Published online Apr 16, 2023. doi: 10.12998/wjcc.v11.i11.2412
Peer-review started: November 12, 2022
First decision: February 14, 2023
Revised: February 22, 2023
Accepted: March 15, 2023
Article in press: March 15, 2023
Published online: April 16, 2023
Processing time: 145 Days and 3 Hours
Dilated cardiomyopathy (DCM) is a genetically heterogeneous cardiac disorder characterized by left ventricular dilation and contractile dysfunction. The substantial genetic heterogeneity evident in patients with DCM contributes to variable disease severity and complicates overall prognosis, which can be very poor.
To identify pathogenic genes in DCM through pedigree analysis.
Our research team identified a patient with DCM in the clinic. Through investigation, we found that the family of this patient has a typical DCM pedigree. High-throughput sequencing technology, next-generation sequencing, was used to sequence the whole exomes of seven samples in the pedigree.
A novel and potentially pathogenic gene mutation-ANK2p.F3067L-was discovered. The mutation was completely consistent with the clinical information for this DCM pedigree. Sanger sequencing was used to further verify the locus of the mutation in pedigree samples. These results were consistent with those of high-throughput sequencing.
ANK2p.F3067L is considered a novel and potentially pathogenic gene mutation in DCM.
Core Tip: Our research team identified a typical dilated cardiomyopathy (DCM) pedigree clinically. High-throughput sequencing technology, namely second-generation sequencing, is used to sequence the entire exon group of seven samples in the pedigree. A new potential pathogenic gene mutation ANK2p.F3067L was found in DCM.
- Citation: Zhang XR, Ren H, Yao F, Liu Y, Song CL. Study of pathogenic genes in a pedigree with familial dilated cardiomyopathy . World J Clin Cases 2023; 11(11): 2412-2422
- URL: https://www.wjgnet.com/2307-8960/full/v11/i11/2412.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i11.2412
Dilated cardiomyopathy (DCM) is a type of genetically heterogeneous cardiomyopathy[1]. It is the primary cause of heart transplantation and the third most common cause of heart failure, malignant arrhythmia, and sudden death[2]. DCM has obvious genetic heterogeneity, with nearly 50% of DCM cases caused by genetic factors that are dominant in pathogenesis[3]. However, research on the pathogenic genes associated with DCM is lacking. With the development of next-generation sequencing technology, it has recently been found that DCM is related to variations in genes encoding sarcomeric, cytoskeletal, nuclear membrane, and desmosomal proteins such as TTN, MYH7, TNNT2, LMNA, and DSP[4,5]. However, only ~40% of familial DCM patients harbor known hereditary changes in pathogenic genes[6], while the etiology of the remaining 60% of familial DCM patients remains unclear. Therefore, the identification of pathogenic genes in DCM through pedigree analysis has become an ardent focus of current research.
We believe that our study makes a significant contribution to the literature and will be of interest to the readership of your journal because we discovered the rs764952487 (ANK2p.F3067L) locus-a novel mutation locus of pathogenic genes-in a DCM family characterized in this study. It would be useful for clinical implications of these results in medicine, especially general medicine.
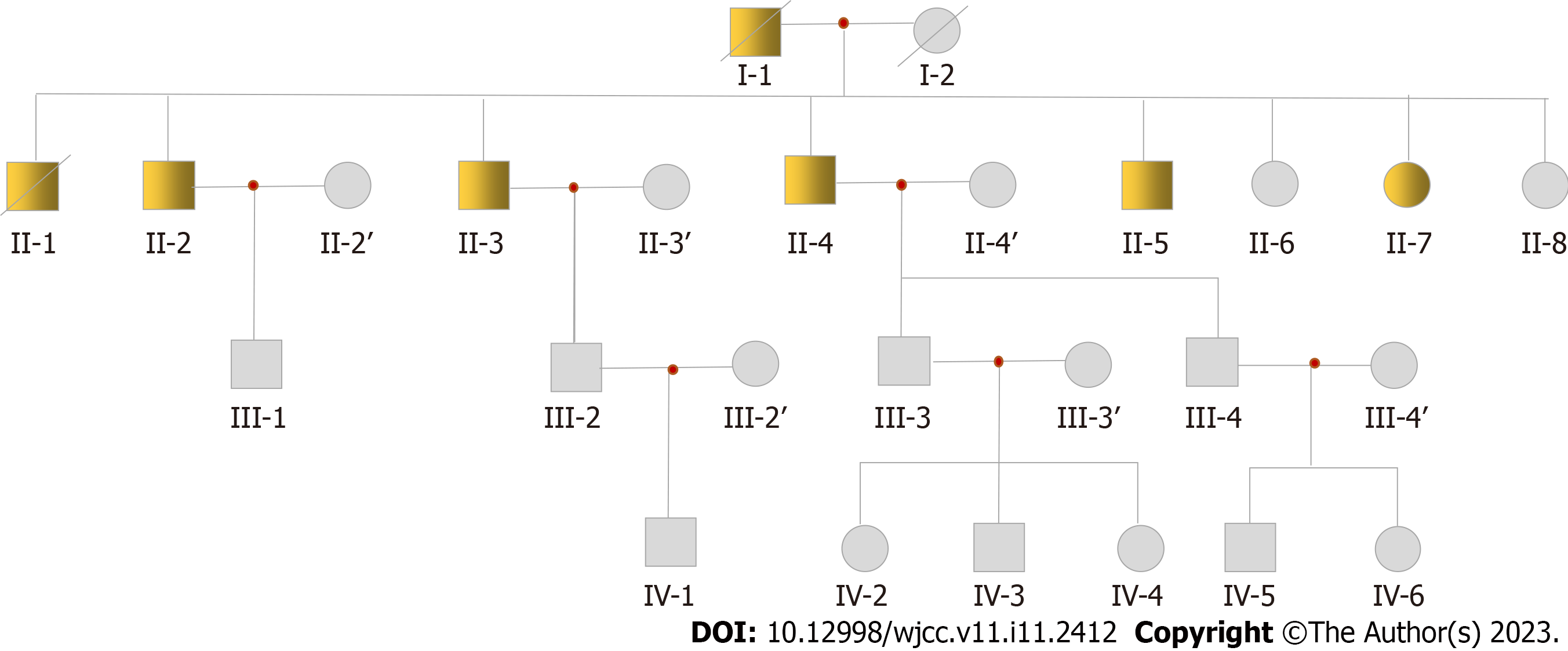
The family with the DCM pedigree resides in Nongan, a county in Jilin Province. The patient’s parents were cousins (both of his parents are deceased-his father having succumbed to DCM). There are five brothers and three sisters in his generation of the family pedigree. Among the eight siblings, six were DCM patients (five males and one female). Based on information from the patient, there are more than 30 living family members in his family. At the time of writing this manuscript, we have obtained 19 samples from this pedigree, including II-2, II-3 and his wife, II-4 and his wife, II-6, II-7, II-8, III-1, III-2 and his wife, III-3, III-4, IV-1, IV-2, IV-3, IV-4, IV-5, and IV-6. According to clinical tests on existing samples, the key clinical information confirmed that these patients are afflicted with DCM. The pedigree chart is presented in Figure 1, from which it may be inferred that DCM in the patient’s pedigree may be autosomal dominant.
