Published online Jun 25, 2025. doi: 10.5501/wjv.v14.i2.101976
Revised: January 26, 2025
Accepted: February 20, 2025
Published online: June 25, 2025
Processing time: 262 Days and 19.3 Hours
Hepatitis C virus (HCV) infection process of progression encompasses multiple stages, commencing with inflammation and culminating in hepatocellular cancer. Numerous invasive and non-invasive procedures exist for diagnosing chronic HCV infection. Though beneficial, invasive procedures can cause morbidity and inadequate representation of the overall degree of fibrosis. Due to these reasons, non-invasive liver fibrosis biomarkers are becoming more prevalent to diagnose and track liver fibrosis without a liver biopsy. These biomarkers can detect liver fibrosis early, improving treatment and preventing cirrhosis and liver failure. Micro ribonucleic acid (MiRNA) dysregulation causes and worsens several diseases including liver disease. MiRNAs can facilitate the diagnosis of liver fibrosis and serve as a predictive tool to enhance patient care by minimizing invasive procedures and enabling more efficient and prompt therapy.
To investigate the diagnostic effectiveness of several miRNAs (miRNA-122, miRNA-21, miRNA-199a, miRNA-155) in assessing the liver fibrosis severity in untreated HCV patients from the Indian Punjab population. We seek to identify the intricate diagnostic relationship of miRNAs with the extent of fibrosis among individuals with HCV.
We considered 100 persons determined as HCV infected by a quantitative Real-Time Polymerase Chain Reaction examination. We employed statistical as well as probabilistic tools to ascertain the diagnostic validity of miRNAs for determining the liver fibrosis stages. We employed Bayesian Networks, to introduce a unique diagnostic paradigm for miRNAs that can be adopted as benchmark to evaluate the liver fibrosis severity in HCV cases.
We found that miRNAs (miR-122, miR-155 and miR-21) showed significant upregulation when compared with control and according to severity of fibrosis (P ≤ 0.05). The area under the curve for miR-122, miR-155, miR-21 and miR-199a in HCV group in relation to Liver Stiffness Measurement was calculated as 0.889, 0.933, 0.912 and 0.035 respectively. MiR-199a was downregulated according to degree of fibrosis.
Depending on the diagnostic accuracy, we have concluded that miR-122, miR-155 and miR-21 are reliable markers to detect fibrosis in Hepatitis C patients.
Core Tip: Early liver fibrosis detection by biomarkers improves treatment and prevents cirrhosis and liver failure. We investigated various microRNAs (miRNA-122, miRNA-21, miRNA-199a, miRNA-155) to determine liver fibrosis severity in untreated Hepatitis C virus (HCV) patients. Our study examined how miRNAs affect HCV fibrosis diagnosis. We tested miRNAs for liver fibrosis stage using statistical and probabilistic approaches for 100 HCV-positive individuals. Bayesian Networks were used to develop a miRNA diagnostic relationship for HCV liver fibrosis severity. miR-122, miR-155, and miR-21 were significantly elevated relative to controls and fibrosis severity (P ≤ 0.05) whereas MiR-199a decreased with fibrosis. Diagnostically, miR-122, miR-155, and miR-21 are accurate fibrosis biomarkers.
- Citation: Kaur N, Garg R, Tapasvi C, Goyal G. Micro RNAs as a potential biomarker for predicting liver fibrosis severity in hepatitis C virus affected patients. World J Virol 2025; 14(2): 101976
- URL: https://www.wjgnet.com/2220-3249/full/v14/i2/101976.htm
- DOI: https://dx.doi.org/10.5501/wjv.v14.i2.101976
Hepatitis C virus (HCV), member of the Flaviviridae family[1], is a bloodborne single-strand ribonucleic acid (RNA) pathogen that predominantly affects the liver. HCV may lead to both acute and chronic hepatitis, ranging from mild disease to a severe lifelong illness, that may end up in liver cirrhosis or malignancy. Figure 1 shows the 9.6-kb HCV genome, highlighting four well-structured regions for the 5' untranslated region (UTR) and stem-loop structures for the 3' UTR[1,2].
