first week, only the GGO score differed between common and critical patients. From the second week onward, the GGO score, solid lesion score, paving stone sign score, and overall extent of involvement score differed between the patients with different degrees of criticality. The spatial distribution of the lesions showed that the upper lung region involvement extent scores were lower for common and severe patients compared to the middle and lower lung regions, while for critically ill patients, there was no statistically significant difference in the CT scores of the upper, middle, and lower lung regions.
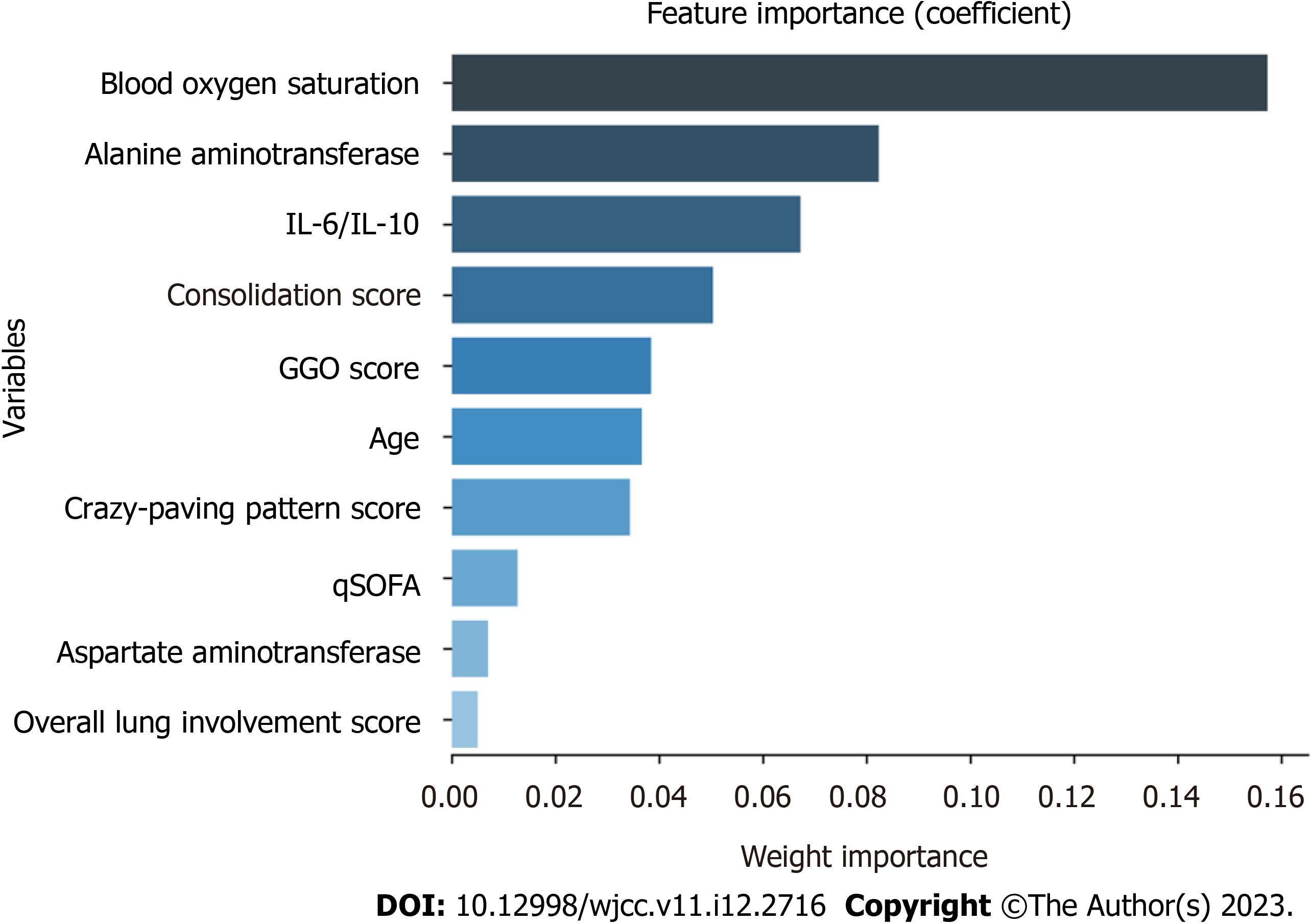
To explore the possible risk factors for severe/critical COVID-19 disease, this study used Lasso regression for the variable importance analysis based on machine learning, and the model parameters were as follows: cv (cross-validation fold): 10; max_iter (number of iterations): 1000; tol (convergence measure): 0.0001; alpha (L1 regularization factor): 0.01. By Lasso regression, the 10 variables with the highest importance (from highest to lowest) were found to be the blood oxygen saturation, ALT, IL-6/IL-10, consolidation score, GGO score, age, crazy-paving pattern score, qSOFA, AST, and overall lung involvement score (Figure 1).
