Published online Mar 16, 2022. doi: 10.12998/wjcc.v10.i8.2474
Peer-review started: June 8, 2021
First decision: September 1, 2021
Revised: September 12, 2021
Accepted: January 29, 2022
Article in press: January 29, 2022
Published online: March 16, 2022
Processing time: 275 Days and 23 Hours
Klebsiella pneumoniae (K. pneumoniae) is an infective microorganism of worldwide concern because of its varied manifestations and life-threatening potential. Genetic analyses have revealed that subspecies of K. pneumoniae exhibit higher virulence and mortality. However, infections with Klebsiella subspecies are often misdiagnosed and underestimated in the clinic because of difficulties in distinguishing K. pneumoniae from its subspecies using routine tests. This case study reports the rapid and fatal effects of K. pneumoniae subspecies.
A 52-year-old male patient was febrile and admitted to hospital. Examinations excluded viral and fungal causes along with mycoplasma/chlamydia and parasitic infections. Bacterial cultures revealed blood-borne K. pneumoniae sensitive to carbapenem antibiotics, although corresponding treatment failed to improve the patient’s symptoms. His condition worsened and death occurred within 72 h of symptom onset from sepsis shock. Application of the PMseq-DNA Pro high throughput gene detection assay was implemented with results obtained after death showing a mixed infection of K. pneumoniae and Klebsiella variicola (K. variicola). Clinical evidence suggested that K. variicola rather than K. pneumoniae contributed to the patient’s poor prognosis.
This is the first case report to show patient death from Klebsiella subspecies infection within a short period of time. This case provides a timely reminder of the clinical hazards posed by Klebsiella subspecies and highlights the limitations of classical laboratory methods in guiding anti-infective therapies for complex cases. Moreover, this report serves as reference for physicians diagnosing similar diseases and provides a recommendation to employ early genetic detection to aid patient diagnosis and management.
Core Tip: Klebsiella pneumoniae infection leads to worldwide concerns with its high mortality and varied manifestation. However, it is difficult to distinguish Klebsiella pneumoniae from its subspecies using classic clinical examinations. We here report a case who died with Klebsiella subspecies infection within 72 h. This case was diagnosed by genetic detection rather than classic laboratory methods. This case suggests that we should be alert to the clinical hazards and fatal effect of Klebsiella subspecies, classic method is limited in guiding the anti-infection therapy for complex cases, and early genetic detection should be performed in the diagnosis and management of complex infection.
- Citation: Long DL, Wang YH, Wang JL, Mu SJ, Chen L, Shi XQ, Li JQ. Fatal community-acquired bloodstream infection caused by Klebsiella variicola: A case report. World J Clin Cases 2022; 10(8): 2474-2483
- URL: https://www.wjgnet.com/2307-8960/full/v10/i8/2474.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i8.2474
Klebsiella pneumoniae (K. pneumoniae) infections are known to be associated with high incidence and mortality. This microorganism causes outbreaks of nosocomial infections and even drug resistance, and can lead to infection in the community among health-care patients or people with underlying immunodeficiency[1]. Based on genetic analysis, K. pneumoniae is divided into three phylogroups: K. pneumoniae (KpI), K. quasipneumoniae (KpII), and K. variicola (KpIII)[2]. KpI is the most frequent group encountered in the clinic, followed by KpIII and KpII[1,3]. KpI is usually defined as classic Klebsiella (Ck) or hypervirulent Klebsiella (Hvkp) according to their invasiveness or virulence, while subspecies of Klebsiella (KpII and KpIII) usually present with higher virulence[1]. Currently, approximately 20% of the human isolates assumed to be K. pneumoniae are in fact K. variicola or K. quasipneumoniae[1]. However, since classical laboratory examinations cannot readily distinguish KpI from the KpII and KpIII phylogroups[4], the clinical hazards and importance of KpII and KpIII are often overlooked.
A 52-year-old man presented with unexplained high fever, abdominal pain, and headache for 1 d.
The patient was subsequently admitted to the intensive care unit with diarrhea and confusion.
The patient reported a 5-year history of type 2 diabetes mellitus (T2DM) and 7 years of suffering gout but had no prior medical history related to the current symptoms.
