Published online Dec 6, 2022. doi: 10.12998/wjcc.v10.i34.12484
Peer-review started: September 7, 2022
First decision: October 20, 2022
Revised: October 25, 2022
Accepted: November 4, 2022
Article in press: November 4, 2022
Published online: December 6, 2022
Processing time: 86 Days and 7.7 Hours
Dysbiosis in the intestinal microflora can affect the gut production of microbial metabolites, and toxic substances can disrupt the barrier function of the intestinal wall, leading to the development of various diseases. Decreased levels of Clostridium subcluster XIVa (XIVa) are associated with the intestinal dysbiosis found in inflammatory bowel disease (IBD) and Clostridium difficile infection (CDI). Since XIVa is a bacterial group responsible for the conversion of primary bile acids (BAs) to secondary BAs, the proportion of intestinal XIVa can be predicted by determining the ratio of deoxycholic acid (DCA)/[DCA + cholic acid (CA)] in feces orserum. For example, serum DCA/(DCA+CA) was significantly lower in IBD patients than in healthy controls, even in the remission period. These results suggest that a low proportion of intestinal XIVa in IBD patients might be a precondition for IBD onset but not a consequence of intestinal inflammation. Another report showed that a reduced serum DCA/(DCA + CA) ratio could predict susceptibility to CDI. Thus, the BA profile, particularly the ratio of secon
Core Tip: Gut dysbiosis, particularly decreased XIVa, correlates strongly with decreased conversion of primary BAs to secondary BAs. Decreased levels of Clostridium subcluster XIVa (XIVa) are associated with the intestinal dysbiosis found in inflammatory bowel disease (IBD) and Clostridium difficile infection (CDI). Since XIVa is a bacterial group responsible for the conversion of primary BAs to secondary BAs, the proportion of intestinal XIVa can be predicted by determining the ratio of deoxycholic acid (DCA)/ [DCA + cholic acid (CA)] in feces or serum. Therefore, the DCA/(DCA+CA) ratio in feces and serum is a valuable marker for detecting dysbiosis without genetic analysis of enterobacteria.
- Citation: Monma T, Iwamoto J, Ueda H, Tamamushi M, Kakizaki F, Konishi N, Yara S, Miyazaki T, Hirayama T, Ikegami T, Honda A. Evaluation of gut dysbiosis using serum and fecal bile acid profiles. World J Clin Cases 2022; 10(34): 12484-12493
- URL: https://www.wjgnet.com/2307-8960/full/v10/i34/12484.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i34.12484
The human gut contains 1014 bacteria, ten times the number of human cells, which constitute 150 times more genes than the human genome[1]. Dysbiosis refers to an alteration of normal healthy state of the microbiota[1]. Alterations in the intestinal microbiota change the metabolites of gut microbiota, and toxic substances can disrupt intestinal barrier function and cause various diseases[2]. Dysbiosis is mainly associated with digestive disorders such as ulcerative colitis (UC)[3-6], Crohn’s disease (CD)[3,4,7,8], irritable bowel syndrome (IBS)[9], non-alcoholic fatty liver disease[10,11], and hepatocellular carcinoma[12]. In addition to digestive disorders, diabetes[13], atherosclerosis[14,15], obesity[16], atopies and asthma[17], and multiple sclerosis[18] have been associated with dysbiosis.
Since the 1990s, molecular biological techniques for the detection of dysbiosis have advanced rapidly since the 1990s, and methods utilizing the bacterial 16S rRNA gene variable region have allowed investigation of the gut microbiota[19]. Furthermore, shotgun metagenomics approaches utilize untargeted sequencing methods to capture all microbial genomes[20]. However, all of these methods require collection of fecal samples, and measurement and data analysis are time-consuming.
Bile acids (BAs) are secreted from the liver into the bile. An active transport system takes up approximately 95% of biliary BAs at the end of the ileum[21], and the remaining 5% is carried to the colon while some are absorbed passively. The absorbed BAs return to the liver through the portal vein called enterohepatic circulation. Intestinal bacteria convert the structure of BAs in the gut, and the converted BAs are present in the feces and enterohepatic circulation. Because a certain quantity t of BAs in the enterohepatic circulation leaks into the peripheral blood, the BA profiles in the feces as well as the peripheral blood may serve as markers of gut microbiota composition.
Dysbiosis has been reported in several gastrointestinal diseases, especially a reduced Clostridium subcluster XIVa (XIVa). XIVa is a major bacterial group that metabolizes BAs in the human gut[22,23]. Therefore we have a new hypothesis that the fecal and serum BA profiles could be a useful biomarker for intestinal XIVa activity. We have demonstrated the new facts that fecal and serum ratios of DCA/(DCA+CA) are useful as surrogate indicators of the gut proportion of XIVa, including the inflammatory bowel diseases (IBD) and Clostridium difficile infection (CDI)[24,25]. The unique insight of this review is that this review focused on the studies using the BA calculated product/(product+substrate) ratio, which is not discussed enough in previous reviews. We believe these results are useful in clinical practice, and it is necessary to investigate various diseases in the future studies.
In this review, we summarize the current literature regarding the relationship between BAs and the gut microbiota and the application of fecal and serum BA profiles as surrogate markers of dysbiosis and associated digestive disorders.
