Published online May 26, 2022. doi: 10.12998/wjcc.v10.i15.5018
Peer-review started: November 19, 2021
First decision: December 27, 2021
Revised: January 10, 2022
Accepted: March 26, 2022
Article in press: March 26, 2022
Published online: May 26, 2022
Processing time: 186 Days and 14.3 Hours
This case report describes a child with Hutchinson-Gilford progeria syndrome (HGPS, OMIM: 176670) caused by LMNA (OMIM: 150330) gene mutation, and we have previously analyzed the clinical manifestations and imaging characteristics of this case. After 1-year treatment and follow-up, we focus on analyzing the changes in the clinical manifestations and genetic diagnosis of the patient.
In April 2020, a 2-year-old boy with HGPS was found to have an abnormal appearance, and growth and development lagged behind those of children of the same age. The child’s weight did not increase normally, the veins of the head were clearly visible, and he had shallow skin color and sparse yellow hair. Peripheral blood DNA samples obtained from the patient and his parents were sequenced using high-throughput whole-exosome sequencing, which was verified by Sanger sequencing. The results showed that there was a synonymous heterozygous mutation of C.1824 C>T (P. G608G) in the LMNA gene.
Mutation of the LMNA gene provides a molecular basis for diagnosis of HGPS and genetic counseling of the family.
Core Tip: The main clinical manifestations of Hutchinson-Gilford progeria syndrome (HGPS) in this 2-year-old boy were abnormal appearance, and growth and development lag behind children of the same age. DNA sequencing showed that there was a synonymous heterozygous mutation in the LMNA gene. This study provides a molecular basis for the etiological diagnosis of HGPS and genetic counseling for the family.
- Citation: Zhang SL, Lin SZ, Zhou YQ, Wang WQ, Li JY, Wang C, Pang QM. Clinical manifestations and gene analysis of Hutchinson-Gilford progeria syndrome: A case report. World J Clin Cases 2022; 10(15): 5018-5024
- URL: https://www.wjgnet.com/2307-8960/full/v10/i15/5018.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i15.5018
Hutchinson-Gilford progeria syndrome (HGPS) is a well-characterized premature aging disorder caused by mutations in LMNA, the gene encoding the nuclear scaffold proteins lamins A and C[1,2]. We have previously analyzed the clinical manifestations and imaging characteristics of this case[3]. HGPS is characterized by the presence of aging-associated symptoms, including lack of subcutaneous fat, alopecia, swollen veins, growth retardation, age spots, joint contractures, osteoporosis, cardiovascular pathology, and death due to heart attacks and strokes in childhood[4]. In this study, a heterozygous mutation of the LMNA gene was found by high-throughput whole-exosome sequencing in a 2-year-old patient with an abnormal appearance, and growth and development lag behind those of children of the same age.
A boy was aged 2 years and 5 mo at the time of diagnosis of HGPS, by which time, he had an abnormal appearance and his growth and development lagged behind those of children of his age.
In the past 6 mo, the weight of the child did not increase and remained at about 8 kg. The veins in the head of the child were clearly visible, his skin was lighter, and his hair was yellow and sparse, which were the main clinical manifestations of the disease. Peripheral blood DNA samples obtained from the patient and his parents were sequenced using high-throughput whole-exosome sequencing, which was verified by Sanger sequencing.
At birth, the parents found that the veins and blood vessels at the trunk of the child were obvious. At the age of 6 mo, the head and trunk veins were more apparent, and the skin pigment in the trunk was partially lost, presenting as spots. Around the age of 1 year, the scalp vein and skin had lost pigment, and his hair was relatively sparse. After 1 year of age, the increase in body weight was not obvious, and he was immediately taken to a local hospital. A diagnosis of rickets was considered, but the relevant treatment seemed useless.
The child was born at 40 wk by cesarean section. He weighed 2650 g at birth and had no history of asphyxia rescue. After birth, he received artificial feeding. Milk and porridge were added as supplementary foods. He could raise up his head at 3 mo, turn over at 4-5 mo, and walk steadily at 11 mo. At the age of 1 year, he could shout “YeYe” and “NaNa”, and at 18 mo, continuous sentences could be spoken. He could express his needs. His neurological and mental development was the same as that of children of the same age. His parents were not consanguineous. There was no family history of familial genetic disease or infectious diseases.