The patient had no particular individual or family history.
Physical assessment revealed a body temperature of 40 ℃ but without other obvious abnormal signs.
Laboratory examinations revealed slightly deteriorated hepatorenal function and clotting function and increases in inflammatory parameters (Tables 1-3). Other laboratory biochemical tests proved negative for signs of viral and mycobacterial infections along with mycoplasma/chlamydia and biomarkers of autoimmune diseases (Table 4). Traditional bacterial culture of the patients’ blood sample showed bacterial infection with K. pneumoniae (Table 5).
| Item | Result (1st) | Reference range |
| TBIL (μmol/L) | 19.4 | 3.6-20.5 |
| TP (μmol/L) | 49.8 | 65-85 |
| ALB (g/L) | 31.4 | 40-55 |
| ALT (U/L) | 156 | 9-50 |
| AST (U/L) | 182 | 15-40 |
| sCrea (μmol/L) | 160 | 57-97 |
| Urea (μmol/L) | 12.40 | 3.1-8 |
| GLU (mmol/L) | 13.89 | 3.9-6.1 |
| K+ (mmol/L) | 3.22 | 3.5-5.5 |
| UA (μmol/L) | 591 | 210-420 |
| Item | Result | Reference range |
| 3P | Negative | Negative |
| D-D (μg/mL) | 60.75↑ | 0-1 |
| FDP (μg/mL) | 128.5↑ | 0-5 |
| PT (s) | 18.3↑ | 9.2-12.2 |
| INR | 1.59↑ | 0.8-1.2 |