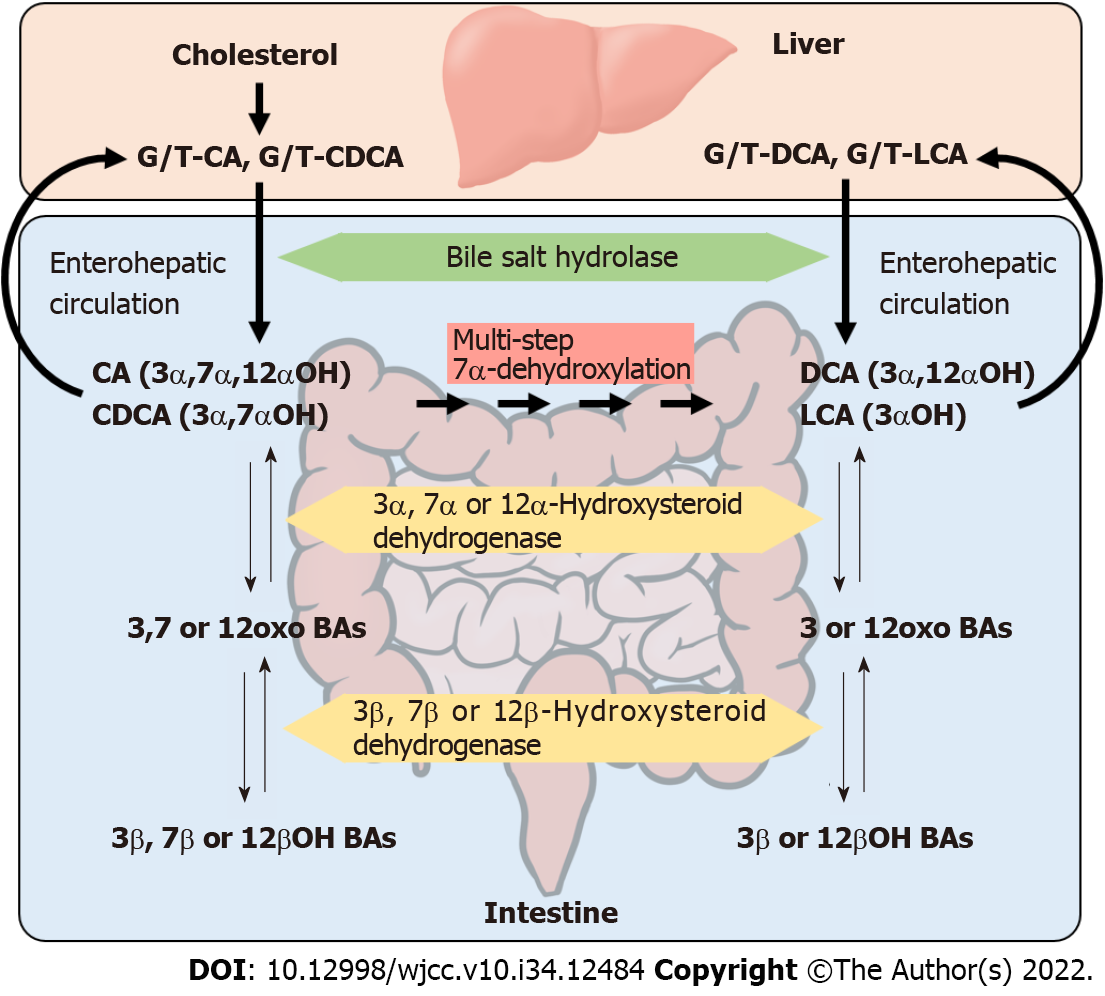
BAs are the end products of cholesterol metabolism. The human liver synthesizes glycine or taurine conjugated cholic acid (CA) and chenodeoxycholic acid (CDCA). These primary BAs are excreted into the bile and transported to the intestine. In the terminal ileum and the large intestine, the intact primary BAs are modified by intestinal bacteria (Figure 1). In this process, glycine and taurine are initially deconjugated by the bile salt hydrolases (BSH) expressed in various bacteria. Then, the hydroxyl group at the C-7α position of CA and CDCA is dehydroxylated, and the secondary BAs, deoxycholic acid (DCA) and lithocholic acid (LCA), are formed. This dehydroxylation step is catalyzed by a multi-step reaction encoded by bile acid-inducible (bai) genes in a single bai operon[21,26]. These bai genes are present only in specific bacteria, which account for nearly 0.0001% of the total colonic flora[21]. In addition, hydroxyl groups at the C-3α, 7α, and 12α positions of both conjugated and unconjugated BAs can be dehydrogenated to carbonyl groups and further epimerized to 3β-, 7β- and 12β-hydroxyl groups by intestinal bacteria[21].
The relationship between the enzymatic transformation of BAs and intestinal bacteria has been studied previously[21]. The deconjugation of amino acids is carried out by a variety of bacteria, including Bacteroides, Peptostreptococcus, Clostridium, Streptococcus, Eubacterium, Lactobacillus, and Bifidobacterium[27]. In contrast, multi-step 7α-dehydroxylation of BAs is mediated by Clostridium cluster IV (C. leptum)[28], cluster XI (C. sordellii, C. hiranonis, and C. bifermentans)[29], and cluster XIVa (C. scindens and C. hylemonae)[30], of which cluster XIVa is reported to play a major central role in this transformation[21,31].
The interaction between gut microbiota and BAs has been implicated in the pathogenesis of various disease states, including IBD, CDI, IBS, asthma, and obesity[32]. In these diseases, alterations of gut microbiota are associated with decreased BA deconjugation (or BSH activity) and/or reduced secondary BA production[32].
A reduced proportion of XIVa and decreased levels of secondary BAs have been reported in dysbiosis-associated gastrointestinal diseases, including IBD[22,23], CDI[33-36], and liver cirrhosis[31,37]. These results suggest that BA composition is markedly affected by the number of XIVa. Conversely, the number of XIVa is affected by intestinal CA amount[31]. Patients with liver cirrhosis have decreased intestinal XIVa and DCA levels due to the reduced size of the CA pool[31,37]. This is in contrast to the findings associated with a high-fat diet, which stimulates the biliary secretion of CA and increases intestinal XIVa and DCA levels[38,39].
Previous reports have also indicated that changes in the intestinal microbiota and increased DCA levels may lead to morbidity, including colon[40] and liver cancers[12]. Epidemiological evidence suggests that colorectal cancer is associated with increased levels of DCA in serum, bile, and stool[40]. Therefore, the benefits of increased XIVa and DCA in patients with gastrointestinal diseases are a topic of debate.