Being a considerable global health issue, HCV requires sustained efforts to avoid transmission and provide appropriate care and treatment for infected individuals. According to the National Centre for Health Statistics, United States around 12717 deaths were caused by HCV in 2022, with an age-standardized mortality rate of 2.89/100000[3]. Liver disease is the predominant reason behind HCV-related deaths. HCV prevalence varies substantially across different ethnicities and countries[4,5]. HCV has been a concerning health problem in India for the past several years, with approximately 0.09%-1.5% of the overall population suffering with the infection[6]. Some regions in India, such as Punjab and the northeastern states, serve as potential hotspots for HCV in contrast to other regions[7].
Most of the HCV-infected individuals may remain asymptomatic during the acute phase and may remain oblivious of their infection until the emergence of chronic liver disease. Chronic HCV infection may develop extreme hepatic adverse effects, such as liver fibrosis, cirrhosis, and hepatocellular carcinoma[8]. Liver fibrosis denotes the unusual proliferation of fibrous connective tissue within the liver, which may compromise its functionality. It commences as a reparative reaction to injury but may result in the buildup of scar tissue in the liver if the injury persists. Liver fibrosis is a progressive disease ranging from mild to severe. The estimation of the liver fibrosis stage becomes crucial to managing the disease progression, as treatment and prognosis differ according to that fibrosis stage[9]. Timely identification and treatment of liver fibrosis can stop disease progression, preventing the development of cirrhosis and its complications.
Liver biopsy is commonly used as a gold standard for diagnosing liver fibrosis and cirrhosis, and it is considered the most potent method of evaluating the degree of disease. However, this method is attributed to significant disadvantages; for instance, it may result in hemorrhage or infection, and it can cause pain and discomfort to the patients[10]. Moreover, this method warrants continuous monitoring and subsequent evaluation of the patient’s post-surgery to track any deterioration in the health conditions. Several studies[11,12] have suggested the possibility of sampling error, leading towards inaccurate reflection of the degree of fibrosis in the liver, leading to wrong diagnosis or incorrect disease staging.
Thus, non-invasive biomarkers are widely seen as substitutes to liver biopsy for evaluating liver fibrosis. Non-invasive biomarkers are either blood tests or imaging techniques that have the potential for detecting and monitoring the progression of liver fibrosis, thereby avoiding liver biopsy. Some examples of non-invasive biomarkers include a blood markers [including hyaluronic acid, fibronectin, laminin, and procollagen-III-peptide, alanine aminotransferase (ALT)-to-platelet ratio index, and the fibrosis index based on four markers], and imaging techniques [including transient elastography, shear wave elastography (SWE), and magnetic resonance imaging]. Several existing studies[11,13,14] have validated the efficacy and diagnostic efficiency of non-invasive biomarkers. The promising findings enabled the extensive utilization of non-invasive biomarkers in clinical practice for evaluating liver fibrosis.
SWE is considered as a reputable non-invasive imaging approach that evaluates liver stiffness among individuals with HCV. It gauges the velocity of a shear wave circulating through tissue, collecting perception on its mechanical characteristics. SWE is especially effective for evaluating liver fibrosis, as fibrotic tissue typically exhibits greater stiffness than healthy tissue. Research[15-17] has demonstrated that SWE can accurately identify liver fibrosis severity among chronic liver patients. These studies exhibited a robust association between liver biopsy and non-invasive SWE in clinical experiments for liver fibrosis evaluation; thus, both can be considered equivalent standards for further studies.
Potential of various micro ribonucleic acid (miRNAs) as biomarkers of severity in chronic liver conditions have been recently explored. Various existing studies[18-20] indicate that miRNAs may function as biomarkers for the finding preliminary evidence of liver fibrosis among people with HCV. The miRNAs, short non-coding RNA molecules, can target and regulate biological processes, hence modifying convoluted gene expression patterns in various cellular functions. miRNAs have a relationship with hepatic stellate cells (HSC) and modulates the migration, differentiation, proliferation, and apoptosis of activated HSCs[21]. Virus host interactions involve various steps for regulating gene expression. The main step is the change of cellular miRNA expression profile. These miRNAs control expression of protein and modulate viral infectivity. HCV infection has a significant effect on cellular miRNA expressions[22,23]. These are discharged into extra cellular space and are conveyed in various manners such as—in alliance alongside Ago 2, exosomes, or high-density lipoprotein in circulation[24].