The child’s weight was 8 kg, head circumference 46.5 cm, and body length 81 cm, which were lower than those of children at the same age. There was no abnormal mental reaction. The skin was thin and had poor elasticity. The skin of the trunk showed spot-like depigmentation, and some superficial lymph nodes were swollen. It could also be seen that the hair was sparse and yellow, the veins of the scalp were clearly visible, and the anterior fontanel was not closed. His eyeballs were slightly prominent, the bridge of the nose was lower, and the face showed mandibuloacral dysplasia. The ear position was not low, and there was no penetrating palm. The chest circumference was 44 cm and the chest showed changes consistent with pigeon breast. There were no abnormalities in the auscultation of his heart and lungs. His knee and ankle joints were stiff, with poor mobility, his fingers and toes were slender, and his joints showed slight swelling, with clubbing fingers. Bilateral Babinski signs were positive.
We carried out tests on the patient’s blood: Leukocyte count 7.9 × 109/L, hemoglobin 130 g/L, platelet count 339 × 109/L, neutrophils 32.4%, lymphocytes 58.3%, and calcitonin 0.00 mg/L. The results of thyroid function tests were: Serum total triiodothyronine (T3) 3.29 nmol/L, total serum thyroxine 86.00 nmol/L, free T3 7.03 pmol/L, thyroid-stimulating hormone 1.93 mIU/L, and parathyroid hormone 0.21 pmol/L. Liver and kidney function tests, electrolytes, blood gas analysis, copper blue analysis, urine and feces analysis, coagulation function tests, and erythrocyte sedimentation rate showed no obvious abnormalities.
The results of electroencephalography and blood/urine tandem mass spectrometry were normal. Color Doppler echocardiography showed mild tricuspid regurgitation. The liver, bile, pancreas, and spleen were examined by color Doppler ultrasonography, and the results showed no abnormalities. The testicular color ultrasound examination showed a fluid dark area around the right testicle, which could be considered as a testicular hydrocele. No abnormal acoustic images were found in the testicle and epididymis on both sides. Chest radiographs showed normal cardiopulmonary septum. Spina bifida of the second sacral vertebral body was possible. Three-dimensional cranial reconstruction showed a low-density lesion in the white matter space of the right parietal lobe. On November 29, 2019, nuclear magnetic resonance imaging of the head revealed a partial perivascular space, with localized keratinosis in bilateral parietal lobes, accompanied by thickening of the maxillary sinus mucosa on the right side, suggesting irregular tooth arrangement.
Whole-exome sequencing (WES) and whole-genome sequencing analyses of copy number variants were performed to seek for genetic counseling and clarify the etiology of the child’s condition. Informed consent was obtained from the parents on behalf of the proband for WES and mitochondrial sequencing and for publication of photographs. DNA was obtained from peripheral blood samples from the patient and his parents. Statistical analysis of the data on single nucleotide polymorphisms (SNPs) and insertion deletion mutations (indels) was conducted. The SNPs and indels were screened using the Database of Single Nucleotide Polymorphisms (dbSNP), 1000 Genomes, and ExAC database. Application of the human Online Mendelian Genetic Database (OMIM) confirmed the reported pathogenic gene locus. The American College of Medical Genetics and Genomics (ACMG) sequence variation interpretation standards and guidelines were used for a comprehensive evaluation of the pathogenicity of mutation sites.