As mentioned above, deconjugation of BAs is easily mediated by the major bacteria, and most of the conjugated BAs in serum reflect the BAs reabsorbed without exposure to these bacteria. Therefore, the unconjugated form of BA is a better marker than total (conjugated + unconjugated) BA to calculate the BA-transformation activity of intestinal microbiota. For estimation of the approximate activity, the product/(product+substrate) ratio was calculated in a previous study[24]. In this approach, 7α-dehydroxylation was estimated by calculating DCA/(DCA+CA) or LCA/(LCA+CDCA). Epimerization of 3αOH BAs to 3βOH BAs is divided into two reactions. The conversion of 3αOH BAs to 3oxo BAs was estimated by 3oxo BAs/(3oxo BAs+3αOH BAs), and that of 3oxo BAs to 3βOH BAs was estimated by 3βOH BAs/(3βOH BAs+3oxo BAs). Epimerization of 7αOH BAs to 7βOH BAs and 12αOH BAs to 12βOH BAs was also estimated in the same way. The highest correlations were obtained between the proportion of fecal XIVa and fecal DCA/(DCA+CA) and serum DCA/(DCA+CA) (Table 1). In addition to DCA/(DCA+CA), LCA/(LCA+CDCA) is another marker for 7α-dehydroxylation, but its correlation coefficient with XIVa was lower than that of DCA/(DCA+CA). In healthy subjects, the ratios of the LCA/(LCA+CDCA) are much smaller than those of DCA/(DCA+CA) in serum but not in feces[24], suggesting that LCA is not easily absorbed from the intestine than other BAs. Therefore, as a serum marker for 7α-dehydroxylation, DCA/(DCA+CA) is better than LCA/(LCA+CDCA). Thus, by measuring the DCA/(DCA+CA) ratio in feces or serum, the abundance of XIVa and presumably the presence of dysbiosis can be estimated[24]. As shown in Table 2, these product/(product+substrate) ratios of BAs are now being applied in several studies, including those involving IBD patients[24] and CDI patients[25], studies on the effects of a high-fat diet in mice[41], and studies on the effects of water-soluble dietary fiber in humans[42]. Furthermore, the product/(product+substrate) ratios of fecal BAs can be calculated from the fecal BA data shown in the previous studies[43,44].
| Fecal microbial taxa | DCA/(DCA+CA) | |
| Serum | Feces | |
| Bifidobacterium | 0.2136 | 0.0683 |
| Lactobacillales | -0.5326a | -0.6830a |
| Bacteroides | 0.1836 | 0.3031c |
| Prevotella | -0.0342 | 0.2693 |
| Clostridium cluster IV | 0.2282 | 0.4307b |
| Clostridium subcluster XIVa | 0.5217a | 0.7659a |
| Clostridium cluster IX | 0.0710 | 0.0208 |
| Clostridium cluster XI | -0.0631 | -0.0053 |
| Clostridium cluster XVIII | 0.0348 | -0.0425 |
| Others | -0.0102 | -0.0637 |
| Ref. | Subjects | Samples | Major findings |
| Murakami et al[24], 2018 | 6 with CD, 6 with UC and 26 HCs | Serum feces | DCA/(DCA+CA)↓; LCA/(LCA+CDCA)↓ |
| Monma et al[25], 2022 | 12 with CDI, 59 without CDI and 46HCs | Serum | DCA/(DCA+CA)↓; LCA/(LCA+CDCA)→ |
| Ushiroda et al[41], 2019 | High-fat diet-fed mice | Serum | DCA/(DCA+CA)↑; LCA/(LCA+CDCA)→ |
| Yasukawa et al[42], 2019 | Healthy volunteers; Effects of Partially hydrolyzed guar gum (PHGG) | Plasma | DCA/(DCA+CA)↓; LCA/(LCA+CDCA)→ |
| Kasai et al[43], 2022 | NAFLD (MF, AF) and 26 HCs | Feces | DCA/(DCA+CA)→; LCA/(LCA+CDCA)→ |
| Misawa et al[44], 2020 | Elobixibat treatment | Feces | DCA/(DCA+CA)→; LCA/(LCA+CDCA)→ |
IBD is a chronic inflammatory condition of the colon and small intestine. The two forms of IBD, UC and CD, overlap clinically and pathologically, but are often very different[45,46]. The etiology of IBD remains unknown, but is believed to be attributable to the interaction of genetic and environmental factors.
The relationship between BAs and IBD has been reported in multiple studies[47-60]. The interaction of BAs and gut microbiota has been suggested to be closely related to the pathogenesis of IBD[48]. The fecal dysmetabolism of BAs observed in IBD is linked to IBD-associated dysbiosis, indicating that BA dysmetabolism could be used as a surrogate marker of IBD[51]. To evaluate the role of BAs in intestinal inflammation, the metabolomic, microbiome, metagenomic, and transcriptomic profiles of stool from the ileal pouches in patients with UC were investigated and revealed that dysbiosis induced secondary BA deficiency, which promotes intestinal inflammation[52].
Bamba et al[61] recently investigated the relationship between the gut microbiota and BA composition in the ileal mucosa of CD. In their study, the proportion of conjugated BAs was significantly higher in CD patients than in controls and was positively correlated with the presence of genera such as Escherichia and Lactobacillus and negatively correlated with the presence of genera such as Roseburia, Intestinibacter, and Faecalibacterium. These results suggested that ileal mucosa-associated dysbiosis and the alteration of BA compositions of fluid in the ileum may influence the pathology of ileal lesions of CD[61].