According to their chief complaints, clinical medical records, and electrocardiogram and echocardiogram results, seven patients were confirmed to have DCM in the pedigree. Among these seven patients, two (I-1, II-1) died of DCM. Four samples of surviving patients were collected (II-2, II-3, II-4, and II-7; sample II-5 was not collected). Although II-2 had no obvious clinical symptoms, echocardiography showed enlargement of the left ventricle. The proband II-3 was first diagnosed at the age of 47, presenting with chest tightness and shortness of breath when fatigued. At the age of 57, mild activity after a common viral illness lead to chest tightness, shortness of breath, cough, and phlegm. These symptoms prompted him to visit a doctor for the first time. Echocardiography revealed a left ventricular end-diastolic diameter of 67 mm and left ventricular ejection fraction (LVEF) of 23%. The results indicated that his heart was significantly enlarged and his LVEF reduced. Coronary angiography was performed, ruling out cardiac enlargement caused by coronary heart disease. The clinical diagnosis was DCM. In the past 5 years, the proband was admitted to the hospital more than 10 times due to heart failure. He was equipped with an implantable cardioverter defibrillator pacemaker at the age of 60. In addition, II-4 and II-7 were also diagnosed with DCM. They also underwent coronary angiography, again ruling out cardiac enlargement caused by coronary heart disease. The clinical characteristics of the patients with DCM in the pedigree are shown in Table 1.
| Pedigree number | Age | Gender | Age of first episode | LVDD (mm) | EF | Arrythmia |
| I-1 | 60 (death) | Male | - | - | - | - |
| II-1 | 63 (death) | Male | - | - | - | - |
| II-2 | 67 | Male | - | 57 | 74 | Tachyarrhythmia |
| II-3 | 62 | Male | 47 | 67 | 23 | Occasional premature beat |
| II-4 | 60 | Male | 53 | 72 | 46 | Occasional premature beat |
| II-7 | 64 | Female | 58 | 60 | 45 | Normal |
The pedigree consisted of 26 family members (13 men and 13 women; Figure 1). They were diagnosed according to the World Health Organization 1995 diagnostic criteria (left ventricle end-diastolic diameter > 2.7 cm/m2; fractional shortening < 25%).
Seven subjects were included in the linkage study. Genomic DNA was extracted from peripheral blood samples. Linkage analysis of the peripheral blood of seven family members was performed.
Five micrograms of DNA from each of the two affected male individuals was used to construct exome libraries using the Agilent SureSelect exome capture system (Agilent Technologies, Santa Clara, CA, United States). The libraries were sequenced on the Illumina Solexa GAIIx platform following the manufacturer’s instructions (Illumina, San Diego, CA, United States). Raw image files were processed using the Illumina pipeline (version 1.3.4) for base calling and generating reads. Reads were aligned to the human reference genome (University of California Santa Cruz, UCSC hg19; Santa Cruz, CA, United States) using Burrows-Wheeler aligner (BWA, version 0.7.5a-r405) software[7]. Single nucleotide polymorphisms (SNPs) and indels (insertions and deletions) were identified using the genome analysis toolkit (GATK, version 4.1.4.1)[8]. SNPs from regions with read depth > 4 and quality scores > 20 (Q20 scores) were reserved for subsequent analysis. SNPs and indels were annotated using ANNOVAR (version 2017-07-17)[9]. Based on the SNP database (dbSNP) and 1000 genomes annotation with PolyPhen prediction, any non-synonymous variants not assigned a ‘benign’ prediction were considered damaging.
Whole-exome sequencing and analysis: To determine the pathogenic mutations of the DCM pedigree, we sequenced the whole exomes of seven samples (II-3, II-4 and his wife, II-7, II-8, III-3, and IV-2) and evaluated the data quality using FastQC (version 0.11.7)[10]. In addition, we removed adapter sequences and filtered low-quality sequences using Trimmomatic (version 0.39)[11]. The results of the filtered exome data are shown in Table 2. Then we aligned the paired-end sequences based on the human reference genome hg38 using BWA (version 0.7.5a-r405)[7]. The results for the comparative analysis of full exon data are presented in Table 3. To evaluate these results, we performed statistical analysis on the coverage of all exon regions. The average exon coverage for each sample was 106×–133× and approximately 95% of the regions reached 20× coverage. The read depth attained during sequencing was deemed sufficient for downstream data processing, with results listed in Table 4.