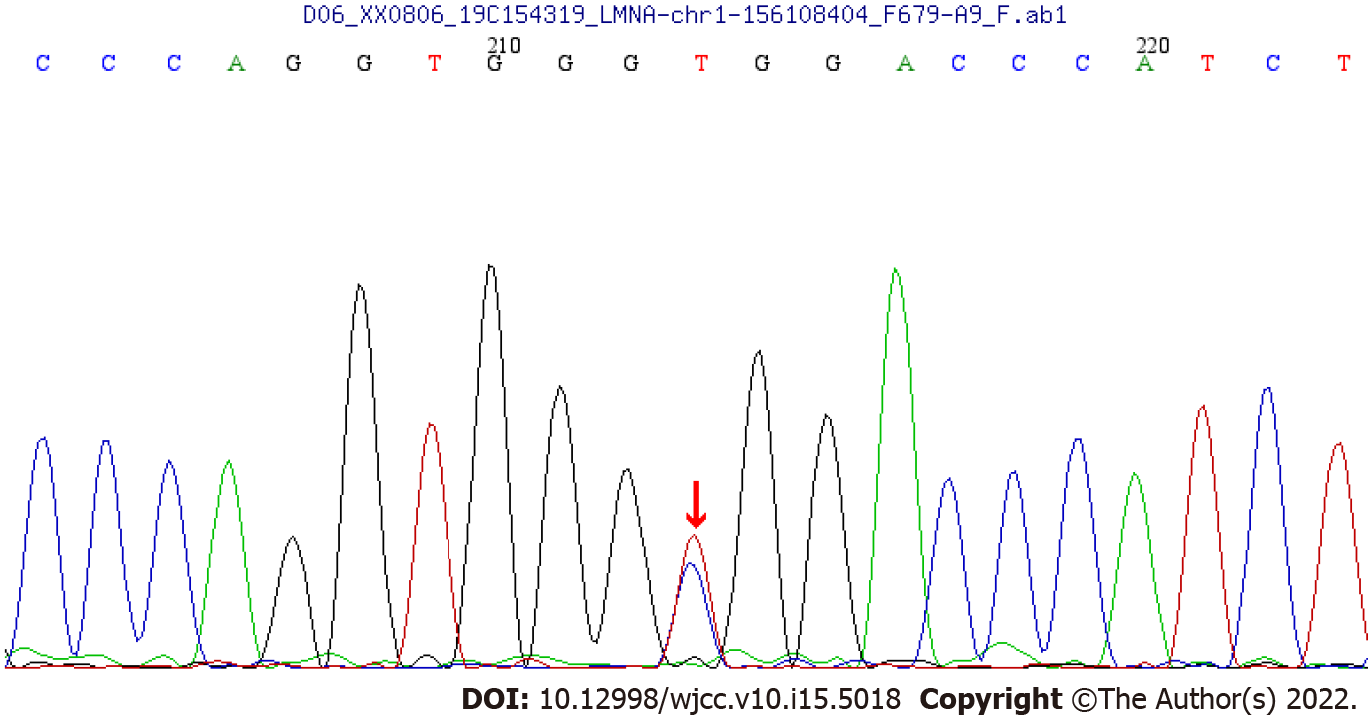
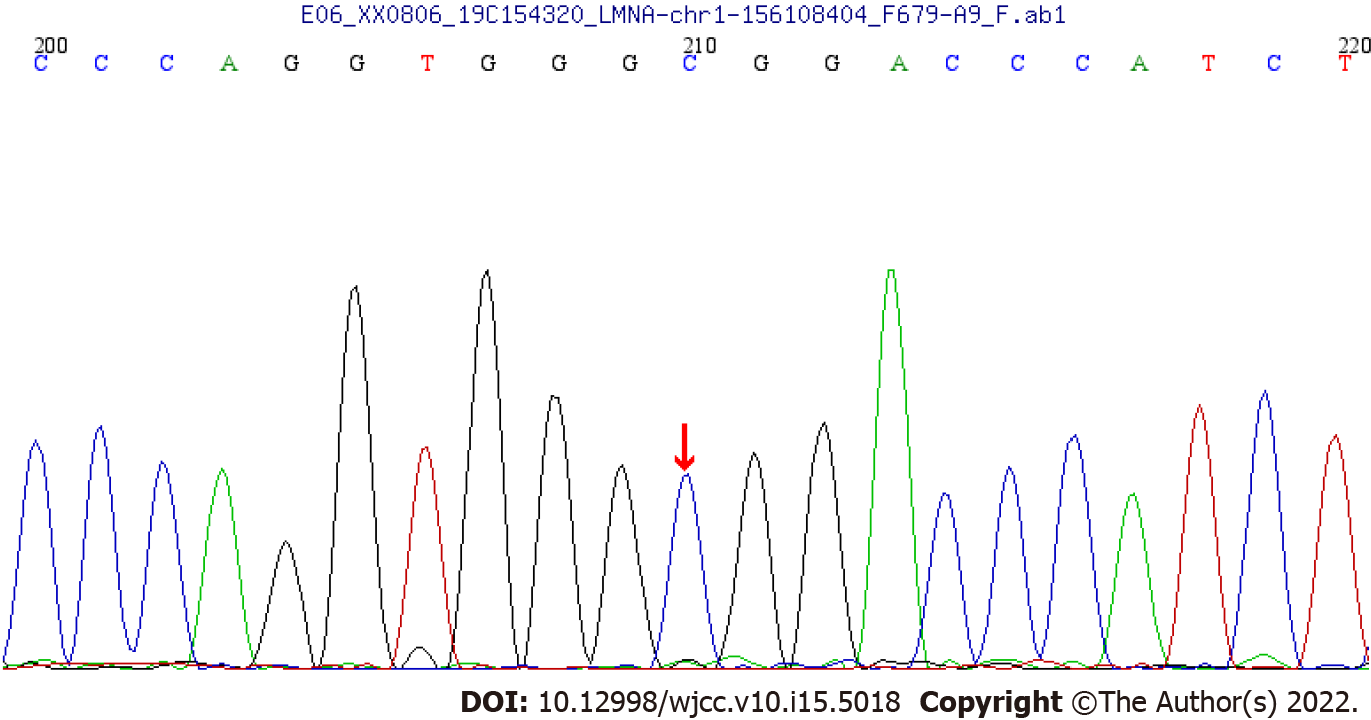
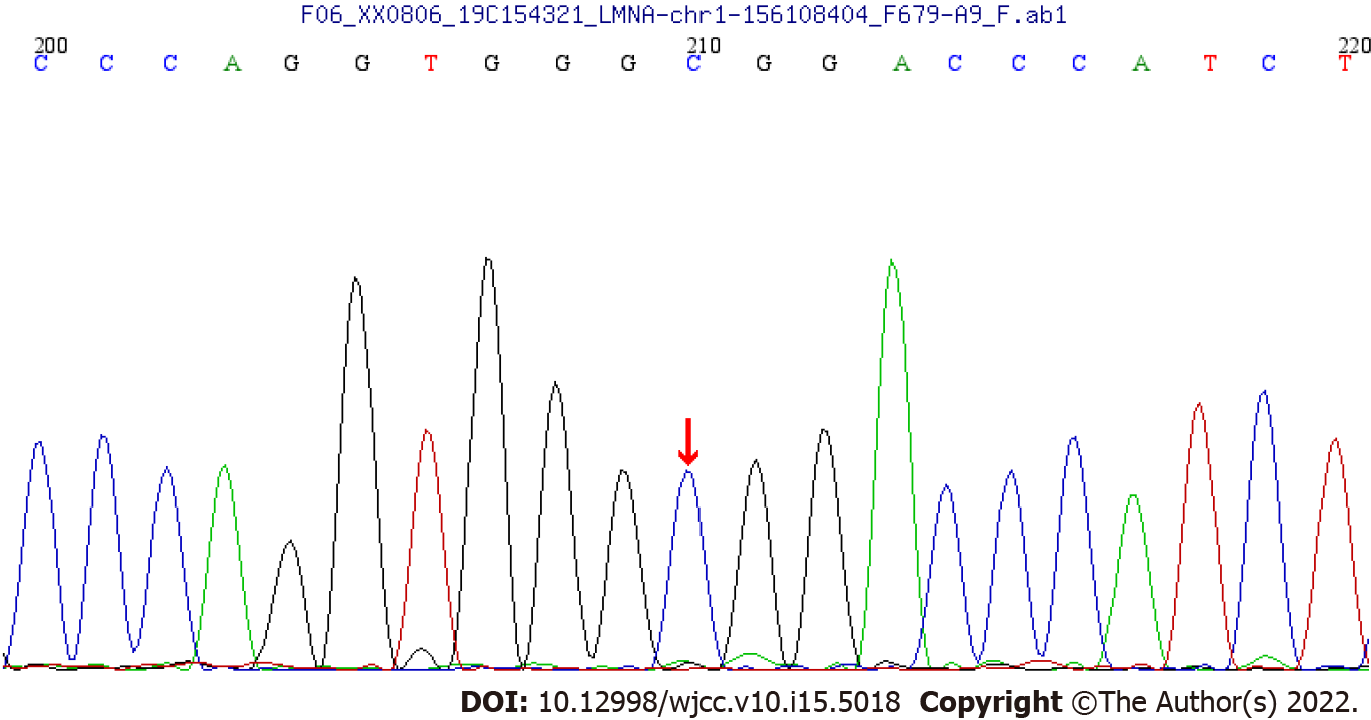
WES showed that there was a synonymous heterozygous variation of the LMNA gene in this patient, one of which was a spontaneous and synonymous mutation C.1824 C>T (P. G608G), which may lead to a change of gene function (Figure 1). No anomalies were found in the child’s father and mother as revealed by WES (Figures 2 and 3). The frequency of the variation in the normal population is unknown, it is a low-frequency variation, and the results of protein function prediction are unknown. According to Sanger sequencing, there was no variation in this locus of parental genes, and the inheritance mode was autosomal dominant inheritance. According to ACMG guidelines, the mutation was suspected to be pathogenic.