| APTT (s) | 67.6↑ | 21.1-36.5 |
| TT (s) | 23.4↑ | 14-21 |
| FBG (g/L) | 22.58 | 1.8-3.5 |
| Item | Result | Reference range |
| WBC (× 109) | 1.2 | 3.5-10 |
| NEUT (× 109) | 0.63 | 1.8-6.3 |
| %NEUT | 52.5 | 40-75 |
| MONO (× 109) | 0.05 | 0.1-0.6 |
| %MONO | 4.2 | 3-10 |
| LYMBP (× 109) | 0.39 | 1.1-3.2 |
| %LYMBP | 32.5 | 20-50 |
| RBC (× 1012) | 1.83 | 3.5-5.5 |
| HGB (g/L) | 52.0 | 114-163 |
| %HCT | 15.6 | 35-50 |
| PLT (× 109) | 10 | 125-350 |
| CRP (mg/L) | 230.33 | 0-5 |
| PCT | > 100 | 0-0.046 |
| Item | Result | Reference |
| RSV-IGM | Negative | Negative |
| ADV-IGM | Negative | Negative |
| IFZA-IGM | Negative | Negative |
| IFZB-IGM | Negative | Negative |
| HPIVs-IGM | Negative | Negative |
| MP-IGM | Negative | Negative |
| CP-IGM | Negative | Negative |
| CBV-IGM | Negative | Negative |
| CAV-IGM | Negative | Negative |
| ECHO-IGM | Negative | Negative |
| LP-IGM | Negative | Negative |
| 2019-nCoV | Negative | Negative |
| EB-DNA (copies/mL) | < 5E + 2 | < 5E + 2 |
| EB-DNA | Negative | Negative |
| CMVDNA DL (copies/mL) | < 5E + 2 | < 5E + 2 |
| CMV DNA DX | Negative | Negative |
| t1 | Test method: Blotting | |