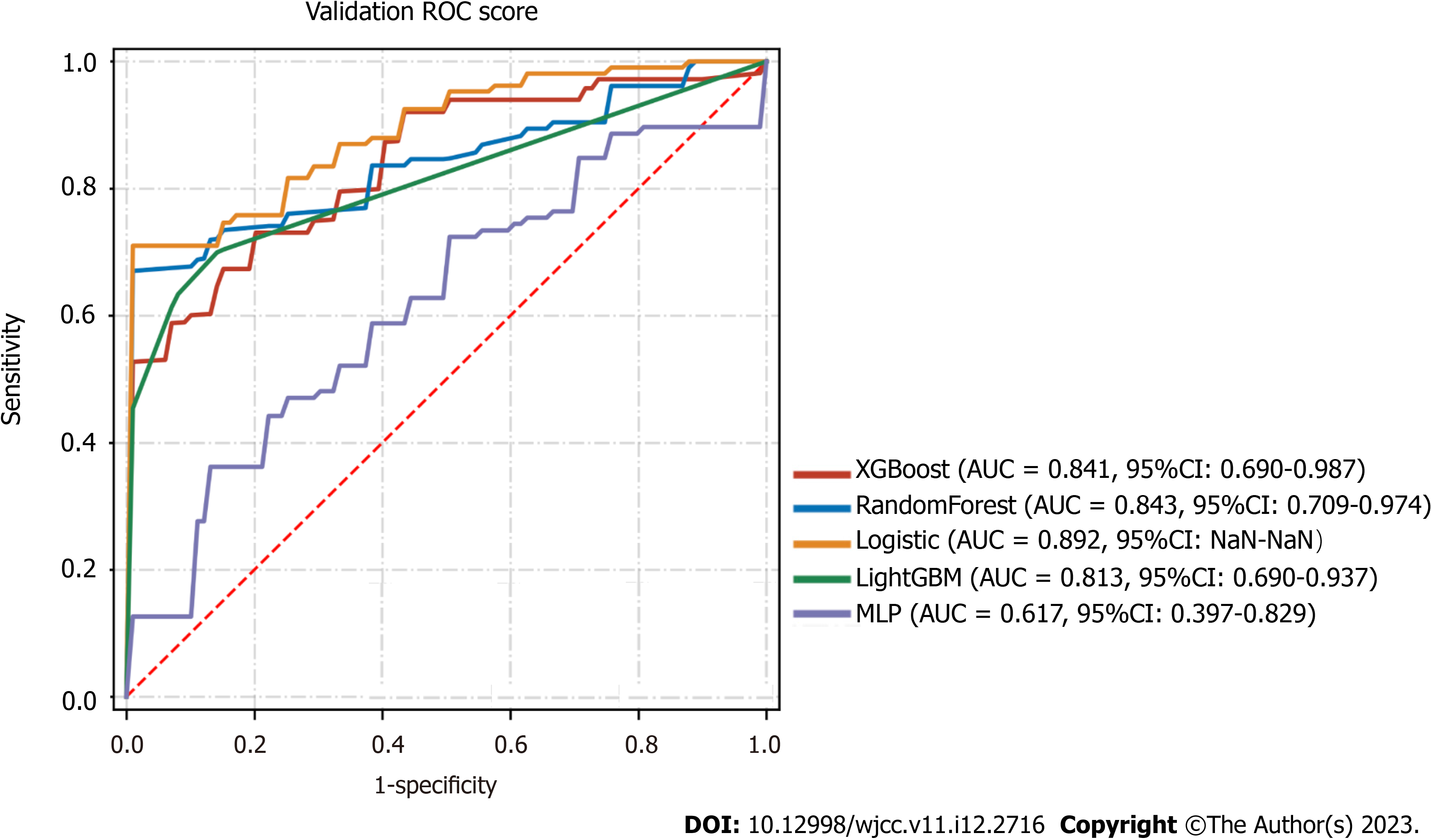
A combination of the results of variable importance analysis and other factors, such as clinical experience, age, IL-6/IL10 Levels, ALT levels, oxygen saturation, qSOFA score, and consolidation score, were selected to be included in the model. A classification task for the data sample was conducted using several machine learning models, including XGBClassifier, RandomForestClassifier, LogisticRegression, LGBMClassifier, and MLPClassifier. A forest plot displays the ROC results of each model for the prediction of severe/critical COVID-19 cases, with the error lines representing the mean and standard deviation of the ROC (Table 3). The means and standard deviations of the ROC were computed by repeating the sampling five times, with each resampled training set accounting for 20% of the overall sample and 80% of the training set. Among the models tested, the best performer in the validation set was the RandomForestClassifier (sorted by AUC), while the best performer in the test set was the LogisticRegression (sorted by AUC). The performance of the algorithms was inconsistent between the training and validation sets. The RandomForestClassifier was more prone to overfitting, while the LogisticRegression appeared to be relatively more stable and was ultimately chosen for the final modeling (Figure 2).