Prediction of liver fibrosis in HCV using miRNA is emerging as one of the most sought out research potentials in recent times. MiRNA may be readily extracted from blood samples and can identify liver fibrosis without the necessity of invasive liver biopsy, hence minimizing the risk of consequences and pain for the patient. These indicators can detect early-stage liver fibrosis, hence increasing the chances of effective therapy and preventing progression to cirrhosis and liver failure. In recent studies[19,25], non-invasive miRNA-based biomarkers have depicted good diagnostic accuracy in contrast to other comparative biomarkers. For example, miR-122 and miR-21 had a substantial link with ALT levels, indicating that they may serve as better biomarkers compared in individuals with chronic hepatitis[19]. Similarly, in[25], it was discovered that miR-126, miR-122, miR-129, miR-199a, miR-203a, and miR-223, in combination with variables that can predict, can function as an original non-invasive procedure for HCV-associated liver fibrosis staging. In[18], serum concentrations of miR-122 and miR-192 were elevated in chronic HCV patients relative to healthy controls. In[21], elevated miR-122 and miR-155 were mostly linked to liver injury brought on by inflammation and alcohol, whereas these miRNAs were primarily found in the protein-rich fraction in liver injury brought on by toxins.
Despite the introduction of several approaches in recent years, their clinical applicability has been constrained by limited data (based on specific ethnicity or location) supporting their exact accuracy and viability in different populations. Thus, in the current investigation, the significance of miRNA was examined to distinguish the degree of liver fibrosis in HCV-infected patients from the Indian Punjab population. The comprehensive analytical and predictive approach assesses the correlation between miRNA and SWE. Additionally, we also uncover the diagnostic relationship of miRNAs with the liver fibrosis severity stages.
One hundred adult untreated HCV patients, comprising both indoor (hospitalized) and outdoor (outpatients) individuals, who visited the Department of Medicine, Guru Gobind Singh Medical College and Hospital, Faridkot, Punjab, India were included in this investigation. The patients identified as HCV-RNA positive were invited to participate in the cross-sectional analysis. The sample size was determined by a mathematical procedure: N = {(Zα+ Zβ)/C}2 + 3, where α represents the Type I error, denoting the likelihood of rejecting the null hypothesis when it is true; β signifies the Type II error, indicating the probability of accepting the null hypothesis when it is false; and C is the correlation coefficient. A practical non-random sampling method was chosen based on the participants' availability and viability. The samples were taken from the Malwa population, which is vulnerable to using injections for therapeutic purposes in an unsafe manner, which is a common occurrence in HCV clusters[6,7]. Patients with an HCV diagnosis were then recruited based on their liver stiffness measurement (LSM) category, which was determined by the SWE measurement, until the limit of the fibrosis category was achieved (about 25 ± 1 persons per category), at which point no further patients in that category were recruited. Moreover, 50 healthy controls (age and sex matched), seronegative for anti-HCV and hepatitis B surface antigen (HBsAg); and having normal liver and kidney function tests were considered for a comparative analysis. All participants who granted informed consent were enrolled in the study, regardless of gender or age. We have considered several confounding factors (such as medication use, diet and lifestyle factors) while sampling. The project initialized after due authorization from the Institutional Ethics Committee.
Criteria for exclusion: The exclusion criteria were adhered to, resulting in the exclusion of participants with comorbidities from the study. Co-infected patients (such as those with hepatitis B, human immunodeficiency virus, or hepatocellular cancer) identified as positive were excluded from the current study. The subjects with ALT flare (readings surpassing five times the upper normal threshold of 45 U/mL), unreliable or failed liver stiffness assessment, and co-infection with multiple HCV genotypes were omitted from the investigation. None of the examined patients were associated with significant alcohol consumption (> 80 g/day). We employed an alcohol consumption questionnaire to exclude significant alcohol usage (more than 50-60 g/d for males/females). Furthermore, pregnant women were exempted from the investigation.
