©The Author(s) 2022.
World J Clin Cases. Jun 26, 2022; 10(18): 5957-5964
Published online Jun 26, 2022. doi: 10.12998/wjcc.v10.i18.5957
Published online Jun 26, 2022. doi: 10.12998/wjcc.v10.i18.5957
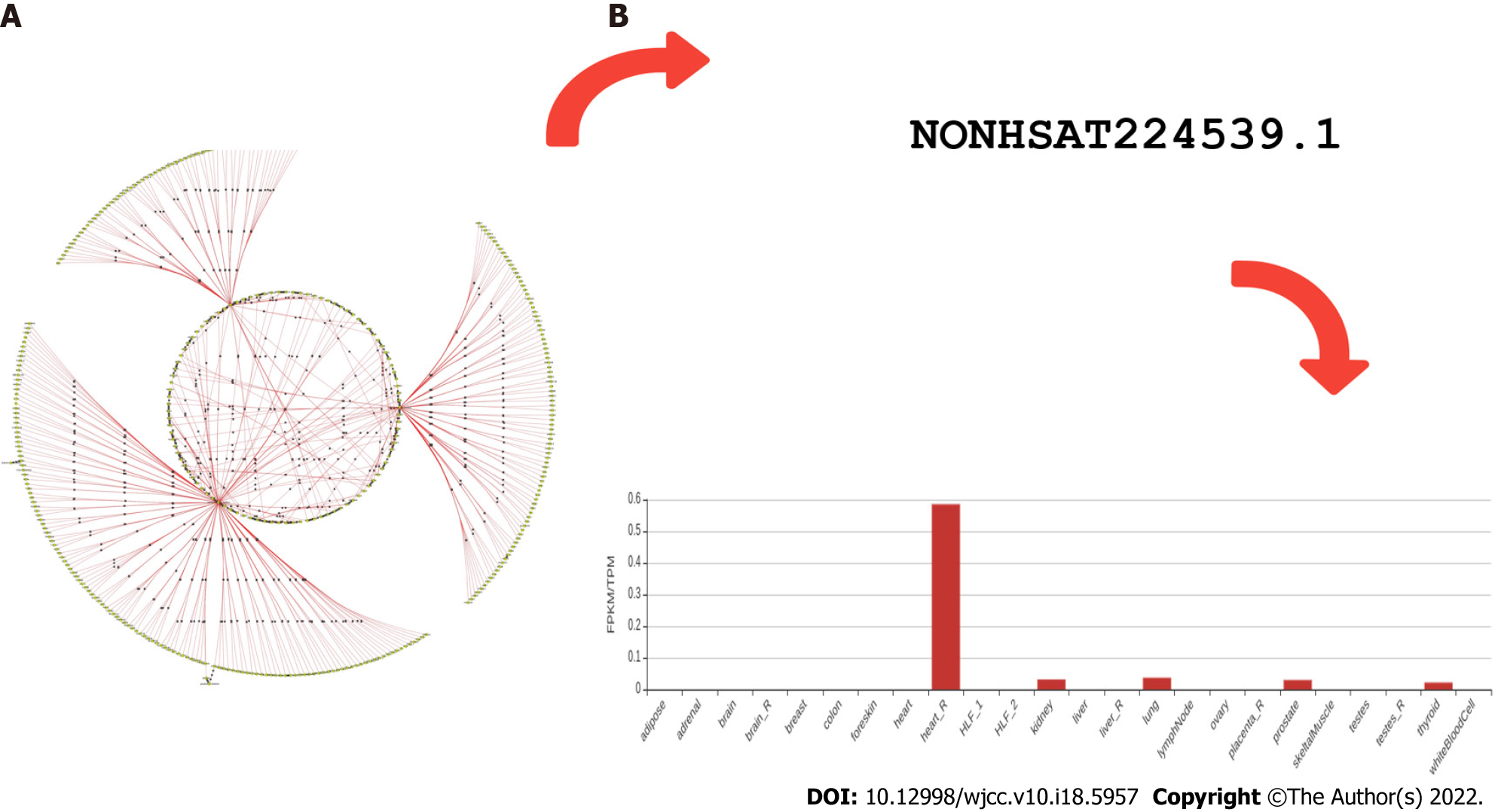
Figure 1 Researchers have chosen interesting genes based on P value, heuristics, and contextuality, and then used CHAT analysis to find high-dimensional gene expression data for confirmation.
Many critical genes, as well as their enriched pathways, were discovered to be involved in the molecular processes of obesity, lupus, adipose tissue, and fatty acid pathways. A: Phenome interactome networks of diabetes represented earlier (Tiwari et al[28], 2018); B: LncRNANONHSAT224539.1 (LINC01128 representative) expression in various tissues, largely seen in the heart, thyroid, kidney, and prostate.
- Citation: Rout M, Kour B, Vuree S, Lulu SS, Medicherla KM, Suravajhala P. Diabetes mellitus susceptibility with varied diseased phenotypes and its comparison with phenome interactome networks. World J Clin Cases 2022; 10(18): 5957-5964
- URL: https://www.wjgnet.com/2307-8960/full/v10/i18/5957.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i18.5957