Sanger sequencing showed that there was a spontaneous and synonymous heterozygous mutation of C.1824 C>T (P. G608G) in the LMNA gene. Due to the mutation of this gene, the child was diagnosed with HGPS.
According to the clinical manifestations, laboratory tests, and gene sequencing results, the clinical phenotype of the patient was HGPS. The patient was treated with vitamin A/D, calcium supplement, and other drugs.
After 18 mo of treatment, the symptoms did not improve. Neither height nor weight have changed significantly since the age of 2 years.
HGPS (OMIM: 176670) is an autosomal dominant genetic disease that causes a remarkable phenotype resembling many aspects of aging[1,5]. The disease was first reported by general practitioner Jonathan Hutchinson in 1886, and was subsequently refined and supplemented[6]. The main first symptoms in typical childhood-onset phenotype include postnatal growth retardation (55%), alopecia (40%), absence of subcutaneous fat (28%), and lipodystrophy (20%)[1,7]. The following symptoms also exist: Midface hypoplasia, micrognathia, osteoporosis, low body weight, decreased joint mobility, and premature aging.
Children affected with HGPS typically appear normal at birth, but within a year the characteristic features of failure to thrive, delayed dentition, alopecia, and sclerodermatous skin changes begin to appear. Mean age at diagnosis is 2.9 years[8,9]. Median life expectancy is about 13 years, with most dying from complications associated with atherosclerosis. The most common mutation in HGPS, C.1824 C>T (P. G608G), is a synonymous substitution.
The LMNA (OMIM: 150330) gene is located on chromosome 1q22, and different sets of mutations in the LMNA and genes coding for interacting proteins, give rise to a variety of genetic disorders collectively called laminopathies[10]. LMNA codes for two major, alternatively spliced transcripts that give rise to lamin A and lamin C proteins, which are generated by use of an alternative 50 splice site in exon 10. Unlike lamin C, lamin A is translated as prelamin A and undergoes post-translational processing steps at the C-terminal CaaX motif[9,11].
We report a case of synonymous heterozygous mutation in the LMNA gene (C.1824 C>T; P.G608G). This variant belongs to a spontaneous mutation, whose parent gene has no variation at this locus, and the genetic mode belongs to the autosomal dominant inheritance[4,5]. As for the function of the mutation, the observed mutations improve the match to a consensus splice donor, suggesting that it activates a cryptic splice site. It shows that 150 nucleotides within exon 11 are missing. This abnormal transcript would be expected to code for a protein with an internal deletion of 50 amino acids near the C terminus of lamin A[10-13].
In the present case, obvious veins and blood vessels at the trunk were found. At the age of 6 mo, these were more apparent, and more typical symptoms appeared: Postnatal growth retardation, lipodystrophy, beak-like nose, micrognathia, and spotty skin pigment loss. Around the age of 1 year, his hair volume was sparse and he had a tendency to alopecia. The joints of the whole body, including the fingers and toes, were stiff, but there was no abnormal function of the circulation and digestive system. The child had normal intelligence and was born with low body weight, but had no history of feeding difficulties. Growth and development before 1 year were basically normal.Initially, the patient’s clinical manifestations were squared skull, pigeon breast, and alopecia, in line with the clinically typical phenotype of rickets, so vitamin A/D and calcium treatment were given, but the ideal treatment effect was not obtained. In order to determine the cause of the disease, after obtaining informed consent from the patients, we used high-throughput WES and identified a synonymous heterozygous mutation of the LMNA gene[2].