| Model | AUC (95%CI) | Accuracy | Sensitivity | Specificity | PPV | NPV | F1 | |
| Mean | XGBoost | 0.841 (0.690-0.987) | 0.745 | 0.768 | 0.88 | 0.92 | 0.572 | 0.823 |
| SD | XGBoost | 0.044 (0.066-0.018) | 0.052 | 0.122 | 0.16 | 0.098 | 0.169 | 0.033 |
| Mean | RandomForest | 0.843 (0.709-0.974) | 0.731 | 0.698 | 0.975 | 0.988 | 0.554 | 0.809 |
| SD | RandomForest | 0.081 (0.139-0.025) | 0.105 | 0.145 | 0.05 | 0.025 | 0.147 | 0.097 |
| Mean | logistic | 0.892 (NaN-NaN) | 0.828 | 0.88 | 0.848 | 0.932 | 0.666 | 0.896 |
| SD | logistic | 0.069 (NaN-NaN) | 0.079 | 0.119 | 0.189 | 0.087 | 0.177 | 0.066 |
| Mean | LightGBM | 0.813 (0.690-0.937) | 0.324 | 0.671 | 0.956 | NaN | 0.324 | NaN |
| SD | LightGBM | 0.067 (0.098-0.038) | 0.071 | 0.092 | 0.058 | NaN | 0.071 | NaN |
| Mean | MLP | 0.617 (0.397-0.829) | 0.655 | 0.634 | 0.764 | NaN | 0.503 | NaN |
| SD | MLP | 0.219 (0.228-0.212) | 0.162 | 0.326 | 0.172 | NaN | 0.099 | NaN |
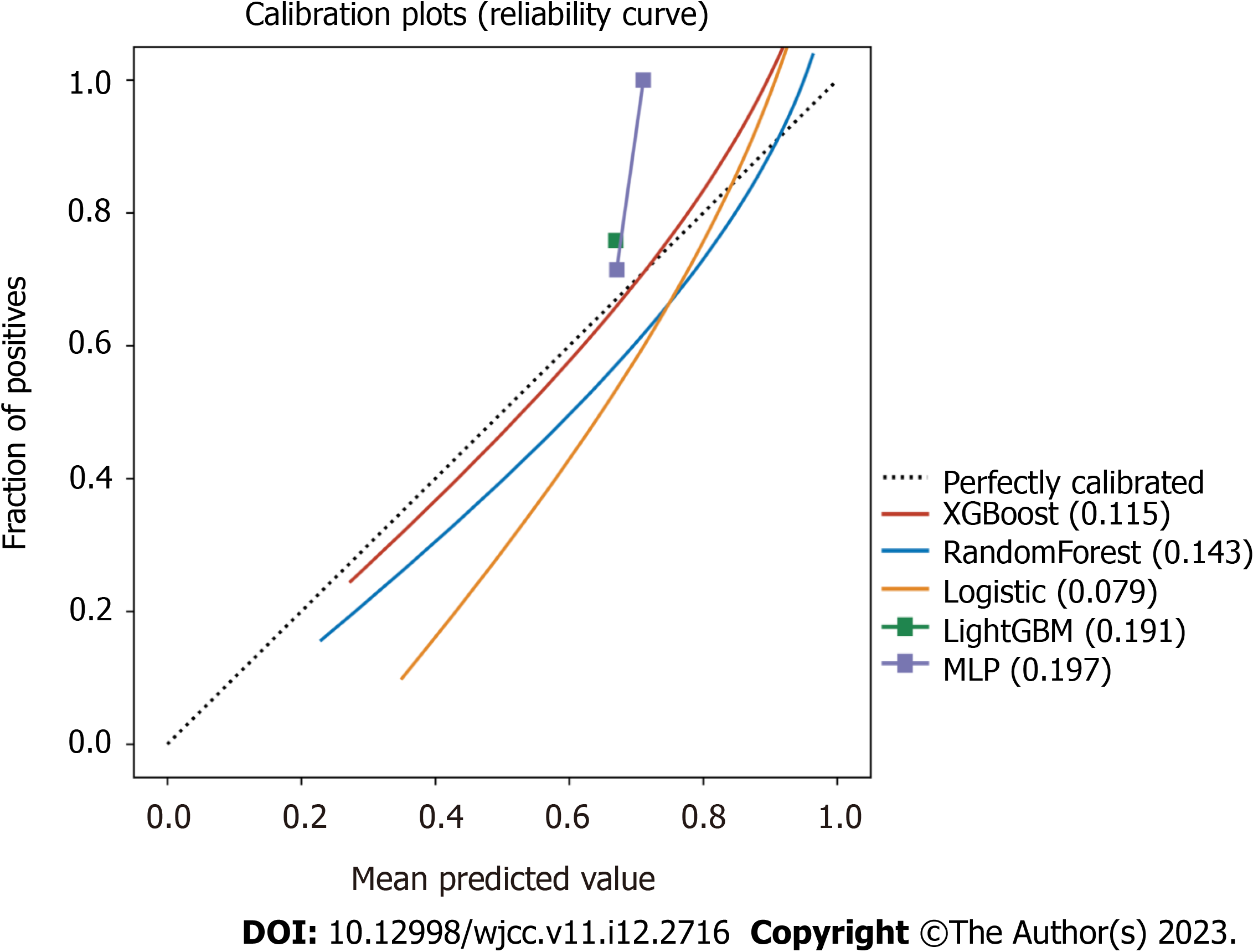
The calibration plots also confirmed good consistency between the “LogisticRegression” algorithm that was predicted and the observed actual risk of severe/critical COVID-19 (Figure 3). A further decision curve analysis was performed, and this analysis showed that the present model provides an excellent net benefit when the clinical decision threshold is between 0% and 100% (Supplementary Figure 1).
Most of the warning scoring systems only use the clinical measurements, such as the level of consciousness and the patient’s vital signs or laboratory testing and do not include the imaging features[1,9,10,12,14]. Since SARS-CoV-2 mainly invades the lungs, a lung CT can often be the best indicator for the severity of the disease and can provide further guidance for the development and prognosis of COVID-19 disease. Several studies have used the CT findings as important indicators to assess the prognosis of COVID-19 patients[15-21]. The prognosis of COVID-19 patients has been predicted by scoring factors, such as the presence of gross glassy shadows on CT images, bronchial and pleural involvement, the presence of a lobular septum, and the lesion morphology, distribution, and size[18,19]. The present study differs from previous studies because the current study used a modified CT severity score that had been previously used in patients with interstitial lung disease, and this score has also been used in patients with COVID-19. This study was the first study that adjusted for complex anatomical localizations by selecting easily recognizable anatomical landmarks for the partitioning and simplification. In this study, the main manifestations of the lung CTs in patients with