5-10 mL of intravenous blood was obtained after the sterilization of the venipuncture site employing an alcohol-based swab. The specimen was transferred into an appropriate vacutainer corresponding to the test category, given enough time to coagulate, and afterwards, the resulting serum was extracted by means of centrifugation and examined. The viral indicators were first evaluated, subsequently followed by the quantification of HCV RNA using quantitative real-time polymerase chain reaction (qRT-PCR) performed by a laboratory accredited at the national level and licensed by the Punjab Government.
The following investigations (routine and special) were conducted on the blood samples collected from HCV affected patients as well as controls.
Routine investigations: The AU-480 automated analyzer (Beckman Coulter) was used for routine examinations. Standard tests were run on each sample, including Hemoglobin (Hb), aspartate transaminase (AST), ALT, alkaline phosphatase (ALP), platelet count, international normalized ratio, total serum proteins, serum albumin, and serum bilirubin. Furthermore, the Hematology Analyzer (Erba Elite 580) served to obtain additional routine assessments, encompassing Hb, blood platelet count, and total leukocyte count (TLC).
Special investigations: Several special investigations conducted are discussed below.
SWE: The Liver stiffness was measured using SWE and compared with results obtaining using Serum Fibronectin. The samples for fibronectin levels were drawn on the day of Elastography. It was performed using ultrasound machine Philips Affiniti 70. The system automatically assessed liver stiffness and expressed the results as the shear wave velocity (versus - meters per second). It also automatically calculated the average elastic modulus (in kPa) for the area of interest. A measuring success rate of > 80% was attained after the liver's fifth segment was shot ten to twelve times. Only after ten of these shots were deemed credible was the result deemed valid. Cutoffs: 7-11 kPa - mild; 12-21 kPa - moderate; and > 21 kPa-severe. Patients were classified according to the degree of fibrosis[26].
MicroRNAs: Four miRNAs (miR-122, miR-155, miR199a, and miR-21) were assessed using RT-PCR. The process for miRNA analysis is explained below.
RNA isolation: Total RNA, comprising microRNAs, was obtained employing the miRNeasy Serum/Plasma Kit (Qiagen Kit, Cat No./ID: 217184). Lyophilized C. elegans (Ce) miR-39 miRNA mimic spike-in control served as the internal control. The kit combined of phenol/guanidine-based lysis of serum/ plasma specimens accompanied by silica-membrane-based purification of total RNA. The quality of RNAs was determined using Nano Drop Spectrophotometer (A260/280). Ce-miR-39 was transferred into the serum samples before RNA extraction for subsequent normalization of miRNAs.
Complementary deoxyribonucleic acid and RT-PCR: Micro RNAs were analyzed by following two step process. First, reverse transcription was carried out later to form complementary deoxyribonucleic acid (cDNA) employing miScript II RT kit (Qiagen Kit, Cat No./ID: 218161). The cycling protocol followed was 37 °C/60 minutes, and 95 °C/5 minutes, and storing undiluted cDNA at -20 °C. Mature miRNAs had been polyadenylated by poly (A) polymerase along with reverse transcribed into cDNA employing oligo-dT primers. Secondly, serum expression of mature microRNAs was evaluated using miScript SYBR Green PCR kit on RT-PCR (Qiagen Kit, Cat No./ID: 218073). The thermocycling protocols for the RT-PCR consisted of the following: The initial denaturation process at 95 °C for 15 minutes, followed by 45 cycles consisting of template denaturation at 94 °C for 15 seconds, annealing at 55 °C for 30 seconds, and extension at 70 °C for 30 seconds. The entire process has been performed in triplicate and in compliance to the supplier's recommendations. Cycle threshold (CT) values were recorded of all the miRNAs of HCV patients and healthy controls. Expression of miRNA was reported as delta CT values but due to inverse relationship between delta CT expression further 2-delta CT was calculated for miRNA expression calculation. This is the relative expression value for miRNA. Also, the fold of change,
Delta CT (test) = CT (miRNA in patient) – CT (cemiR39 in patient).
Delta CT (Healthy control) = CT (miRNA in control) – CT (cemiR39 in control).
Delta-delta CT = Delta CT (test) – Delta CT (healthy control).
NER = 2- delta-delta CT.