| A-PR3 | Negative | Negative |
| A-MP0 | Negative | Negative |
| A-GBM | Negative | Negative |
| t2 | Test method: Fluorescence | |
| cANCA | Negative | Negative |
| pANCA | Negative | Negative |
| Specimen | Blood | ||
| Equipment | Phoenix100 | ||
| Items | Bacterial culture + antimicrobial susceptibility | ||
| Results | Klebsiella pneumoniae | ||
| Antibiotics | MIC | Result interpretation | Cutoff |
| Cefotaxime | S | S ≤ 1; R ≥ 4 | |
| Cotrimoxazole | ≤ 20 | S | S ≤ 2/38; R ≥ 4/76 |
| Tigecycline | ≤ 0.5 | S | |
| Levofloxacin | ≤ 0.12 | S | S ≤ 0.5; R ≥ 2 |
| Amikacin | ≤ 2 | S | S ≤ 16; R ≥ 64 |
| Imipenem | ≤ 0.25 | S | S ≤ 1; R ≥ 4 |
| Er ertapenem | ≤ 0.12 | S | S ≤ 0.5; R ≥ 2 |
| Cefepime | ≤ 0.12 | S | S ≤ 2; R ≥ 16 |
| Ce foperazone/sulbactam | ≤ 8 | S | S ≤ 16; R≥ 64 |
| Ceftriaxone | ≤ 0.25 | S | S ≤ 1; R ≥ 4 |
| Ceftazidime | ≤ 0.12 | S | S ≤ 4; R ≥ 16 |
| Cefoxitin | ≤ 4 | S | S ≤ 8; R ≥ 32 |
| Cefuroxime axetil | 4 | S | |
| Cefuroxime | 4 | S | S ≤ 4; R ≥ 32 |
| Piperacillin/tatabatam | ≤ 4 | S | S ≤ 16/4; R ≥ 128/4 |
| Amoxicillin/clavulanate | ≤ 2 | S | S ≤ 8/4; R ≥ 32/16 |
| ESBL | Neg | - | |