COVID-19 included GGOs in both lungs, thickening of the lobular septa in the form of the “pavement sign”, solid changes, fibrous cords, etc. Multiple signs could exist simultaneously, and a “white lung” could be seen in severe cases. In contrast, pleural effusion, lymph node enlargement, cavitation, and nodular lesions were less common. Therefore, only the typical lesion types, such as GGOs, solid lesions and paving stone signs, were selected for the modified score, and this simplified the lesion types. Furthermore, the modified imaging score was not time-consuming, and the reliability of the two reviewers' scores for each type of lesion was high. Thus, the improved scoring increased the ability of clinicians to make rapid judgments. In addition, the overall extent of the involvement score downplays the identification of the lesion type, and the upper lung regional involvement score narrows the target area. Both of these scores have a statistically similar predictive validity, as compared to the GGO score, and both of the former scores reduce the complexity and sensitivity of the assessment. Therefore, these novel scores could be a relatively simple alternative to the GGO score. Regarding the predictive value of the lung CT score, this study found that a GGO score of more than 5 on the lung CT at week 2 could be used as a significant indictor for severe COVID-19, even before the development of oxygen reserve depletion, clinical decompensation, and sudden deterioration. In conclusion, clinicians should perform a comprehensive evaluation in COVID-19 patients, which should be used in conjunction with the patient's lung CT score, in order to provide enough respiratory support to the patient as early as possible. The inclusion of the CT score in the clinical severity grading criteria may be considered in the future. In addition to the imaging findings, a variety of clinical features and laboratory indicators have been included in in the models of different studies to develop appropriate predictive models. These factors mainly included age, sex, the patient’s vital signs (temperature, arterial systolic blood pressure, respiration, heart rate), and the patient’s laboratory indicators (neutrophil count, platelet count, CRP, arterial partial pressure of oxygen, blood creatinine value, eGFR, serum albumin value)[16,17,20,22,23]; in addition, the patients’ underlying diseases (hypertension, COPD, etc.) have also been included in some risk prediction models[17,23]. In this study, qSOFA, aspartate aminotransferase, oxygenation, and dyspnea were found to have a good predictive value for the prediction of patients with severe/critical COVID-19.