| Sample | Total fragment | Both surviving read | Both surviving percent (%) | Forward surviving read | Forward surviving percent (%) | Reverse surviving read | Reverse surviving percent (%) |
| NKHS180096608-1A | 46819669 | 44206494 | 94.42 | 1744245 | 3.73 | 479022 | 1.02 |
| NKHS180096609-1A | 49533235 | 46949404 | 94.78 | 1678343 | 3.39 | 520287 | 1.05 |
| NKHS180096610-1A | 48923103 | 46768485 | 95.60 | 1261086 | 2.58 | 556552 | 1.14 |
| BDYE190000049-1A-A15-D709 | 40876211 | 38862445 | 95.07 | 1166146 | 2.85 | 500985 | 1.23 |
| BDYE190000049-1A-A16-D712 | 41083445 | 37681342 | 91.72 | 2509536 | 6.11 | 328352 | 0.80 |
| BDYE190000049-1A-A18-N710 | 48628579 | 45254586 | 93.06 | 2318870 | 4.77 | 604022 | 1.24 |
| BDYE190000049-1A-A94-N709 | 40940470 | 38630007 | 94.36 | 1406632 | 3.44 | 533856 | 1.30 |
| Sample | Total read | Map read | Map percent (%) | Pair read | Pair percent (%) | Unmap read | Unmap percent (%) | Error rate | Insert size |
| NKHS180096608-1A | 88412988 | 88409570 | 100.00 | 88406934 | 99.99 | 3418 | 0.00 | 1.423552e-03 | 240.0 |
| NKHS180096609-1A | 93898808 | 93894502 | 100.00 | 93891688 | 99.99 | 4306 | 0.00 | 1.443503e-03 | 251.1 |
| NKHS180096610-1A | 93536970 | 93533588 | 100.00 | 93530978 | 99.99 | 3382 | 0.00 | 1.420605e-03 | 263.9 |
| BDYE190000049-1A-A15-D709 | 77724890 | 77721381 | 100.00 | 77719324 | 99.99 | 3509 | 0.00 | 1.472474e-03 | 268.5 |
| BDYE190000049-1A-A16-D712 | 75362684 | 75358337 | 99.99 | 75354470 | 99.99 | 4347 | 0.01 | 1.745109e-03 | 242.0 |
| BDYE190000049-1A-A18-N710 | 90509172 | 90505290 | 100.00 | 90502656 | 99.99 | 3882 | 0.00 | 1.417089e-03 | 228.7 |
| BDYE190000049-1A-A94-N709 | 77260014 | 77257377 | 100.00 | 77255286 | 99.99 | 2637 | 0.00 | 1.423070e-03 | 247.2 |
| Sample | Target length | Coverage length | Coverage ratio (%) | 20× coverage length | 20× coverage ratio (%) | Average coverage ratio |
| NKHS180096608-1A | 60448148 | 60132794 | 99.48 | 57685743 | 95.43 | 120.31 |
| NKHS180096609-1A | 60448148 | 60141586 | 99.49 | 57964001 | 95.89 | 125.84 |
| NKHS180096610-1A | 60448148 | 60001829 | 99.26 | 57821317 | 95.65 | 121.60 |
| BDYE190000049-1A-A15-D709 | 60448148 | 60134381 | 99.48 | 57334579 | 94.85 | 106.07 |
| BDYE190000049-1A-A16-D712 | 60448148 | 60119415 | 99.46 | 57181880 | 94.60 | 109.16 |
| BDYE190000049-1A-A18-N710 | 60448148 | 59997575 | 99.25 | 57728734 | 95.50 | 132.73 |
| BDYE190000049-1A-A94-N709 | 60448148 | 59993121 | 99.25 | 57223253 | 94.67 | 110.74 |
Identification of potentially pathogenic gene mutations: To identify potentially pathogenic gene mutations, we first identified sequence mutations using GATK software based on sequence alignment results. A total of 790998 sequence variation sites were obtained from seven samples. Subsequently, the sequence variations were annotated using ANNOVAR (version 2017-07-17)[9]. These annotations refer to a number of public databases, including RefSeq gene, cytoBand, phast ConsElements46way, genomic Super Dups, gwas Catalog, avsnp147, cosmic70, esp6500siv2_all, nci60, exac03, 1000g2015aug_all, 1000g2015aug_afr, 1000g2015aug_amr, 1000g2015 aug_eur, 1000g2015aug_eas, 1000g2015aug_sas, dbSNP, SIFT, Polyphen2, LRT, Mutation Taster, Mutation Assessor, FATHMM, PROVEAN, VEST3, CADD, DANN, fath mm-MKL, Meta SVM, Meta LR, integrated fit Cons, integrated confidence, GERP++_RS, phyloP7way_vertebrat, phyloP20way_mammalian, phastCons7way_vertebrate, phastCons20way_mammalian, SiPhy_29way_log Odds, and clinvar _20170130. Next, the sequence variations and annotation results were multilayer-filtered, based primarily on the allele frequency of the locus in the population [the locus with mutation allele frequency (MAF) ≥ 0.05 was filtered according to the MAF value of the site in sequence variation databases, such as esp6500siv2_all, ExAC ALL, ExAC_EAS, ExAC_SAS, 1000g2015aug_all, 1000g2015aug_eas, and 1000g2015aug_sas], locus annotation information, locus harmfulness prediction, clinical information, comparison of sample genotype, and comparison with the Online Mendelian Inheritance in Man database.
In addition, we identified CNVs using HMZDelFinder (version 2016)[12] and identified variations in chromosome structure using Meerkat. Finally, based on an investigation of the relevant literature and comprehensive analysis of the filtered mutation sites, rs764952487 (ANK2p.F3067L) emerged as a potential new DCM pathogenic gene mutation site.