Previous studies have confirmed that intestinal XIVa, as well as cluster IV, are significantly decreased in patients with CD[7,8] and UC[5,6]. Serum DCA/(DCA+CA) was examined in controls and IBD patients in remission and exacerbation periods, and was significantly lower in IBD patients than in healthy controls, even in the remission periods. These results show that the low proportion of intestinal XIVa proportion in IBD patients is not a consequence of intestinal inflammation but a precursor to the development of IBD[24].
Bile acid malabsorption (BAM) is one of the hallmarks of CD, and BAs are potential activators of PXR. Therefore, the relationship between BAM and PXR activity in CD patients was investigated. Serum concentrations of 7α-hydroxy-4-cholesten-3-one (C4), a marker for hepatic bile acid biosynthesis[62], and FGF19, a marker for intestinal BA flux[63], were compared among patients with CD and UC and control participants. C4 Levels in CD patients were significantly higher than those in controls. In particular, the C4 values of CD patients with a history of ileal resection were markedly elevated and significantly higher than those of CD patients without a history of surgery. In contrast, serum FGF19 Levels in CD patients were significantly lower than those in UC patients, and tended to be lower than those in control individuals. CD patients with a history of ileal resection showed a marked decrease in the serum FGF19 concentration, which was significantly lower than those in CD patients without a history of surgery[47]. In addition, a significant negative correlation between 4β-hydroxycholesterol, a known marker for CYP3A4 activity, and C4 concentration was observed in CD patients. Since CYP3A4 is a target gene of PXR, the degree of BAM in CD patients was closely related to the deactivation of PXR. Thus, BA is a critical factor for the preservation of baseline activity of hepato-intestinal PXR in CD patients[47].
CDI is a common infection associated with hospitals and antibiotics. It causes a variety of clinical manifestations of colitis in healthcare facilities and the community[64-67]. Because CDI can be life-threatening, especially in the elderly, methods for screening high-risk hospitalized patients and preventing and treating CDI are desirable. Many reports have described the relationship between BAs and CDI. The secondary BAs, DCA and LCA, are more hydrophobic than the original primary BAs and have strong antimicrobial effects due to their high affinity with the lipids of cell membranes[68]. In addition to the bactericidal action, secondary BAs inhibits the proliferation of CD, the pathogen causing intractable diarrhea[33]. Previous reports have shown that DCA and LCA inhibit CD growth in vitro[69,70] and in vivo[71-74], and the levels of these secondary BAs in stool are reduced in CDI patients[32,75].
Regarding the relationship between indigenous enterobacteria and CDI, Clostridium scindens, one of the BA 7α-dehydroxylating bacteria, is associated with resistance to CDI[33,34]. In addition, fecal samples from CDI patients more frequently show negative results for bile acid-inducible (bai) genes than samples from control subjects, indicating that bai gene-positive species are involved in resistance to CD colonization[35]. More interestingly, the bile acid 7α-dehydroxylating bacteria, C. scindens and C. sordellii, secrete tryptophan-derived antibiotics and inhibit CD growth. These antibiotics inhibit cell division of CD, and the secondary BAs such as DCA and LCA, but not CA, enhance the inhibitory activity of these antibiotics[36].
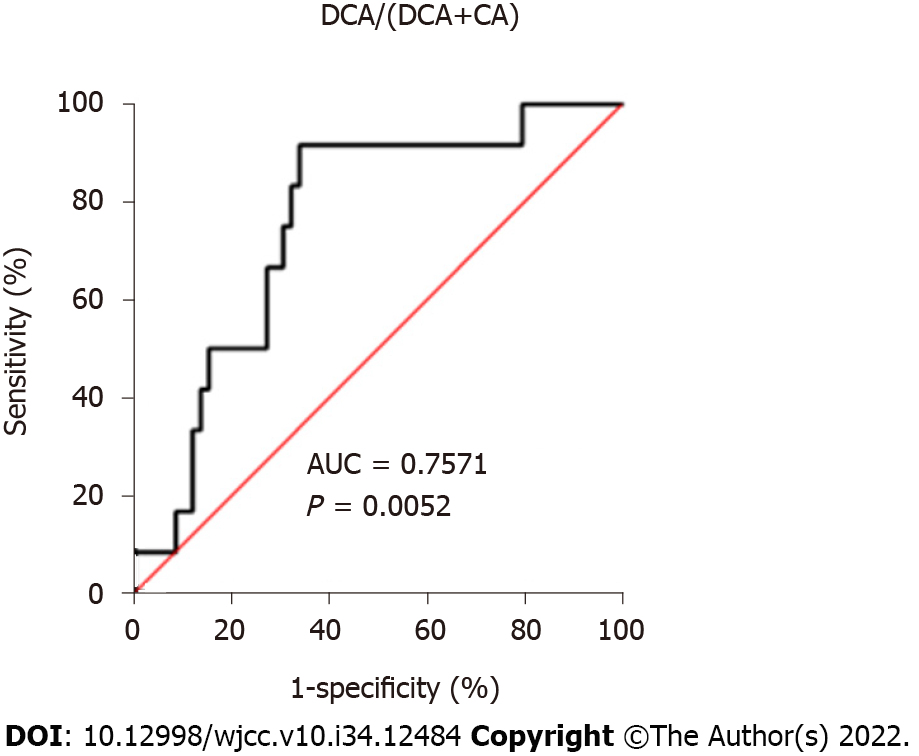
Although many reports have shown the relationships between BAs and CDI, studies using BA composition as a predictive surrogate marker to CDI susceptibility are limited[25,75]. Allegretti et al[75] showed that the fecal DCA to glycoursodeoxycholate (GUDCA) ratio was the best predictor and a potential biomarker for the recurrence of CDI. However, GUDCA is not a substrate of DCA, and GUDCA concentration is influenced by a number of factors other than BA 7α-dehydroxylation activity, including glycine/taurine conjugation ratio, deconjugation activity, the conversion rate of CDCA to ursodeoxycholate (UDCA) by 7-epimerization, and the possibility of UDCA administration to patients with hepatobiliary diseases. On the other hand, we showed that the serum DCA/(DCA+CA) ratio at the time of admission (before the use of antibiotics and CDI onset), was significantly low in patients who developed CDI while in the hospital compared to those in patients who did not develop CDI or in healthy controls[25]. In this study, DCA/(DCA+CA) < 0.349 was the cut-off value for discriminating patients at high risk of CDI before treatment with antibiotics, and the sensitivity and specificity of this threshold were 91.67% and 66.10%, respectively (Figure 2). The use of antibiotics represents the greatest risk factor for the development of CDI. However, patients who develop CDI already have a gut microbiota with significantly reduced diversity prior to antibiotic therapy[76].