Based on the patient’s symptoms and laboratory data, sepsis and septic shock were diagnosed according to the diagnostic criteria.
The initial treatment prescribed after the availability of the laboratory results involved a broad-spectrum regimen for bacterial and fungal infections (intravenous meropenem 1 g per 6 h + intravenous caspofungin 70 mg/initial dose).
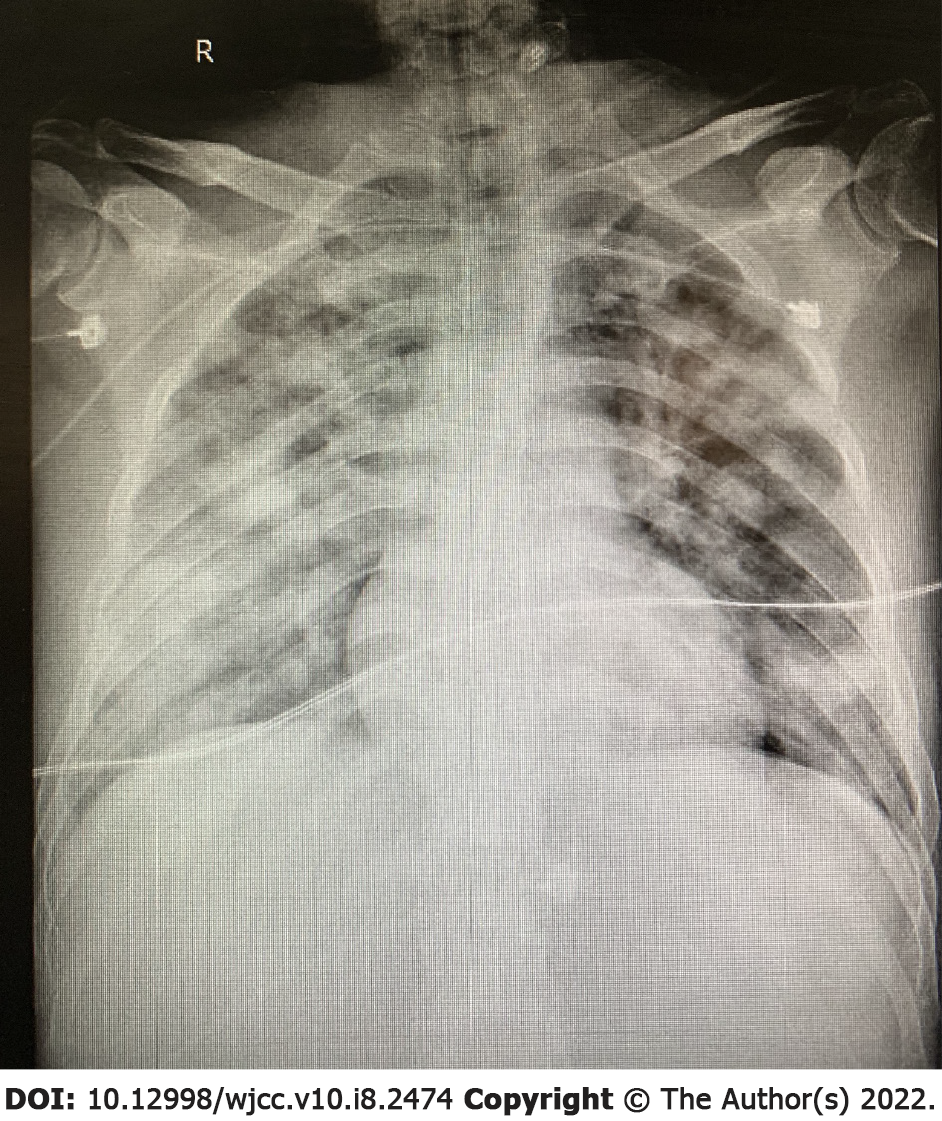
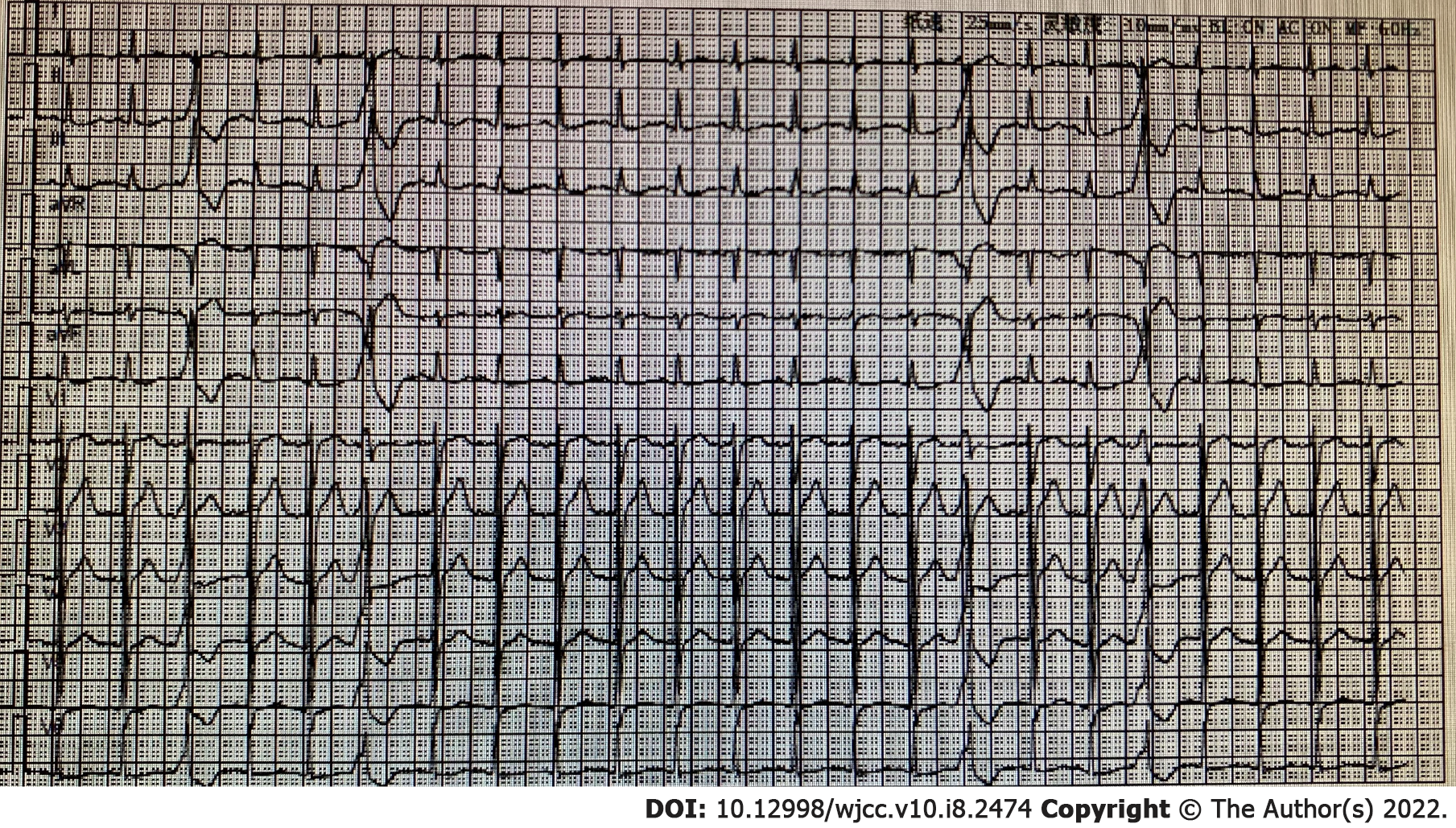
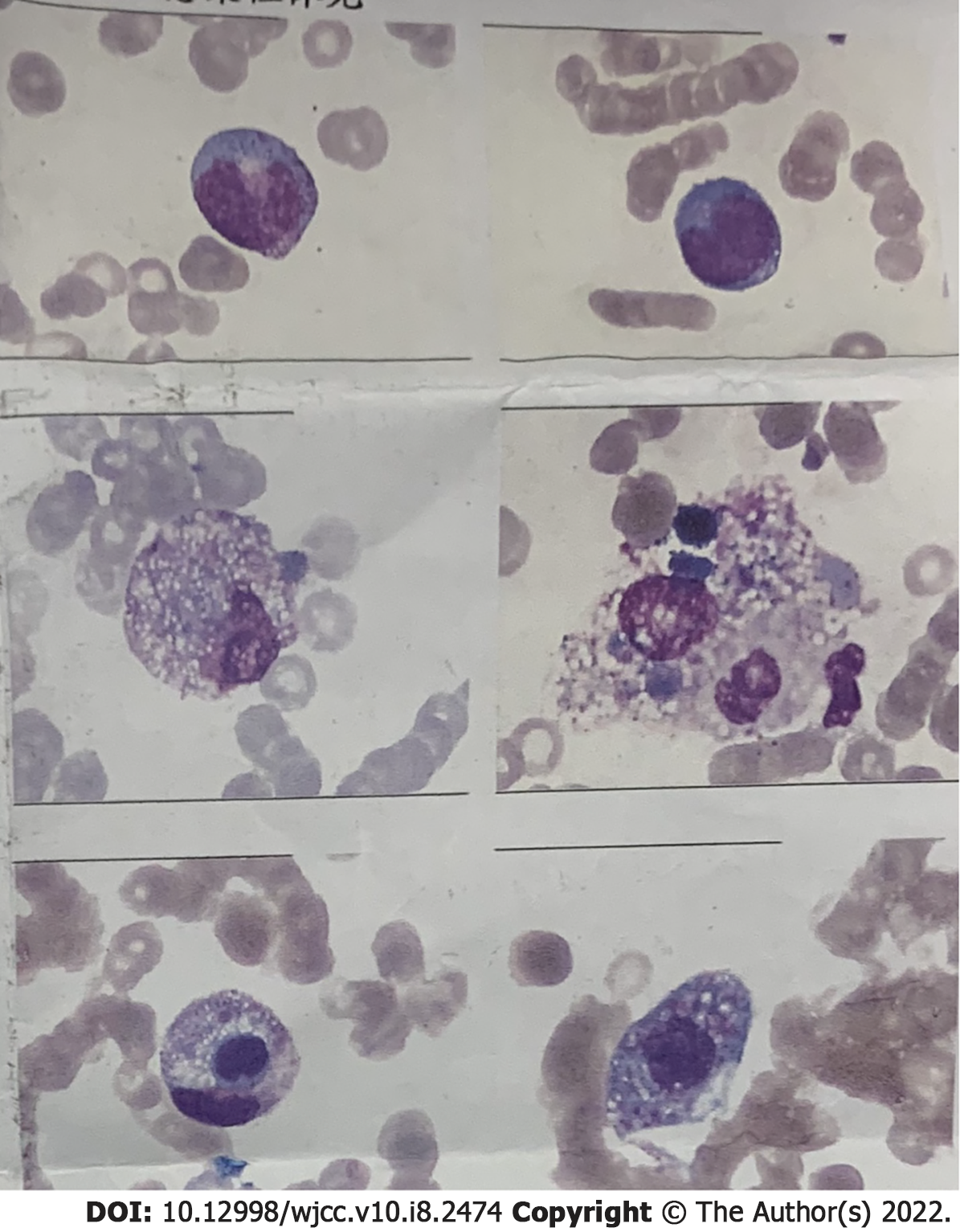
The patient’s condition rapidly deteriorated within hours of admission with decreased blood pressure and reduced oxygen saturation. Life support therapies including mechanical ventilation and vasoactive drugs did not improve the patient’s condition. He suffered cardiac arrest on the second day of admission and was declared clinically dead after several rescue efforts. The PMseq-DNA Pro high throughput gene detection assay was initiated on the second day after admission with the results acquired 2 d later. This analysis revealed infection with K. pneumoniae and K. variicola (Table 6). A bone marrow biopsy supported the findings of severe bacterial infection (Figure 4).