The frequency of the rs764952487 mutation genotype in the population was less than 0.0001 (1/10000) in each public database. The mutation genotype is located in a conserved region of the genome. Its conservation score in the phastConsElements100way database is 499. It is predicted to be a harmful mutation by LRT, Mutation Taster, PROVEAN, MetaSVM, MetaLR, CAP, fathmm-MKL, and other software. This site is located in the 38th exon of the ankyrin-B gene (ANK2) (reference sequence NM_001148)-an arrhythmia pathogenic gene. The codon changes to c.T9199C and the amino acid changes to p.F3067L (phenylalanine mutates to leucine), characterized as non-synonymous mutations (missense mutations). Information on the mutation sites is shown in Table 5.
| Site | Mutation | Gene | Reference sequence | Gene subregion | Codon | Amino acid |
| Chr4: 113357817 (rs764952487) | T/C | ANK2 | NM_001148 | Exon38 | c.T9199C | p.F3067L |
According to a study on the protein structure of the ANK2 gene, rs764952487 is located on the helix of ZU5-ZU5-UPA-DD tandem, the neighbor of the death domain (DD). Studies have confirmed that the ZU5-ZU5-UPA domain can form a tightly packed structural supermodel, while DD can enter freely. The formation of the ZZU supermodel does not affect the spectral binding of the anchorin. However, it does change the interface mutation of the ZZU domain and further impairs the function of ankyrin-B, which has become the target of many pathogenic mutations.
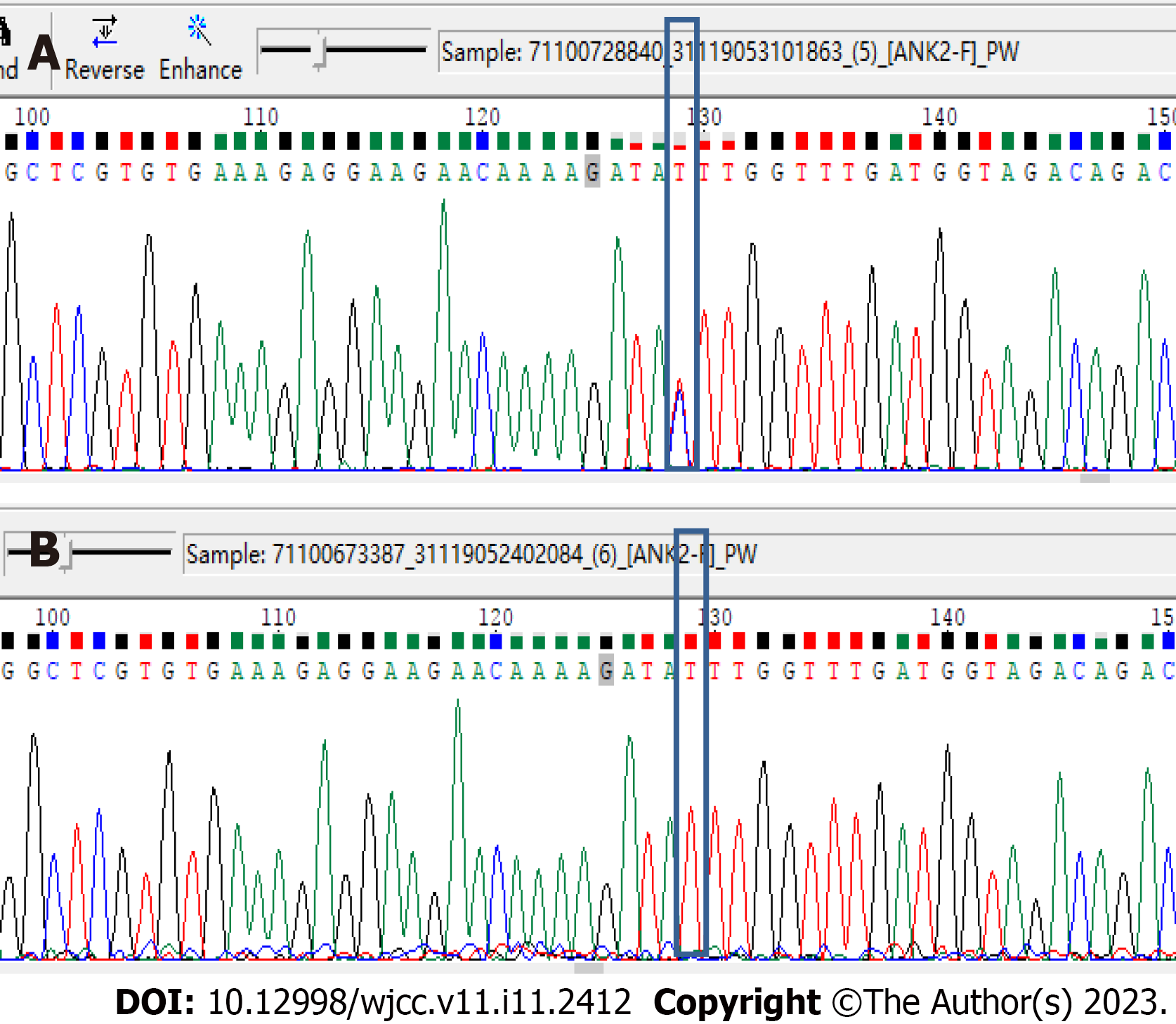
To further determine the reliability of gene mutation identification, we obtained the sequence alignment results for rs764952487 in seven samples. As shown in Figure 2, mutations were detected in samples from all three afflicted patients (II-3, II-4, and II-7), while mutations were not detected in the other four samples. In all seven samples, the sequence coverage of the site location was greater than 100×, ensuring the accuracy of the identification of variant sites.
At the same time, we verified the locus of rs764952487 in all 19 samples, including the above seven samples, using Sanger sequencing. The Sanger sequencing results were consistent with the results for whole-exome sequencing. Moreover, the Sanger results for the rs764952487 Locus in all samples were consistent with the clinical information. This further indicated that the rs764952487 Locus may constitute a novel pathogenic gene mutation in DCM. The clinical information and mutation site genotype of the rs764952487 Locus in all samples are shown in Table 4. The Sanger sequencing results for some of the samples are shown in Figure 3. As shown in Figure 3A, the rs764952487 Locus for sample II-3 is a heterozygous mutant. As shown in Figure 3B, the rs764952487 Locus for sample III--3 is wild-type.