Gut dysbiosis, particularly decreased XIVa, correlates strongly with decreased conversion of primary BAs to secondary BAs. Therefore, the DCA/(DCA+CA) ratio in feces and serum is a valuable marker for detecting dysbiosis caused by decreased XIVa without genetic analysis of enterobacteria.
| 1. | Lepage P, Leclerc MC, Joossens M, Mondot S, Blottière HM, Raes J, Ehrlich D, Doré J. A metagenomic insight into our gut's microbiome. Gut. 2013;62:146-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 246] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 2. | Ohno H. Impact of commensal microbiota on the host pathophysiology: focusing on immunity and inflammation. Semin Immunopathol. 2015;37:1-3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 3. | Matsuoka K, Kanai T. The gut microbiota and inflammatory bowel disease. Semin Immunopathol. 2015;37:47-55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 407] [Cited by in RCA: 602] [Article Influence: 50.2] [Reference Citation Analysis (1)] |
| 4. | Knights D, Lassen KG, Xavier RJ. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut. 2013;62:1505-1510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 323] [Cited by in RCA: 354] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 5. | Nemoto H, Kataoka K, Ishikawa H, Ikata K, Arimochi H, Iwasaki T, Ohnishi Y, Kuwahara T, Yasutomo K. Reduced diversity and imbalance of fecal microbiota in patients with ulcerative colitis. Dig Dis Sci. 2012;57:2955-2964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 121] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 6. | Rossen NG, Fuentes S, van der Spek MJ, Tijssen JG, Hartman JH, Duflou A, Löwenberg M, van den Brink GR, Mathus-Vliegen EM, de Vos WM, Zoetendal EG, D'Haens GR, Ponsioen CY. Findings From a Randomized Controlled Trial of Fecal Transplantation for Patients With Ulcerative Colitis. Gastroenterology. 2015;149:110-118.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 616] [Cited by in RCA: 703] [Article Influence: 63.9] [Reference Citation Analysis (0)] |
| 7. | Andoh A, Tsujikawa T, Sasaki M, Mitsuyama K, Suzuki Y, Matsui T, Matsumoto T, Benno Y, Fujiyama Y. Faecal microbiota profile of Crohn's disease determined by terminal restriction fragment length polymorphism analysis. Aliment Pharmacol Ther. 2009;29:75-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 8. | Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, Vandamme P, Vermeire S. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. 2011;60:631-637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 692] [Cited by in RCA: 819] [Article Influence: 54.6] [Reference Citation Analysis (0)] |
| 9. | Kassinen A, Krogius-Kurikka L, Mäkivuokko H, Rinttilä T, Paulin L, Corander J, Malinen E, Apajalahti J, Palva A. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 2007;133:24-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 711] [Cited by in RCA: 729] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 10. | Mouzaki M, Comelli EM, Arendt BM, Bonengel J, Fung SK, Fischer SE, McGilvray ID, Allard JP. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013;58:120-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 496] [Cited by in RCA: 571] [Article Influence: 43.9] [Reference Citation Analysis (1)] |
| 11. | Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo-Perez F, Guy CD, Seed PC, Rawls JF, David LA, Hunault G, Oberti F, Calès P, Diehl AM. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63:764-775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 763] [Cited by in RCA: 1114] [Article Influence: 111.4] [Reference Citation Analysis (1)] |
| 12. | Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, Honda K, Ishikawa Y, Hara E, Ohtani N. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1318] [Cited by in RCA: 1725] [Article Influence: 132.7] [Reference Citation Analysis (2)] |
| 13. | Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3971] [Cited by in RCA: 5017] [Article Influence: 358.4] [Reference Citation Analysis (1)] |
| 14. | Karlsson FH, Fåk F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, Bäckhed F, Nielsen J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. 2012;3:1245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 711] [Cited by in RCA: 992] [Article Influence: 76.3] [Reference Citation Analysis (0)] |
| 15. | Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575-1584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2130] [Cited by in RCA: 2524] [Article Influence: 194.2] [Reference Citation Analysis (0)] |
| 16. | Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480-484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6397] [Cited by in RCA: 5785] [Article Influence: 340.3] [Reference Citation Analysis (1)] |
| 17. | Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, Panzer AR, LaMere B, Rackaityte E, Lukacs NW, Wegienka G, Boushey HA, Ownby DR, Zoratti EM, Levin AM, Johnson CC, Lynch SV. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22:1187-1191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 595] [Cited by in RCA: 860] [Article Influence: 86.0] [Reference Citation Analysis (0)] |
| 18. | Miyake S, Kim S, Suda W, Oshima K, Nakamura M, Matsuoka T, Chihara N, Tomita A, Sato W, Kim SW, Morita H, Hattori M, Yamamura T. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS One. 2015;10:e0137429. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 463] [Cited by in RCA: 595] [Article Influence: 54.1] [Reference Citation Analysis (0)] |
| 19. | Olsen GJ, Woese CR, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 632] [Cited by in RCA: 557] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 20. | Quince C, Walker AW, Simpson JT, Loman NJ, Segata N. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017;35:833-844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 761] [Cited by in RCA: 1150] [Article Influence: 127.8] [Reference Citation Analysis (0)] |