| Type | Genus (number of sequences) | Species (number of sequences) | ||
| G | Klebsiella | 68405 | Klebsiella pneumoniae | 243747 |
| Klebsiella variicola | 543 | |||
Klebsiella is a genus of Gram-negative bacterium within the Enterobacteriaceae family. It usually causes opportunistic nosocomial infections among hospitalized patients or outbreaks of community-acquired infections. Klebsiella mainly colonizes human gut but it has also been isolated from the skin surfaces such as hands and face, and can be isolated from various environmental sources including water, plants, and soil[1,5]. The genus contains several subspecies that manifest varied clinical outcomes, even death, leading to significant concerns about the accurate and timely identification of the Klebsiella subspecies involved together with a better understanding of the patient risk factors involved to reduce mortality risks[6].
Recent research has revealed that diabetes is a significant risk factor for hypervirulent K. pneumoniae infection and for causing serious complications[7-9]. In our patient’s case, a medical history of T2DM could have contributed to an underlying immunodeficiency that was responsible for the fatal systemic infection. Although both K. pneumoniae and K. variicola were detected in the patients’ blood sample, K. variicola may have played a more decisive role in the resulting outcome since treatment to target K. pneumoniae failed to improve the patient’s condition. Moreover, this is consistent with the notion that K. variicola is a frequent cause of bloodstream infections and higher mortality[3,4].
It is difficult to distinguish K. pneumoniae and its subspecies by classic bacterial culture methods. This may lead to misdiagnosis or delayed diagnosis and incorrect treatment[4]. As shown in this case, K. pneumoniae was found in blood culture and although the clinical isolate was shown to be sensitive to the carbapenem class of antibiotics, the patient did not respond to treatment with meropenem. Similar to K. pneumoniae, drug-resistant plasmids in the bacterial structure of K. variicola contribute to its virulence and resistance, but the K. variicola has the higher-risk antibiotic resistance-related genes sequences, thus giving it higher virulence and resistance[10,11]. Recent clinical observations have shown that tigecycline and polymyxin display higher rates of treatment success in hypervirulent Klebsiella infection than other antibacterial drugs such as carbapenem[12]. Moreover, a combination of treatments is preferred to monotherapy in cases of severe infections[13,14]. Unfortunately, treatments to target K. variicola infection were not prescribed here because the patients’ illness rapidly progressed before genotyping results were available.
As well illustrated by our case, K. pneumoniae subspecies can be rapidly fatal although their presence may often be overlooked due to the limitations of routine clinical examinations. This case should raise awareness among clinicians to consider Klebsiella subspecies infections, especially in cases of unexplained fever or other suspicious clinical presentations that may indicate this condition. Moreover, this case highlights the need to introduce genetic techniques into current clinical practices, especially for the early diagnosis of severe infections.
In summary, we have reported a patient dead with fatal infection caused by K. variicola. This fatal infection was identified by PMseq-DNA Pro high throughput gene detection assay. This case calls attention to Klebsiella subspecies infections and the need for early introduction of genetic technology in critically ill patients.