Table 6 shows that among the samples collected in the pedigree so far, all DCM patients carry the ANK2p.F3067L locus. No abnormalities were found in family members who did not carry ANK2p.F3067L. Younger participants carrying ANK2p.F3067L had not yet reported clinical symptoms or received abnormal ultrasound results. In summary, ANK2p.F3067L-a novel and potentially pathogenic mutation, was discovered by genetic investigation of this DCM pedigree. Considering the influence of ANK mutations on prolonged QT intervals, all DCM patients in the pedigree were reviewed. The mutation did not cause arrhythmia in patient II-3.
| Pedigree number | Gender | Age | DCM | ANK2 (T>C) |
| II-2 | Male | 67 | Yes | T/C |
| II-3 | Male | 62 | Yes | T/C |
| II-4 | Male | 60 | Yes | T/C |
| II-6 | Female | 71 | No | T/T |
| II-7 | Female | 65 | Yes | T/C |
| II-8 | Female | 56 | No | T/T |
| II-3’ | Female | 56 | No | T/T |
| II-4’ | Female | 55 | No | T/T |
| III-1 | Male | 40 | Unknow | T/T |
| III-2 | Male | 41 | Unknow | T/C |
| III-3 | Male | 38 | Unknow | T/T |
| III-4 | Male | 32 | Unknow | T/C |
| III-3’ | Female | 37 | No | T/T |
| IV-3 | Female | 15 | Unknow | T/T |
| IV-4 | Female | 13 | Unknow | T/T |
| IV-1 | Male | 11 | Unknow | T/T |
| IV-2 | Male | 11 | Unknow | T/T |
| IV-5 | Male | 6 | Unknow | T/C |
| IV-6 | Female | 2 | Unknow | T/T |
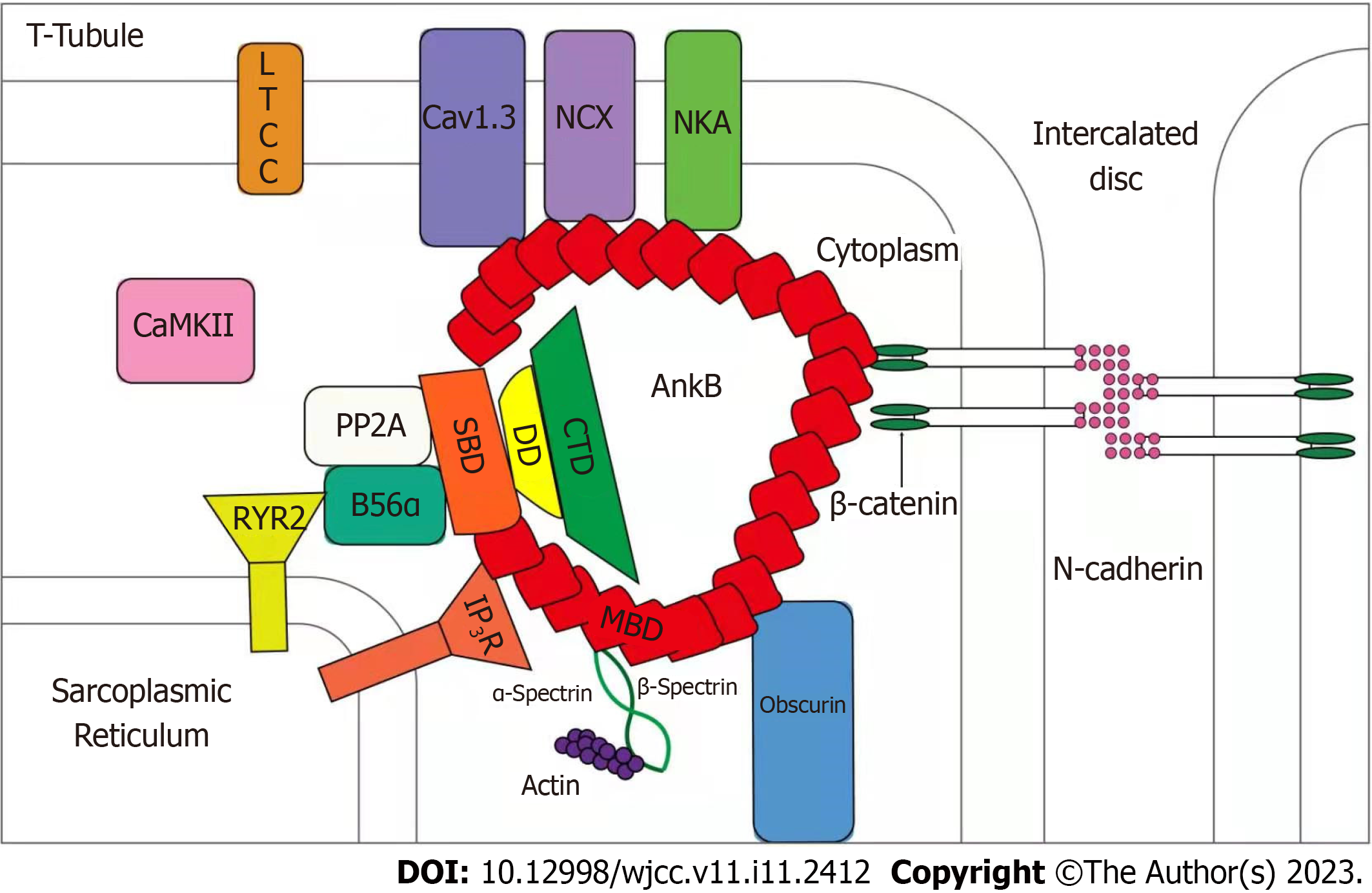
Ankyrin-B is a member of the anchor protein family. As a multifunctional membrane adapter, ankyrin-B is an important ion channel protein auxiliary. It is widely expressed in human tissues and highly expressed in cardiomyocytes, playing an important role in a variety of cardiac physiological functions. Ankyrin-B plays a vital role in the localization of ion transporters, ion channel localization, calcium homeostasis regulation, and membrane stability of cardiomyocytes (Figure 4). As shown in Figure 1, Na+/K+-ATPase, Na+/Ca2+ exchanger (NCX1), and inositol 1,4,5-trisphosphate receptor in the sarcoplasmic reticulum and transverse tubules of cardiomyocytes, maintain normal contractile function and signal transduction function in cardiomyocytes[13]. The targeting and post-translational stability of NCX1 in cardiomyocytes depends on the expression of ankyrin-B, while elevated expression of NCX1 is directly related to myocardial contractile function[14]. The elevated expression of NCX1 leads to an imbalance in intracellular calcium homeostasis and activates calpains. The activation of calpains partially degrades contractile proteins and inhibits myocardial systolic function, resulting in cardiac enlargement and eventually leading to DCM[15,16]. Therefore, the expression of ankyrin-B is important for maintaining the normal contractile function of cardiomyocytes.