| 21. | Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1644] [Cited by in RCA: 2145] [Article Influence: 102.1] [Reference Citation Analysis (0)] |
| 22. | Takahashi K, Nishida A, Fujimoto T, Fujii M, Shioya M, Imaeda H, Inatomi O, Bamba S, Sugimoto M, Andoh A. Reduced Abundance of Butyrate-Producing Bacteria Species in the Fecal Microbial Community in Crohn's Disease. Digestion. 2016;93:59-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 469] [Cited by in RCA: 545] [Article Influence: 54.5] [Reference Citation Analysis (0)] |
| 23. | Nishida A, Inoue R, Inatomi O, Bamba S, Naito Y, Andoh A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol. 2018;11:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 533] [Cited by in RCA: 983] [Article Influence: 109.2] [Reference Citation Analysis (3)] |
| 24. | Murakami M, Iwamoto J, Honda A, Tsuji T, Tamamushi M, Ueda H, Monma T, Konishi N, Yara S, Hirayama T, Miyazaki T, Saito Y, Ikegami T, Matsuzaki Y. Detection of Gut Dysbiosis due to Reduced Clostridium Subcluster XIVa Using the Fecal or Serum Bile Acid Profile. Inflamm Bowel Dis. 2018;24:1035-1044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 25. | Monma T, Iwamoto J, Honda A, Ueda H, Kakizaki F, Yara S, Miyazaki T, Ikegami T. Evaluation of the Risk of Clostridium difficile Infection Using a Serum Bile Acid Profile. Metabolites. 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 26. | Funabashi M, Grove TL, Wang M, Varma Y, McFadden ME, Brown LC, Guo C, Higginbottom S, Almo SC, Fischbach MA. A metabolic pathway for bile acid dehydroxylation by the gut microbiome. Nature. 2020;582:566-570. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 255] [Cited by in RCA: 432] [Article Influence: 72.0] [Reference Citation Analysis (0)] |
| 27. | Macdonald IA, Bokkenheuser VD, Winter J, McLernon AM, Mosbach EH. Degradation of steroids in the human gut. J Lipid Res. 1983;24:675-700. [PubMed] |
| 28. | Stellwag EJ, Hylemon PB. 7alpha-Dehydroxylation of cholic acid and chenodeoxycholic acid by Clostridium leptum. J Lipid Res. 1979;20:325-333. [PubMed] |
| 29. | Kitahara M, Takamine F, Imamura T, Benno Y. Clostridium hiranonis sp. nov., a human intestinal bacterium with bile acid 7alpha-dehydroxylating activity. Int J Syst Evol Microbiol. 2001;51:39-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 152] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 30. | Kitahara M, Takamine F, Imamura T, Benno Y. Assignment of Eubacterium sp. VPI 12708 and related strains with high bile acid 7alpha-dehydroxylating activity to Clostridium scindens and proposal of Clostridium hylemonae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2000;50 Pt 3:971-978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 106] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 31. | Ridlon JM, Alves JM, Hylemon PB, Bajaj JS. Cirrhosis, bile acids and gut microbiota: unraveling a complex relationship. Gut Microbes. 2013;4:382-387. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 227] [Cited by in RCA: 282] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 32. | Joyce SA, Gahan CG. Disease-Associated Changes in Bile Acid Profiles and Links to Altered Gut Microbiota. Dig Dis. 2017;35:169-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 92] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 33. | Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205-208. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1172] [Cited by in RCA: 1415] [Article Influence: 117.9] [Reference Citation Analysis (0)] |
| 34. | Studer N, Desharnais L, Beutler M, Brugiroux S, Terrazos MA, Menin L, Schürch CM, McCoy KD, Kuehne SA, Minton NP, Stecher B, Bernier-Latmani R, Hapfelmeier S. Functional Intestinal Bile Acid 7α-Dehydroxylation by Clostridium scindens Associated with Protection from Clostridium difficile Infection in a Gnotobiotic Mouse Model. Front Cell Infect Microbiol. 2016;6:191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 157] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 35. | Solbach P, Chhatwal P, Woltemate S, Tacconelli E, Buhl M, Gerhard M, Thoeringer CK, Vehreschild MJGT, Jazmati N, Rupp J, Manns MP, Bachmann O, Suerbaum S. BaiCD gene cluster abundance is negatively correlated with Clostridium difficile infection. PLoS One. 2018;13:e0196977. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 36. | Kang JD, Myers CJ, Harris SC, Kakiyama G, Lee IK, Yun BS, Matsuzaki K, Furukawa M, Min HK, Bajaj JS, Zhou H, Hylemon PB. Bile Acid 7α-Dehydroxylating Gut Bacteria Secrete Antibiotics that Inhibit Clostridium difficile: Role of Secondary Bile Acids. Cell Chem Biol. 2019;26:27-34.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 169] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 37. | Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, Takei H, Muto A, Nittono H, Ridlon JM, White MB, Noble NA, Monteith P, Fuchs M, Thacker LR, Sikaroodi M, Bajaj JS. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58:949-955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 503] [Cited by in RCA: 656] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 38. | Yokota A, Fukiya S, Islam KB, Ooka T, Ogura Y, Hayashi T, Hagio M, Ishizuka S. Is bile acid a determinant of the gut microbiota on a high-fat diet? Gut Microbes. 2012;3:455-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 156] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 39. | David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559-563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8550] [Cited by in RCA: 7186] [Article Influence: 598.8] [Reference Citation Analysis (1)] |
| 40. | Ridlon JM, Wolf PG, Gaskins HR. Taurocholic acid metabolism by gut microbes and colon cancer. Gut Microbes. 2016;7:201-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 246] [Article Influence: 24.6] [Reference Citation Analysis (9)] |