The qSOFA score is an important diagnostic tool for organ failure. Current studies on the role of inflammatory factors in organ failure have suggested that, compared with COVID-19 patients who were not admitted to the ICU for treatment, critically ill COVID-19 patients in the ICU have higher levels of IP-10, macrophage inflammatory protein 1A (MIP-1A), serum granulocyte colony-stimulating factor (GSCF), GCSF and TNF-α expression levels, suggesting a positive correlation between the inflammatory storm and the disease severity[5,24,25]. It is known that the levels of cytokines, including interleukin, play a crucial role in the progression of COVID-19. During an inflammatory storm, a sustained increase in the expression of proinflammatory factors, produced by the body's immune system, can exacerbate the disease progression, while anti-inflammatory factors can promote pathogen clearance and tissue repair. Monitoring the levels of both pro- and anti-inflammatory cytokines early in the course of COVID-19 is important for determining the patient's condition, treatment plan, and prognosis. Multiple studies have shown that the trends of relevant cytokines are essential for early identification and treatment of critical COVID-19 cases[5,6,24,26]. Above all, this study not only simplifies the complex anatomical location of the lesions that are seen on lung CT but also combines the findings on medical imaging with the patient’s clinical features and laboratory indicators, and the inclusion of all of these factors will more accurately predict the prognosis of COVID-19 patients during the early course of the disease. In this study, there are several limitations to consider. Firstly, it is a retrospective study, which may introduce bias in the results and difficulties in the statistical analysis due to the absence of CT data from the first examination and from critical patients. Secondly, not all of the CT scans included in the analysis were high-resolution scans, which could affect the accuracy of the readings. Finally, the low proportion of deaths in the sample size limited the ability to analyze the predictive value of CT on in-hospital patient outcomes. To address these limitations, future studies could consider expanding the sample size and conducting prospective studies.In the future, it is necessary to evaluate data and perform statistics from multiple medical centers to further evaluate adult COVID-19 confirmed cases, to establish a “COVID-19 clinical-imaging database”, and to systematically analyze the patients’ clinical information, laboratory tests and imaging data of admitted and discharged patients with the help of imaging and histological analysis methods. These methods can also help to more accurately assess the lesion progression, establish a quantitative assessment criteria, determine the early warning signals for severe/critical COVID-19 disease, and establish a predictive model for early warning for the pro
In conclusion, CT scores provide a valuable and objective measure of the progression of COVID-19 in patients. The trends of CT scores differed between common, severe, and critical patients, and monitoring these scores over time can help reduce unnecessary exposure to radiation and cost. The 2-wk CT scores of patients can also be useful in predicting disease deterioration in hospital patients with an average admission severity. Factors such as qSOFA score, aspartate aminotransferase, oxygen saturation, and dyspnea were found to be significant predictors of severe or critical COVID-19.
coronavirus disease 2019 (COVID-19) is a global pandemic that requires early identification and intervention to reduce morbidity and mortality. Chest computed tomography (CT) score has been shown to be a factor in the diagnosis and treatment of COVID-19 pneumonia. However, there is currently a lack of effective early warning systems for severe/critical COVID-19.
To develop a severe/critical COVID-19 prediction model using a combination of imaging scores, clinical features, and biomarker levels.
To identify key factors in predicting severe/critical COVID-19 cases using improved chest CT scores and machine learning algorithms.
The study used an improved scoring system to extract chest CT characteristics of COVID-19 patients, and considered general clinical indicators such as dyspnea, oxygen saturation, alanine aminotransferase, and aspartate aminotransferase. Lasso regression was employed to evaluate the significance of different disease characteristics.
A COVID-19 severe/critical early warning system was established using machine learning algorithms including XGBClassifier, Logistic Regression, MLPClassifier, RandomForestClassifier, and AdaBoost Classifier.
The prediction model based on improved CT scores and machine learning algorithms is effective in detecting early warning signals of severe/critical COVID-19.
The findings suggest that this method is a feasible solution for early detection of severe/critical COVID-19 evolution and may help reduce morbidity and mortality.