ANK2 is a gene coding for ankyrin-B. Its size is approximately 560 kb, consisting of 53 exons on human chromosome 4. In the present study, we found that ANK2 is a common pathogenic gene in arrhythmia syndrome. Its genetic mutation can lead to a broad-spectrum arrhythmia phenotype called "anchor protein-B syndrome".
Although arrhythmia syndrome and hereditary cardiomyopathy are considered to be different genetic diseases, there is a significant phenotypic overlap between them. Clinically, cardiomyopathy is often associated with arrhythmias and abnormal cardiac conduction. Conversely, gene mutations associated with arrhythmia syndrome are also linked to morphological and structural abnormalities in certain types of cardiomyopathy[17-19]. It has been confirmed that common arrhythmia genes such as PLN, SCN5A, KCNQ1, KCNH2, and KCNE2 are associated with hereditary cardiomyopathy[20]. Recent studies have found that there are multiple co-pathogenic genes in arrhythmia syndrome and DCM (such as ABCC9 and SCN5A)[18,21]. A number of researchers have found that variations in the SCN5A gene can cause calcium homeostasis imbalance, resulting in myocardial injury and cardiac enlargement. This eventually leads to DCM[19-23]. In vitro studies have shown that most ANK2 gene mutations can cause abnormal expression and distribution of ankyrin-B and its binding proteins, resulting in an imbalance of calcium homeostasis in cardiomyocytes[24,25].
In recent years, an increasing number of researchers have found that ANK2 gene mutations not only cause arrhythmia syndrome but are also related to the occurrence of cardiomyopathy. As early as 2003, Mohler et al[26] found that ANK2+/- myocardial cells exhibited spontaneous contraction capacity decline and intracellular calcium dynamic disorder. In 2015, Lopes et al[20] found that mutations in ANK2 can modify the phenotypic expression of hypertrophic cardiomyopathy. In 2017, Swayne et al[24] reported the case of a severe idiopathic DCM patient (sudden death at the age of 36) and a DCM patient with pulmonary hypertension. Both of these patients carried the ANK2 gene mutation. In the same year, Forleo et al[27] also found ANK2 gene variation in patients with DCM. Although there is a host of ankyrin-B isoforms in the heart, the primary isoform is canonical 220-kD ankyrin-B[28]. Ankyrin-B-188 and ankyrin-B-212 are two ankyrin-B isoforms present in the heart. Ankyrin-B-188 is expressed in human ventricular cardiomyocytes, which regulates NCX expression, whereas ankyrin-B-212 is expressed in cardiomyocytes and skeletal muscle, is localized to the M-line, and exclusively interacts with obscurin[29]. A cardiomyocyte-specific ankyrin-B-knockout mouse was designed (as ankyrin-B-null mice die shortly after birth) that developed a phenotype including dramatic structural abnormalities, biventricular dilation, reduced ejection fraction, cardiac fibrosis, premature death, and exercise-induced death[30].
In summary, our research team discovered the rs764952487 (ANK2p.F3067L) locus-a novel mutation locus of pathogenic genes-in the DCM family characterized in this study. The codon changes to c.T9199C and the amino acid changes to p.F3067L (phenylalanine mutates to leucine). It is a non-synonymous mutation (i.e., missense mutation). The frequency of the rs764952487 mutation genotype in the population of each public database is less than 0.0001 (1/10 000). The mutation genotype is located in a conserved region of the genome. Its conservation score in the phastCons-Elements100way database is 499. It is predicted to be a harmful mutation by LRT, MutationTaster, PROVEAN, MetaSVM, MetaLR, CAP, fathmm-MKL, and other software. Sanger sequencing of the rs764952487 locus in 19 pedigrees was performed by our team to validate the whole-exome sequencing results. The clinical information of this DCM family is consistent with that of the former. Among all samples collected thus far, all DCM patients carried ANK2p.F3067L, while no abnormalities were found in non-ANK2p.F3067L carriers. Young people carrying ANK2p.F3067L did not report clinical symptoms or receive abnormal ultrasound results. This indicates that ANK2p.F3067L is likely a pathogenic gene mutation in DCM and that its influence on the onset of DCM is age-dependent. Our team will continue to monitor the pedigree, especially the young ANK2p.F3067L carriers who have not yet developed DCM. We will further verify the function of ANK2p.F3067L in vitro and in vivo, as a novel gene mutation in this pedigree.
In summary, ANK2p.F3067L is considered a novel and potentially pathogenic gene mutation in DCM.