| 41. | Ushiroda C, Naito Y, Takagi T, Uchiyama K, Mizushima K, Higashimura Y, Yasukawa Z, Okubo T, Inoue R, Honda A, Matsuzaki Y, Itoh Y. Green tea polyphenol (epigallocatechin-3-gallate) improves gut dysbiosis and serum bile acids dysregulation in high-fat diet-fed mice. J Clin Biochem Nutr. 2019;65:34-46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 114] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 42. | Yasukawa Z, Inoue R, Ozeki M, Okubo T, Takagi T, Honda A, Naito Y. Effect of Repeated Consumption of Partially Hydrolyzed Guar Gum on Fecal Characteristics and Gut Microbiota: A Randomized, Double-Blind, Placebo-Controlled, and Parallel-Group Clinical Trial. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 43. | Kasai Y, Kessoku T, Tanaka K, Yamamoto A, Takahashi K, Kobayashi T, Iwaki M, Ozaki A, Nogami A, Honda Y, Ogawa Y, Kato S, Imajo K, Higurashi T, Hosono K, Yoneda M, Usuda H, Wada K, Kawanaka M, Kawaguchi T, Torimura T, Kage M, Hyogo H, Takahashi H, Eguchi Y, Aishima S, Kobayashi N, Sumida Y, Honda A, Oyamada S, Shinoda S, Saito S, Nakajima A. Association of Serum and Fecal Bile Acid Patterns With Liver Fibrosis in Biopsy-Proven Nonalcoholic Fatty Liver Disease: An Observational Study. Clin Transl Gastroenterol. 2022;13:e00503. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 44. | Misawa N, Higurashi T, Takatsu T, Iwaki M, Kobayashi T, Yoshihara T, Ashikari K, Kessoku T, Fuyuki A, Matsuura T, Ohkubo H, Usuda H, Wada K, Naritaka N, Takei H, Nittono H, Matsumoto M, Honda A, Nakajima A, Camilleri M. The benefit of elobixibat in chronic constipation is associated with faecal deoxycholic acid but not effects of altered microbiota. Aliment Pharmacol Ther. 2020;52:821-828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 45. | Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2693] [Cited by in RCA: 2772] [Article Influence: 115.5] [Reference Citation Analysis (3)] |
| 46. | Baumgart DC, Sandborn WJ. Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet. 2007;369:1641-1657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1247] [Cited by in RCA: 1385] [Article Influence: 72.9] [Reference Citation Analysis (0)] |
| 47. | Iwamoto J, Saito Y, Honda A, Miyazaki T, Ikegami T, Matsuzaki Y. Bile acid malabsorption deactivates pregnane X receptor in patients with Crohn's disease. Inflamm Bowel Dis. 2013;19:1278-1284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 48. | Yang M, Gu Y, Li L, Liu T, Song X, Sun Y, Cao X, Wang B, Jiang K, Cao H. Bile Acid-Gut Microbiota Axis in Inflammatory Bowel Disease: From Bench to Bedside. Nutrients. 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 139] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 49. | Gallagher K, Catesson A, Griffin JL, Holmes E, Williams HRT. Metabolomic Analysis in Inflammatory Bowel Disease: A Systematic Review. J Crohns Colitis. 2021;15:813-826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 93] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 50. | Wilson A, Almousa A, Teft WA, Kim RB. Attenuation of bile acid-mediated FXR and PXR activation in patients with Crohn's disease. Sci Rep. 2020;10:1866. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 51. | Duboc H, Rajca S, Rainteau D, Benarous D, Maubert MA, Quervain E, Thomas G, Barbu V, Humbert L, Despras G, Bridonneau C, Dumetz F, Grill JP, Masliah J, Beaugerie L, Cosnes J, Chazouillères O, Poupon R, Wolf C, Mallet JM, Langella P, Trugnan G, Sokol H, Seksik P. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531-539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 513] [Cited by in RCA: 701] [Article Influence: 53.9] [Reference Citation Analysis (0)] |
| 52. | Sinha SR, Haileselassie Y, Nguyen LP, Tropini C, Wang M, Becker LS, Sim D, Jarr K, Spear ET, Singh G, Namkoong H, Bittinger K, Fischbach MA, Sonnenburg JL, Habtezion A. Dysbiosis-Induced Secondary Bile Acid Deficiency Promotes Intestinal Inflammation. Cell Host Microbe. 2020;27:659-670.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 610] [Article Influence: 101.7] [Reference Citation Analysis (0)] |
| 53. | Yang ZH, Liu F, Zhu XR, Suo FY, Jia ZJ, Yao SK. Altered profiles of fecal bile acids correlate with gut microbiota and inflammatory responses in patients with ulcerative colitis. World J Gastroenterol. 2021;27:3609-3629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 97] [Cited by in RCA: 99] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 54. | Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, Vatanen T, Hall AB, Mallick H, McIver LJ, Sauk JS, Wilson RG, Stevens BW, Scott JM, Pierce K, Deik AA, Bullock K, Imhann F, Porter JA, Zhernakova A, Fu J, Weersma RK, Wijmenga C, Clish CB, Vlamakis H, Huttenhower C, Xavier RJ. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4:293-305. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 888] [Cited by in RCA: 1359] [Article Influence: 194.1] [Reference Citation Analysis (0)] |
| 55. | Jacobs JP, Goudarzi M, Singh N, Tong M, McHardy IH, Ruegger P, Asadourian M, Moon BH, Ayson A, Borneman J, McGovern DP, Fornace AJ Jr, Braun J, Dubinsky M. A Disease-Associated Microbial and Metabolomics State in Relatives of Pediatric Inflammatory Bowel Disease Patients. Cell Mol Gastroenterol Hepatol. 2016;2:750-766. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 176] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 56. | Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, Casero D, Courtney H, Gonzalez A, Graeber TG, Hall AB, Lake K, Landers CJ, Mallick H, Plichta DR, Prasad M, Rahnavard G, Sauk J, Shungin D, Vázquez-Baeza Y, White RA 3rd; IBDMDB Investigators, Braun J, Denson LA, Jansson JK, Knight R, Kugathasan S, McGovern DPB, Petrosino JF, Stappenbeck TS, Winter HS, Clish CB, Franzosa EA, Vlamakis H, Xavier RJ, Huttenhower C. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655-662. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2122] [Reference Citation Analysis (0)] |