We followed this patient for 18 mo, and found that his height and weight did not improve. This shows that most of the disease manifestations are determined by genetic mutations, so the support therapy does not improve quality of life[4,9,14].
People's cognition of the disease has been gradually deepened, prompting further exploration of the treatment for progeria. The main treatment strategies include gene therapy, biological therapy, and small molecule treatment[15,16]. Gene therapy can directly repair pathogenic mutations; although this therapy still has many limitations and is not commonly used currently, it is anticipated to be a valuable treatment in the future.
Modern studies suggest that lonafarnib, although unable to cure the disease, is able to improve some clinical symptoms of the disease and, therefore, can be a target for further research. Kang et al[17] reported that progerinin may be an efficient, stable, and safe treatment.
With respect to the small molecule treatment, significant progress has been made in the enzyme isoprenylcysteine carboxylmethyltransferase inhibitors and the interaction between progerin and lamin A[18]. Regarding the biological therapy, antisense oligonucleotides have shown encouraging results in progeria.
ABE variants were reported to have higher editing activity than ABE7.10max by Koblan et al[19]. They suggest that ABE editing has the potential to cooperate with emerging premature treatments, including farnesyltransferase inhibitors, other small molecule drugs, or antisense oligonucleotides targeting the mutant LMNA allele.
We report a case of synonymous heterozygous mutation in the LMNA gene (C.1824 C>T; P.G608G). This variant belongs to a spontaneous mutation, whose parent gene has no variation at this locus, and the genetic mode belongs to the autosomal dominant inheritance. After 18 mo of treatment, the symptoms did not improve. Neither height nor weight have changed significantly. Up to now, the gene therapy is still in the developmental stage, so there are many unknown areas of the disease to be further investigated.
Taken together, although there are multiple obstacles to overcome before HGPS can be cured, effective treatments are only a matter of time as the molecular mechanisms of the disease are continuously understood and the exploration of therapeutic strategies continues[20].
We would like to thank the child and his family members for agreeing to participate in this study.
| 1. | Hennekam RC. Hutchinson-Gilford progeria syndrome: review of the phenotype. Am J Med Genet A. 2006;140:2603-2624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 363] [Cited by in RCA: 390] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 2. | Piekarowicz K, Machowska M, Dzianisava V, Rzepecki R. Hutchinson-Gilford Progeria Syndrome-Current Status and Prospects for Gene Therapy Treatment. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 3. | Zhang SL, Pang QM, Wu SL. Clinical Manifestation and Imaging Analysis of One Case of Infantile Progeria. Zhongguo Quanke Yixue. 2021;24(S1):153-155. |
| 4. | Beyret E, Liao HK, Yamamoto M, Hernandez-Benitez R, Fu Y, Erikson G, Reddy P, Izpisua Belmonte JC. Single-dose CRISPR-Cas9 therapy extends lifespan of mice with Hutchinson-Gilford progeria syndrome. Nat Med. 2019;25:419-422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 107] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 5. | McCord RP, Nazario-Toole A, Zhang H, Chines PS, Zhan Y, Erdos MR, Collins FS, Dekker J, Cao K. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res. 2013;23:260-269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 256] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 6. | Hutchinson J. Congenital Absence of Hair and Mammary Glands with Atrophic Condition of the Skin and its Appendages, in a Boy whose Mother had been almost wholly Bald from Alopecia Areata from the age of Six. Med Chir Trans. 1886;69:473-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 97] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 7. | Aguado J, Sola-Carvajal A, Cancila V, Revêchon G, Ong PF, Jones-Weinert CW, Wallén Arzt E, Lattanzi G, Dreesen O, Tripodo C, Rossiello F, Eriksson M, d'Adda di Fagagna F. Inhibition of DNA damage response at telomeres improves the detrimental phenotypes of Hutchinson-Gilford Progeria Syndrome. Nat Commun. 