We would like to thank the medical team members of fighting against coronavirus disease 2019 of Peking University.
| 1. | Gao Y, Cai GY, Fang W, Li HY, Wang SY, Chen L, Yu Y, Liu D, Xu S, Cui PF, Zeng SQ, Feng XX, Yu RD, Wang Y, Yuan Y, Jiao XF, Chi JH, Liu JH, Li RY, Zheng X, Song CY, Jin N, Gong WJ, Liu XY, Huang L, Tian X, Li L, Xing H, Ma D, Li CR, Ye F, Gao QL. Machine learning based early warning system enables accurate mortality risk prediction for COVID-19. Nat Commun. 2020;11:5033. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 185] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 2. | Wu Z, McGoogan JM. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239-1242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11409] [Cited by in RCA: 11617] [Article Influence: 1936.2] [Reference Citation Analysis (2)] |
| 3. | Yu M, Liu Y, Xu D, Zhang R, Lan L, Xu H. Prediction of the Development of Pulmonary Fibrosis Using Serial Thin-Section CT and Clinical Features in Patients Discharged after Treatment for COVID-19 Pneumonia. Korean J Radiol. 2020;21:746-755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 168] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 4. | Wynants L, Van Calster B, Collins GS, Riley RD, Heinze G, Schuit E, Bonten MMJ, Dahly DL, Damen JAA, Debray TPA, de Jong VMT, De Vos M, Dhiman P, Haller MC, Harhay MO, Henckaerts L, Heus P, Kammer M, Kreuzberger N, Lohmann A, Luijken K, Ma J, Martin GP, McLernon DJ, Andaur Navarro CL, Reitsma JB, Sergeant JC, Shi C, Skoetz N, Smits LJM, Snell KIE, Sperrin M, Spijker R, Steyerberg EW, Takada T, Tzoulaki I, van Kuijk SMJ, van Bussel B, van der Horst ICC, van Royen FS, Verbakel JY, Wallisch C, Wilkinson J, Wolff R, Hooft L, Moons KGM, van Smeden M. Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. BMJ. 2020;369:m1328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1833] [Cited by in RCA: 1800] [Article Influence: 300.0] [Reference Citation Analysis (0)] |
| 5. | Xiong Y, Liu Y, Cao L, Wang D, Guo M, Jiang A, Guo D, Hu W, Yang J, Tang Z, Wu H, Lin Y, Zhang M, Zhang Q, Shi M, Zhou Y, Lan K, Chen Y. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9:761-770. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 877] [Cited by in RCA: 867] [Article Influence: 144.5] [Reference Citation Analysis (0)] |
| 6. | Burgos-Blasco B, Güemes-Villahoz N, Santiago JL, Fernandez-Vigo JI, Espino-Paisán L, Sarriá B, García-Feijoo J, Martinez-de-la-Casa JM. Hypercytokinemia in COVID-19: Tear cytokine profile in hospitalized COVID-19 patients. Exp Eye Res. 2020;200:108253. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 7. | Bhatraju PK, Ghassemieh BJ, Nichols M, Kim R, Jerome KR, Nalla AK, Greninger AL, Pipavath S, Wurfel MM, Evans L, Kritek PA, West TE, Luks A, Gerbino A, Dale CR, Goldman JD, O'Mahony S, Mikacenic C. Covid-19 in Critically Ill Patients in the Seattle Region - Case Series. N Engl J Med. 2020;382:2012-2022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1819] [Cited by in RCA: 1871] [Article Influence: 311.8] [Reference Citation Analysis (0)] |
| 8. | Goyal P, Choi JJ, Pinheiro LC, Schenck EJ, Chen R, Jabri A, Satlin MJ, Campion TR Jr, Nahid M, Ringel JB, Hoffman KL, Alshak MN, Li HA, Wehmeyer GT, Rajan M, Reshetnyak E, Hupert N, Horn EM, Martinez FJ, Gulick RM, Safford MM. Clinical Characteristics of Covid-19 in New York City. N Engl J Med. 2020;382:2372-2374. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1644] [Cited by in RCA: 1629] [Article Influence: 271.5] [Reference Citation Analysis (0)] |
| 9. | Lai X, Liu J, Zhang T, Feng L, Jiang P, Kang L, Liu Q, Gao Y. Clinical, laboratory and imaging predictors for critical illness and mortality in patients with COVID-19: protocol for a systematic review and meta-analysis. BMJ Open. 2020 Dec 24;10(12):e039813.. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 10. | Ma B, Gong J, Yang Y, Yao X, Deng X, Chen X. Applicability of MuLBSTA scoring system as diagnostic and prognostic role in early warning of severe COVID-19. Microb Pathog. 2021;150:104706. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Covino M, Sandroni C, Santoro M, Sabia L, Simeoni B, Bocci MG, Ojetti V, Candelli M, Antonelli M, Gasbarrini A, Franceschi F. Predicting intensive care unit admission and death for COVID-19 patients in the emergency department using early warning scores. Resuscitation. 2020;156:84-91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 89] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 12. | Maves RC, Richard SA, Lindholm DA, Epsi N, Larson DT, Conlon C, Everson K, Lis S, Blair PW, Chi S, Ganesan A, Pollett S, Burgess TH, Agan BK, Colombo RE, Colombo CJ; EPICC COVID-19 Cohort Study Group. Predictive Value of an Age-Based Modification of the National Early Warning System in Hospitalized Patients With COVID-19. Open Forum Infect Dis. 2021;8:ofab421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 13. | Hansell DM, Bankier AA, MacMahon H, McLoud TC, Müller NL, Remy J. Fleischner Society: glossary of terms for thoracic imaging. Radiology. 2008;246:697-722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2471] [Cited by in RCA: 2674] [Article Influence: 148.6] [Reference Citation Analysis (0)] |
| 14. | Abers MS, Delmonte OM, Ricotta EE, Fintzi J, Fink DL, de Jesus AAA, Zarember KA, Alehashemi S, Oikonomou V, Desai JV, Canna SW, Shakoory B, Dobbs K, Imberti L, Sottini A, Quiros-Roldan E, Castelli F, Rossi C, Brugnoni D, Biondi A, Bettini LR, D'Angio' M, Bonfanti P, Castagnoli R, Montagna D, Licari A, Marseglia GL, Gliniewicz EF, Shaw E, Kahle DE, Rastegar AT, Stack M, Myint-Hpu K, Levinson SL, DiNubile MJ, Chertow DW, Burbelo PD, Cohen JI, Calvo KR, Tsang JS; NIAID COVID-19 Consortium, Su HC, Gallin JI, Kuhns DB, Goldbach-Mansky R, Lionakis MS, Notarangelo LD. An immune-based biomarker signature is associated with mortality in COVID-19 patients. JCI Insight. 2021;6:e144455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 265] [Cited by in RCA: 258] [Article Influence: 51.6] [Reference Citation Analysis (0)] |
| 15. | Hu H, Du H, Li J, Wang Y, Wu X, Wang C, Zhang Y, Zhang G, Zhao Y, Kang W, Lian J. Early prediction and identification for severe patients during the pandemic of COVID-19: A severe COVID-19 risk model constructed by multivariate logistic regression analysis. J Glob Health. 2020;10:020510. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 16. | Dai Z, Zeng D, Cui D, Wang D, Feng Y, Shi Y, Zhao L, Xu J, Guo W, Yang Y, Zhao X, Li D, Zheng Y, Wang A, Wu M, Song S, Lu H. Prediction of COVID-19 Patients at High Risk of Progression to Severe Disease. Front Public Health. 2020;8:574915. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 17. | Ma X, Li A, Jiao M, Shi Q, An X, Feng Y, Xing L, Liang H, Chen J, Li H, Li J, Ren Z, Sun R, Cui G, Zhou Y, Cheng M, Jiao P, Wang Y, Xing J, Shen S, Zhang Q, Xu A, Yu Z. Characteristic of 523 COVID-19 in Henan Province and a Death Prediction Model. Front Public Health. 2020;8:475. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (1)] |
| 18. | Zhang B, Wang X, Tian X, Zhao X, Liu B, Wu X, Du Y, Huang G, Zhang Q. Differences and prediction of imaging characteristics of COVID-19 and non-COVID-19 viral pneumonia: A multicenter study. Medicine. 2020;99:e22747. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (1)] |
| 19. | Li D, Zhang Q, Tan Y, Feng X, Yue Y, Bai Y, Li J, Xu Y, Chen S, Xiao SY, Sun M, Li X, Zhu F. Prediction of COVID-19 Severity Using Chest Computed Tomography and Laboratory Measurements: Evaluation Using a Machine Learning Approach. JMIR Med Inform. 2020;8:e21604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 20. | Allenbach Y, Saadoun D, Maalouf G, Vieira M, Hellio A, Boddaert J, Gros H, Salem JE, Resche Rigon M, Menyssa C, Biard L, Benveniste O, Cacoub P; DIMICOVID. Development of a multivariate prediction model of intensive care unit transfer or death: A French prospective cohort study of hospitalized COVID-19 patients. PLoS One. 2020;15:e0240711. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 21. | Che Azemin MZ, Hassan R, Mohd Tamrin MI, Md Ali MA. COVID-19 Deep Learning Prediction Model Using Publicly Available Radiologist-Adjudicated Chest X-Ray Images as Training Data: Preliminary Findings. Int J Biomed Imaging. 2020;2020:8828855. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 22. | Feng C, Wang L, Chen X, Zhai Y, Zhu F, Chen H, Wang Y, Su X, Huang S, Tian L, Zhu W, Sun W, Zhang L, Han Q, Zhang J, Pan F, Chen L, Zhu Z, Xiao H, Liu Y, Liu G, Chen W, Li T. A novel artificial intelligence-assisted triage tool to aid in the diagnosis of suspected COVID-19 pneumonia cases in fever clinics. Ann Transl Med. 2021;9:201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 23. | Guan X, Zhang B, Fu M, Li M, Yuan X, Zhu Y, Peng J, Guo H, Lu Y. Clinical and inflammatory features based machine learning model for fatal risk prediction of hospitalized COVID-19 patients: results from a retrospective cohort study. Ann Med. 2021;53:257-266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 92] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 24. | Hue S, Beldi-Ferchiou A, Bendib I, Surenaud M, Fourati S, Frapard T, Rivoal S, Razazi K, Carteaux G, Delfau-Larue MH, Mekontso-Dessap A, Audureau E, de Prost N. Uncontrolled Innate and Impaired Adaptive Immune Responses in Patients with COVID-19 Acute Respiratory Distress Syndrome. Am J Respir Crit Care Med. 2020;202:1509-1519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 162] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 25. | Chen Y, Wang J, Liu C, Su L, Zhang D, Fan J, Yang Y, Xiao M, Xie J, Xu Y, Li Y, Zhang S. IP-10 and MCP-1 as biomarkers associated with disease severity of COVID-19. Mol Med. 2020;26:97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 217] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 26. | Tan Y, Tang F. SARS-CoV-2-mediated immune system activation and potential application in immunotherapy. Med Res Rev. 2021;41:1167-1194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 27. | Liang W, Liang H, Ou L, Chen B, Chen A, Li C, Li Y, Guan W, Sang L, Lu J, Xu Y, Chen G, Guo H, Guo J, Chen Z, Zhao Y, Li S, Zhang N, Zhong N, He J; China Medical Treatment Expert Group for COVID-19. Development and Validation of a Clinical Risk Score to Predict the Occurrence of Critical Illness in Hospitalized Patients With COVID-19. JAMA Intern Med. 2020;180:1081-1089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 858] [Cited by in RCA: 1012] [Article Influence: 168.7] [Reference Citation Analysis (0)] |
| 28. | Mei X, Lee HC, Diao KY, Huang M, Lin B, Liu C, Xie Z, Ma Y, Robson PM, Chung M, Bernheim A, Mani V, Calcagno C, Li K, Li S, Shan H, Lv J, Zhao T, Xia J, Long Q, Steinberger S, Jacobi A, Deyer T, Luksza M, Liu F, Little BP, Fayad ZA, Yang Y. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat Med. 2020;26:1224-1228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 737] [Cited by in RCA: 572] [Article Influence: 95.3] [Reference Citation Analysis (0)] |
| 29. | Bai HX, Wang R, Xiong Z, Hsieh B, Chang K, Halsey K, Tran TML, Choi JW, Wang DC, Shi LB, Mei J, Jiang XL, Pan I, Zeng QH, Hu PF, Li YH, Fu FX, Huang RY, Sebro R, Yu QZ, Atalay MK, Liao WH. Artificial Intelligence Augmentation of Radiologist Performance in Distinguishing COVID-19 from Pneumonia of Other Origin at Chest CT. Radiology. 2020;296:E156-E165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 260] [Cited by in RCA: 254] [Article Influence: 42.3] [Reference Citation Analysis (1)] |
| 30. | Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q, Cao K, Liu D, Wang G, Xu Q, Fang X, Zhang S, Xia J. Using Artificial Intelligence to Detect COVID-19 and Community-acquired Pneumonia Based on Pulmonary CT: Evaluation of the Diagnostic Accuracy. Radiology. 2020;296:E65-E71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1260] [Cited by in RCA: 945] [Article Influence: 157.5] [Reference Citation Analysis (13)] |
| 31. | Bai HX, Hsieh B, Xiong Z, Halsey K, Choi JW, Tran TML, Pan I, Shi LB, Wang DC, Mei J, Jiang XL, Zeng QH, Egglin TK, Hu PF, Agarwal S, Xie FF, Li S, Healey T, Atalay MK, Liao WH. Performance of Radiologists in Differentiating COVID-19 from Non-COVID-19 Viral Pneumonia at Chest CT. Radiology. 2020;296:E46-E54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 726] [Cited by in RCA: 729] [Article Influence: 121.5] [Reference Citation Analysis (1)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Respiratory system
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gica N, Romania; Shahria MT, United States S-Editor: Xing YX L-Editor: A P-Editor: Xing YX