Dilated cardiomyopathy (DCM) is a type of genetically heterogeneous cardiomyopathy; it is the primary cause of heart transplantation and the third most common cause of heart failure, malignant arrhythmia, and sudden death. Although DCM has obvious genetic heterogeneity, with nearly 50% of DCM cases caused by genetic factors that are dominant in pathogenesis, research on the pathogenic genes associated with DCM is lacking.
With the development of next-generation sequencing technology, it has recently been found that DCM is related to variations in a number of genes encoding sarcomeric, cytoskeletal, nuclear membrane, and desmosomal proteins. However, only ~40% of familial DCM patients harbor known hereditary changes in pathogenic genes, while the etiology of the remaining 60% of familial DCM patients remains unclear. Therefore, the identification of pathogenic genes in DCM through pedigree analysis has become critical in the field of cardiovascular disease.
Our research team identified a typical DCM pedigree clinically. This study aimed to identify pathogenic genes in DCM through pedigree analysis.
Our research team sequenced the whole exomes of seven samples in the pedigree using high-throughput sequencing technology, namely next-generation sequencing, and verified the potential candidate gene mutations in nineteen samples of the pedigree using Sanger sequencing. Then, we identified the sample sequence mutations using bioinformatics methods, and identified the family pathogenic gene mutations according to the family sample clinical information and gene variation annotation information.
A novel and potentially pathogenic gene mutation-ANK2p.F3067L-was discovered. The codon changes to c.T9199C and the amino acid changes to p.F3067L (phenylalanine mutates to leucine). It is a non-synonymous mutation (i.e., missense mutation).
The mutation-ANK2p.F3067L was completely consistent with the clinical information for this DCM pedigree. It is considered a novel and potentially pathogenic gene mutation in DCM.
Our study adds new members to DCM pathogenic genes, and makes a significant contribution to the literature. It would be useful for clinical implications of these results in medicine, especially general medicine.
The authors thank those who contributed to this paper, as well as all participants and the reviewers for their careful reading and valuable.
| 1. | Ramchand J, Wallis M, Macciocca I, Lynch E, Farouque O, Martyn M, Phelan D, Chong B, Lockwood S, Weintraub R, Thompson T, Trainer A, Zentner D, Vohra J, Chetrit M, Hare DL, James P. Prospective Evaluation of the Utility of Whole Exome Sequencing in Dilated Cardiomyopathy. J Am Heart Assoc. 2020;9:e013346. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 2. | Liu X, Yu H, Pei J, Chu J, Pu J, Zhang S. Clinical characteristics and long-term prognosis in patients with chronic heart failure and reduced ejection fraction in China. Heart Lung Circ. 2014;23:818-826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 3. | McNally EM, Golbus JR, Puckelwartz MJ. Genetic mutations and mechanisms in dilated cardiomyopathy. J Clin Invest. 2013;123:19-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 390] [Cited by in RCA: 354] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 4. | Zhao Y, Feng Y, Zhang YM, Ding XX, Song YZ, Zhang AM, Liu L, Zhang H, Ding JH, Xia XS. Targeted next-generation sequencing of candidate genes reveals novel mutations in patients with dilated cardiomyopathy. Int J Mol Med. 2015;36:1479-1486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 5. | Pérez-Serra A, Toro R, Sarquella-Brugada G, de Gonzalo-Calvo D, Cesar S, Carro E, Llorente-Cortes V, Iglesias A, Brugada J, Brugada R, Campuzano O. Genetic basis of dilated cardiomyopathy. Int J Cardiol. 2016;224:461-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 64] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 6. | Section of Precision Cardiovascular Medicine of Chinese Society of Cardiology, Precision Cardiovascular Medicine Branch of China International Exchange; Promotive Association for Medical; Health Care Editorial Board of Chinese Journal of Cardiology. [Guideline for the genetic diagnosis of monogenic cardiovascular diseases]. Zhonghua Xin Xue Guan Bing Za Zhi. 2019;47:175-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 7. | Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589-595. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7598] [Cited by in RCA: 9106] [Article Influence: 569.1] [Reference Citation Analysis (0)] |
| 8. | McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16037] [Cited by in RCA: 20086] [Article Influence: 1255.4] [Reference Citation Analysis (0)] |
| 9. | Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7976] [Cited by in RCA: 11087] [Article Influence: 692.9] [Reference Citation Analysis (0)] |
| 10. | de Sena Brandine G, Smith AD. Falco: high-speed FastQC emulation for quality control of sequencing data. F1000Res. 2019;8:1874. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 143] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 11. | Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114-2120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30322] [Cited by in RCA: 44734] [Article Influence: 3727.8] [Reference Citation Analysis (2)] |