2019;10:4990. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 8. | Ahmed MS, Ikram S, Bibi N, Mir A. Hutchinson-Gilford Progeria Syndrome: A Premature Aging Disease. Mol Neurobiol. 2018;55:4417-4427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 46] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 9. | Sun S, Qin W, Tang X, Meng Y, Hu W, Zhang S, Qian M, Liu Z, Cao X, Pang Q, Zhao B, Wang Z, Zhou Z, Liu B. Vascular endothelium-targeted Sirt7 gene therapy rejuvenates blood vessels and extends life span in a Hutchinson-Gilford progeria model. Sci Adv. 2020;6:eaay5556. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 10. | Maraldi NM, Capanni C, Cenni V, Fini M, Lattanzi G. Laminopathies and lamin-associated signaling pathways. J Cell Biochem. 2011;112:979-992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 88] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 11. | Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, Dutra A, Pak E, Durkin S, Csoka AB, Boehnke M, Glover TW, Collins FS. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1602] [Cited by in RCA: 1618] [Article Influence: 70.3] [Reference Citation Analysis (2)] |
| 12. | Zaremba-Czogalla M, Dubińska-Magiera M, Rzepecki R. Laminopathies: the molecular background of the disease and the prospects for its treatment. Cell Mol Biol Lett. 2011;16:114-148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 13. | Navarro CL, De Sandre-Giovannoli A, Bernard R, Boccaccio I, Boyer A, Geneviève D, Hadj-Rabia S, Gaudy-Marqueste C, Smitt HS, Vabres P, Faivre L, Verloes A, Van Essen T, Flori E, Hennekam R, Beemer FA, Laurent N, Le Merrer M, Cau P, Lévy N. Lamin A and ZMPSTE24 (FACE-1) defects cause nuclear disorganization and identify restrictive dermopathy as a lethal neonatal laminopathy. Hum Mol Genet. 2004;13:2493-2503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 257] [Article Influence: 11.7] [Reference Citation Analysis (2)] |
| 14. | Kodachi T, Matsumoto S, Mizuguchi M, Osaka H, Kanai N, Nanba E, Ohno K, Yamagata T. Severe demyelination in a patient with a late infantile form of Niemann-Pick disease type C. Neuropathology. 2017;37:426-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 15. | Macicior J, Marcos-Ramiro B, Ortega-Gutiérrez S. Small-Molecule Therapeutic Perspectives for the Treatment of Progeria. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 16. | Rahman MM, Ferdous KS, Ahmed M, Islam MT, Khan MR, Perveen A, Ashraf GM, Uddin MS. Hutchinson-Gilford Progeria Syndrome: An Overview of the Molecular Mechanism, Pathophysiology and Therapeutic Approach. Curr Gene Ther. 2021;21:216-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 17. | Kang SM, Yoon MH, Ahn J, Kim JE, Kim SY, Kang SY, Joo J, Park S, Cho JH, Woo TG, Oh AY, Chung KJ, An SY, Hwang TS, Lee SY, Kim JS, Ha NC, Song GY, Park BJ. Progerinin, an optimized progerin-lamin A binding inhibitor, ameliorates premature senescence phenotypes of Hutchinson-Gilford progeria syndrome. Commun Biol. 2021;4:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 18. | Marcos-Ramiro B, Gil-Ordóñez A, Marín-Ramos NI, Ortega-Nogales FJ, Balabasquer M, Gonzalo P, Khiar-Fernández N, Rolas L, Barkaway A, Nourshargh S, Andrés V, Martín-Fontecha M, López-Rodríguez ML, Ortega-Gutiérrez S. Isoprenylcysteine Carboxylmethyltransferase-Based Therapy for Hutchinson-Gilford Progeria Syndrome. ACS Cent Sci. 2021;7:1300-1310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 19. | Koblan LW, Erdos MR, Wilson C, Cabral WA, Levy JM, Xiong ZM, Tavarez UL, Davison LM, Gete YG, Mao X, Newby GA, Doherty SP, Narisu N, Sheng Q, Krilow C, Lin CY, Gordon LB, Cao K, Collins FS, Brown JD, Liu DR. In vivo base editing rescues Hutchinson-Gilford progeria syndrome in mice. Nature. 2021;589:608-614. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 259] [Cited by in RCA: 328] [Article Influence: 65.6] [Reference Citation Analysis (0)] |
| 20. | Lai WF, Wong WT. Progress and trends in the development of therapies for Hutchinson-Gilford progeria syndrome. Aging Cell. 2020;19:e13175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ortega-Gutiérrez S, Spain; Rodrigues AT, Brazil S-Editor: Gao CC L-Editor: Wang TQ P-Editor: Gao CC