A statistical analysis was undertaken on the collected data employing SPSS software version 21 for Windows (IBM Corp., Armonk, NY, United States) for determining the significant nature of the investigations carried out. A P value of less than 0.05 was deemed to be highly significant in statistical terms. The comparison of quantitative variables among the research groups was carried out employing the analysis of variance-Kruskal-Wallis technique.
We employed a Complex Bayesian network to do a probabilistic analysis of the data, revealing the intricate link that exists between miRNAs and severity of the condition. Joint probability distributions can be represented graphically using the Bayesian network formalism. They are factorizations of joint probability distributions represented by acyclic directed graphs. Each joint probability distribution over n random variables can be expressed as a product of the probability distributions of each variable conditional on the others and factorized in n! ways[28,29]. A Complex Bayesian model based on the following Bayes theorem, i.e., P (×1, ×2) = P (×1|×2) P (×2) = P (×2|×1) P (×1) = > P (×1|×2) = P (×2|×1) P (×1)/P (×2) was adopted to analyze the probabilistic relationship between special investigations and the disease severity. The model was deployed on the Genie tool (https://www.bayesfusion.com/), a graphical user interface which enables interactive model building and learning.
The outcomes achieved through the clinical assessment and analytical statistics comprising HCV patients are explained below.
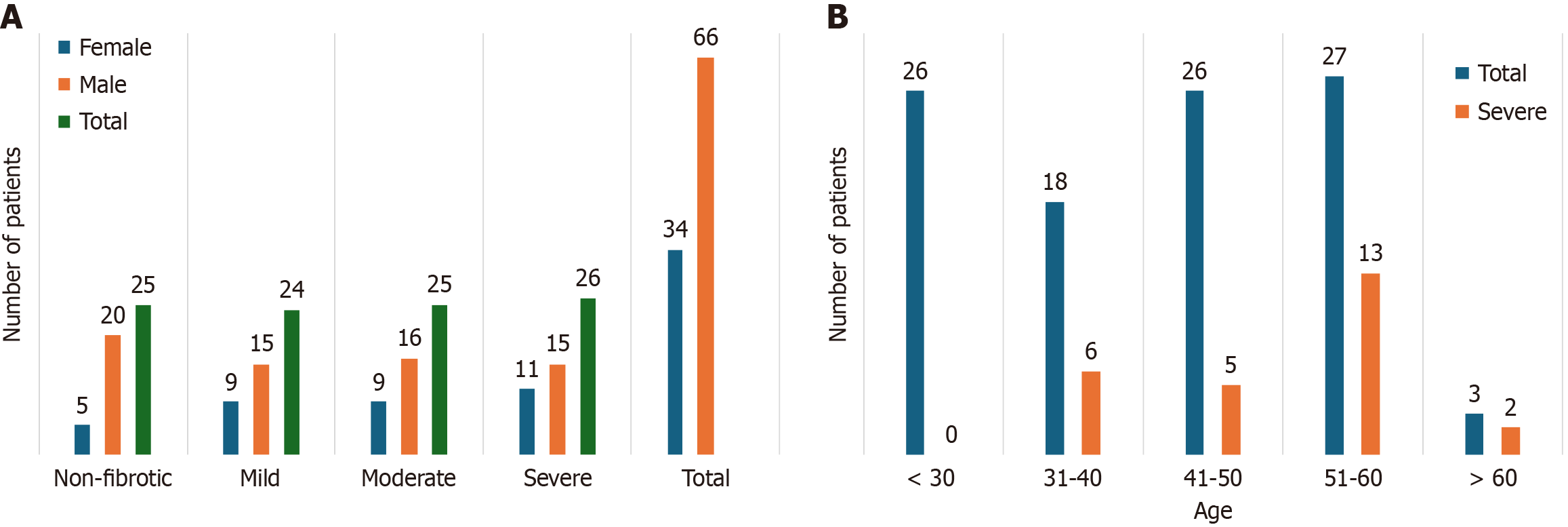
The current investigation encompassed 100 untreated HCV patients, which included 66 males as well as 34 females, having a mean age of 42.7 ± 13 years. In accordance with SWE, determined by LSM values, the study sample was categorized into fibrotic (75 patients) and non-fibrotic classes (25 patients). The patients were grouped into four categories, non-fibrotic, mild, moderate and severe fibrosis. The distribution of females and males across each category is displayed in Figure 2A. The middle-aged patients (51-60 years) depicted higher frequency of severe fibrosis as compared to other age groups (depicted in Figure 2B). LSM values by SWE in HCV group were 14.06 ± 8.05. The fibrotic cases (n = 75) were further divided into three groups on the basis of LSM values, i.e., mild (24), moderate (25), and severe (26).