| 12. | Gambin T, Akdemir ZC, Yuan B, Gu S, Chiang T, Carvalho CMB, Shaw C, Jhangiani S, Boone PM, Eldomery MK, Karaca E, Bayram Y, Stray-Pedersen A, Muzny D, Charng WL, Bahrambeigi V, Belmont JW, Boerwinkle E, Beaudet AL, Gibbs RA, Lupski JR. Homozygous and hemizygous CNV detection from exome sequencing data in a Mendelian disease cohort. Nucleic Acids Res. 2017;45:1633-1648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 13. | Koenig SN, Mohler PJ. The evolving role of ankyrin-B in cardiovascular disease. Heart Rhythm. 2017;14:1884-1889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 14. | Skogestad J, Aronsen JM, Tovsrud N, Wanichawan P, Hougen K, Stokke MK, Carlson CR, Sjaastad I, Sejersted OM, Swift F. Coupling of the Na+/K+-ATPase to Ankyrin B controls Na+/Ca2+ exchanger activity in cardiomyocytes. Cardiovasc Res. 2020;116:78-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 15. | Primessnig U, Bracic T, Levijoki J, Otsomaa L, Pollesello P, Falcke M, Pieske B, Heinzel FR. Long-term effects of Na(+) /Ca(2+) exchanger inhibition with ORM-11035 improves cardiac function and remodelling without lowering blood pressure in a model of heart failure with preserved ejection fraction. Eur J Heart Fail. 2019;21:1543-1552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 16. | Song SS, Tang G, Tang LX, Si LY, Xiong W. NAKα2 inhibits fibrosis formation and protects against cardiomyocyte hypertrophy by suppressing hypertrophy associated molecules and activating LTCC/NCX signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:3021-3033. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 17. | Boczek NJ, Ye D, Jin F, Tester DJ, Huseby A, Bos JM, Johnson AJ, Kanter R, Ackerman MJ. Identification and Functional Characterization of a Novel CACNA1C-Mediated Cardiac Disorder Characterized by Prolonged QT Intervals With Hypertrophic Cardiomyopathy, Congenital Heart Defects, and Sudden Cardiac Death. Circ Arrhythm Electrophysiol. 2015;8:1122-1132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 18. | Bienengraeber M, Olson TM, Selivanov VA, Kathmann EC, O'Cochlain F, Gao F, Karger AB, Ballew JD, Hodgson DM, Zingman LV, Pang YP, Alekseev AE, Terzic A. ABCC9 mutations identified in human dilated cardiomyopathy disrupt catalytic KATP channel gating. Nat Genet. 2004;36:382-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 277] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 19. | Olson TM, Michels VV, Ballew JD, Reyna SP, Karst ML, Herron KJ, Horton SC, Rodeheffer RJ, Anderson JL. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA. 2005;293:447-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 403] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 20. | Lopes LR, Syrris P, Guttmann OP, O'Mahony C, Tang HC, Dalageorgou C, Jenkins S, Hubank M, Monserrat L, McKenna WJ, Plagnol V, Elliott PM. Novel genotype-phenotype associations demonstrated by high-throughput sequencing in patients with hypertrophic cardiomyopathy. Heart. 2015;101:294-301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 138] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 21. | McNair WP, Ku L, Taylor MR, Fain PR, Dao D, Wolfel E, Mestroni L; Familial Cardiomyopathy Registry Research Group. SCN5A mutation associated with dilated cardiomyopathy, conduction disorder, and arrhythmia. Circulation. 2004;110:2163-2167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 337] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 22. | Bezzina CR, Lahrouchi N, Priori SG. Genetics of sudden cardiac death. Circ Res. 2015;116:1919-1936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 221] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 23. | Ge J, Sun A, Paajanen V, Wang S, Su C, Yang Z, Li Y, Jia J, Wang K, Zou Y, Gao L, Fan Z. Molecular and clinical characterization of a novel SCN5A mutation associated with atrioventricular block and dilated cardiomyopathy. Circ Arrhythm Electrophysiol. 2008;1:83-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 62] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 24. | Swayne LA, Murphy NP, Asuri S, Chen L, Xu X, McIntosh S, Wang C, Lancione PJ, Roberts JD, Kerr C, Sanatani S, Sherwin E, Kline CF, Zhang M, Mohler PJ, Arbour LT. Novel Variant in the ANK2 Membrane-Binding Domain Is Associated With Ankyrin-B Syndrome and Structural Heart Disease in a First Nations Population With a High Rate of Long QT Syndrome. Circ Cardiovasc Genet. 2017;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 25. | Choi CSW, Souza IA, Sanchez-Arias JC, Zamponi GW, Arbour LT, Swayne LA. Ankyrin B and Ankyrin B variants differentially modulate intracellular and surface Cav2.1 levels. Mol Brain. 2019;12:75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 26. | Mohler PJ, Schott JJ, Gramolini AO, Dilly KW, Guatimosim S, duBell WH, Song LS, Haurogné K, Kyndt F, Ali ME, Rogers TB, Lederer WJ, Escande D, Le Marec H, Bennett V. Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death. Nature. 2003;421:634-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 723] [Cited by in RCA: 669] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 27. | Forleo C, D'Erchia AM, Sorrentino S, Manzari C, Chiara M, Iacoviello M, Guaricci AI, De Santis D, Musci RL, La Spada A, Marangelli V, Pesole G, Favale S. Targeted next-generation sequencing detects novel gene-phenotype associations and expands the mutational spectrum in cardiomyopathies. PLoS One. 2017;12:e0181842. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (1)] |
| 28. | Sucharski HC, Dudley EK, Keith CBR, El Refaey M, Koenig SN, Mohler PJ. Mechanisms and Alterations of Cardiac Ion Channels Leading to Disease: Role of Ankyrin-B in Cardiac Function. Biomolecules. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 29. | Wu HC, Yamankurt G, Luo J, Subramaniam J, Hashmi SS, Hu H, Cunha SR. Identification and characterization of two ankyrin-B isoforms in mammalian heart. Cardiovasc Res. 2015;107:466-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 30. | Roberts JD, Murphy NP, Hamilton RM, Lubbers ER, James CA, Kline CF, Gollob MH, Krahn AD, Sturm AC, Musa H, El-Refaey M, Koenig S, Aneq MÅ, Hoorntje ET, Graw SL, Davies RW, Rafiq MA, Koopmann TT, Aafaqi S, Fatah M, Chiasson DA, Taylor MR, Simmons SL, Han M, van Opbergen CJ, Wold LE, Sinagra G, Mittal K, Tichnell C, Murray B, Codima A, Nazer B, Nguyen DT, Marcus FI, Sobriera N, Lodder EM, van den Berg MP, Spears DA, Robinson JF, Ursell PC, Green AK, Skanes AC, Tang AS, Gardner MJ, Hegele RA, van Veen TA, Wilde AA, Healey JS, Janssen PM, Mestroni L, van Tintelen JP, Calkins H, Judge DP, Hund TJ, Scheinman MM, Mohler PJ. Ankyrin-B dysfunction predisposes to arrhythmogenic cardiomyopathy and is amenable to therapy. J Clin Invest. 2019;129:3171-3184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cardiac and cardiovascular systems
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gupta P, United States S-Editor: Zhang H L-Editor: A P-Editor: Zhang H