We thank Dr. Zuo MM for proofreading of the manuscript.
| 1. | Martin RM, Bachman MA. Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Front Cell Infect Microbiol. 2018;8:4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 504] [Cited by in RCA: 646] [Article Influence: 80.8] [Reference Citation Analysis (0)] |
| 2. | Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112:E3574-E3581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 682] [Cited by in RCA: 921] [Article Influence: 83.7] [Reference Citation Analysis (0)] |
| 3. | Shi Q, Lan P, Huang D, Hua X, Jiang Y, Zhou J, Yu Y. Diversity of virulence level phenotype of hypervirulent Klebsiella pneumoniae from different sequence type lineage. BMC Microbiol. 2018;18:94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 4. | Seki M, Gotoh K, Nakamura S, Akeda Y, Yoshii T, Miyaguchi S, Inohara H, Horii T, Oishi K, Iida T, Tomono K. Fatal sepsis caused by an unusual Klebsiella species that was misidentified by an automated identification system. J Med Microbiol. 2013;62:801-803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 5. | Podschun R, Pietsch S, Höller C, Ullmann U. Incidence of Klebsiella species in surface waters and their expression of virulence factors. Appl Environ Microbiol. 2001;67:3325-3327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 121] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 6. | Paczosa MK, Mecsas J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol Mol Biol Rev. 2016;80:629-661. [PubMed] |
| 7. | Liu C, Shi J, Guo J. High prevalence of hypervirulent Klebsiella pneumoniae infection in the genetic background of elderly patients in two teaching hospitals in China. Infect Drug Resist. 2018;11:1031-1041. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 8. | Li J, Ren J, Wang W, Wang G, Gu G, Wu X, Wang Y, Huang M, Li J. Risk factors and clinical outcomes of hypervirulent Klebsiella pneumoniae induced bloodstream infections. Eur J Clin Microbiol Infect Dis. 2018;37:679-689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 88] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 9. | Hodgson K, Morris J, Bridson T, Govan B, Rush C, Ketheesan N. Immunological mechanisms contributing to the double burden of diabetes and intracellular bacterial infections. Immunology. 2015;144:171-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 265] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 10. | Potter RF, Lainhart W, Twentyman J, Wallace MA, Wang B, Burnham CA, Rosen DA, Dantas G. Population Structure, Antibiotic Resistance, and Uropathogenicity of Klebsiella variicola. mBio. 2018;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 64] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 11. | Rodríguez-Medina N, Martínez-Romero E, De la Cruz MA, Ares MA, Valdovinos-Torres H, Silva-Sánchez J, Lozano-Aguirre L, Martínez-Barnetche J, Andrade V, Garza-Ramos U. A Klebsiella variicola Plasmid Confers Hypermucoviscosity-Like Phenotype and Alters Capsule Production and Virulence. Front Microbiol. 2020;11:579612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 12. | Lat A, Clock SA, Wu F, Whittier S, Della-Latta P, Fauntleroy K, Jenkins SG, Saiman L, Kubin CJ. Comparison of polymyxin B, tigecycline, cefepime, and meropenem MICs for KPC-producing Klebsiella pneumoniae by broth microdilution, Vitek 2, and Etest. J Clin Microbiol. 2011;49:1795-1798. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 52] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 13. | Bassetti M, Giacobbe DR, Giamarellou H, Viscoli C, Daikos GL, Dimopoulos G, De Rosa FG, Giamarellos-Bourboulis EJ, Rossolini GM, Righi E, Karaiskos I, Tumbarello M, Nicolau DP, Viale PL, Poulakou G; Critically Ill Patients Study Group of the European Society of Clinical Microbiology and Infectious Disease (ESCMID); Hellenic Society of Chemotherapy (HSC) and Società Italiana di Terapia Antinfettiva (SITA). Management of KPC-producing Klebsiella pneumoniae infections. Clin Microbiol Infect. 2018;24:133-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 135] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 14. | Agyeman AA, Bergen PJ, Rao GG, Nation RL, Landersdorfer CB. A systematic review and meta-analysis of treatment outcomes following antibiotic therapy among patients with carbapenem-resistant Klebsiella pneumoniae infections. Int J Antimicrob Agents. 2020;55:105833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 103] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Infectious diseases
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Apiratwarakul K S-Editor: Yan JP L-Editor: Wang TQ P-Editor: Yan JP