| 57. | Wang Y, Gao X, Zhang X, Xiao F, Hu H, Li X, Dong F, Sun M, Xiao Y, Ge T, Li D, Yu G, Liu Z, Zhang T. Microbial and metabolic features associated with outcome of infliximab therapy in pediatric Crohn's disease. Gut Microbes. 2021;13:1-18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 66] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 58. | Diederen K, Li JV, Donachie GE, de Meij TG, de Waart DR, Hakvoort TBM, Kindermann A, Wagner J, Auyeung V, Te Velde AA, Heinsbroek SEM, Benninga MA, Kinross J, Walker AW, de Jonge WJ, Seppen J. Exclusive enteral nutrition mediates gut microbial and metabolic changes that are associated with remission in children with Crohn's disease. Sci Rep. 2020;10:18879. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 90] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 59. | Labbé A, Ganopolsky JG, Martoni CJ, Prakash S, Jones ML. Bacterial bile metabolising gene abundance in Crohn's, ulcerative colitis and type 2 diabetes metagenomes. PLoS One. 2014;9:e115175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 115] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 60. | Ogilvie LA, Jones BV. Dysbiosis modulates capacity for bile acid modification in the gut microbiomes of patients with inflammatory bowel disease: a mechanism and marker of disease? Gut. 2012;61:1642-1643. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 61. | Bamba S, Inatomi O, Nishida A, Ohno M, Imai T, Takahashi K, Naito Y, Iwamoto J, Honda A, Inohara N, Andoh A. Relationship between the gut microbiota and bile acid composition in the ileal mucosa of Crohn's disease. Intest Res. 2022;20:370-380. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 19] [Reference Citation Analysis (1)] |
| 62. | Sauter G, Berr F, Beuers U, Fischer S, Paumgartner G. Serum concentrations of 7alpha-hydroxy-4-cholesten-3-one reflect bile acid synthesis in humans. Hepatology. 1996;24:123-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 19] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 63. | Lundåsen T, Gälman C, Angelin B, Rudling M. Circulating intestinal fibroblast growth factor 19 has a pronounced diurnal variation and modulates hepatic bile acid synthesis in man. J Intern Med. 2006;260:530-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 306] [Cited by in RCA: 329] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 64. | Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ. Clostridium difficile infection. Nat Rev Dis Primers. 2016;2:16020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 439] [Cited by in RCA: 638] [Article Influence: 63.8] [Reference Citation Analysis (0)] |
| 65. | Czepiel J, Dróżdż M, Pituch H, Kuijper EJ, Perucki W, Mielimonka A, Goldman S, Wultańska D, Garlicki A, Biesiada G. Clostridium difficile infection: review. Eur J Clin Microbiol Infect Dis. 2019;38:1211-1221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 503] [Article Influence: 71.9] [Reference Citation Analysis (1)] |
| 66. | Ooijevaar RE, van Beurden YH, Terveer EM, Goorhuis A, Bauer MP, Keller JJ, Mulder CJJ, Kuijper EJ. Update of treatment algorithms for Clostridium difficile infection. Clin Microbiol Infect. 2018;24:452-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 100] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 67. | Oksi J, Anttila VJ, Mattila E. Treatment of Clostridioides (Clostridium) difficile infection. Ann Med. 2020;52:12-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 68. | Kurdi P, Kawanishi K, Mizutani K, Yokota A. Mechanism of growth inhibition by free bile acids in lactobacilli and bifidobacteria. J Bacteriol. 2006;188:1979-1986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 265] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 69. | Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190:2505-2512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 607] [Cited by in RCA: 571] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 70. | Sorg JA, Sonenshein AL. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol. 2010;192:4983-4990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 285] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 71. | Theriot CM, Koenigsknecht MJ, Carlson PE Jr, Hatton GE, Nelson AM, Li B, Huffnagle GB, Z Li J, Young VB. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 635] [Cited by in RCA: 748] [Article Influence: 68.0] [Reference Citation Analysis (0)] |
| 72. | Theriot CM, Bowman AA, Young VB. Antibiotic-Induced Alterations of the Gut Microbiota Alter Secondary Bile Acid Production and Allow for Clostridium difficile Spore Germination and Outgrowth in the Large Intestine. mSphere. 2016;1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 239] [Cited by in RCA: 374] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 73. | Winston JA, Theriot CM. Impact of microbial derived secondary bile acids on colonization resistance against Clostridium difficile in the gastrointestinal tract. Anaerobe. 2016;41:44-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 115] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 74. | Cheng S, Zhu L, Faden HS. Interactions of bile acids and the gut microbiota: learning from the differences in Clostridium difficile infection between children and adults. Physiol Genomics. 2019;51:218-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 75. | Allegretti JR, Kearney S, Li N, Bogart E, Bullock K, Gerber GK, Bry L, Clish CB, Alm E, Korzenik JR. Recurrent Clostridium difficile infection associates with distinct bile acid and microbiome profiles. Aliment Pharmacol Ther. 2016;43:1142-1153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 160] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 76. | Berkell M, Mysara M, Xavier BB, van Werkhoven CH, Monsieurs P, Lammens C, Ducher A, Vehreschild MJGT, Goossens H, de Gunzburg J, Bonten MJM, Malhotra-Kumar S; ANTICIPATE study group. Microbiota-based markers predictive of development of Clostridioides difficile infection. Nat Commun. 2021;12:2241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (1)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Japan
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Ji G, China; Šarenac TM, Serbia S-Editor: Liu JH L-Editor: A P-Editor: Liu JH