All the investigations were performed in both groups. The comparison of median and interquartile ranges (IQR) for routine investigations in HCV cases and healthy controls is mentioned in Table 1. The Liver function tests (AST, ALT and ALP) were found as statically significant. Also, Total Proteins, platelet count, and Serum Albumin were significant in HCV Cases with regard to the control group (P ≤ 0.05).
| Parameter | HCV cases | Controls | P value | ||
| Median | IQR | Median | IQR | ||
| Hb | 13.00 | 11.15-14 | 12.90 | 11.6-13.775 | 0.936 |
| INR | 1.08 | 1-1.1925 | 1.00 | 1-1.1 | 0.001 |
| TSP | 7.00 | 6.6-7.2 | 7.65 | 7.3-8 | 0.001 |
| Albumin | 3.90 | 3.5-4.3 | 5.00 | 4.5-5.2 | 0.001 |
| Bilirubin | 0.70 | 0.4125-0.9 | 0.70 | 0.4125-0.9 | 0.988 |
| AST | 72.00 | 43.25-110 | 27.00 | 22-30 | 0.001 |
| ALT | 80.00 | 44.5-126.25 | 21.00 | 16.25-26 | 0.001 |
| ALP | 95.50 | 82-133.5 | 77.00 | 70-96.5 | 0.001 |
| TLC | 7700.00 | 6800-8900 | 7700.00 | 6500-8700 | 0.497 |
| Plt Cnt. (X1000MM3) | 163.50 | 120.75-221.25 | 270.00 | 202.5-347.5 | 0.001 |
miR-122: MiRNA-122 expression (normalized) was calculated following qRT-PCR for both the groups i.e. controls and HCV patients’ group. It was found that mean ± SD of patients and controls were 1.31 ± 1.81 and 0.25 ± 0.19 respectively. This clearly depicts that expression of miR-122 was upregulated in HCV cases relative to healthy controls (P ≤ 0.05). Also, the miRNA expression of control group was compared with fibrotic group (HCV) (Table 2). Also, Fold of change value, i.e., NER for miR-122 was found to be 6.62 ± 9.52 and it showed significant up regulation in comparison to control group.
| HCV cases | Controls | P value | |||
| Median | IQR | Median | IQR | ||
| miR-122 | 0.69 | 0.18-1.54 | 0.18 | 0.125-0.33 | 0.000 |
| miR-155 | 0.66 | 0.22-1.54 | 0.18 | 0.125-0.287 | 0.000 |
| miR-199a | 0.29 | 0.064-1 | 2.64 | 1.89-4 | 0.000 |
| miR-21 | 0.79 | 0.225-1.79 | 0.21 | 0.154-0.35 | 0.000 |
miR-199a: MiRNA-199a was found to be downregulated when we compare HCV study group with control group (Table 2). It was seen that mean ± SD of patients and controls were 1.04 ± 2.07 and 3.33 ± 2.27 respectively Also, Fold of change value, i.e., NER for miR-15 was found to be 0.49 ± 1.07, this shows a significant down regulation when contrasted to control group (P ≤ 0.05).
miR-155: MiRNA-155 expression was calculated following qRT-PCR for both the groups i.e. controls and HCV patients’ group. It was found that mean ± SD of patients and controls were 1.03 ± 1.05 and 0.23 ± 0.17 respectively, which clearly depicts that the expression of miR-155 was upregulated in HCV Fibrotic cases in relation to healthy controls (P ≤ 0.05) (Table 2). Also, Fold of change value i.e. NER for miR-155 was found to be 5.82 ± 6.51 and it showed significant up regulation in comparison to control group.
miR-21: The mean ± SD of miRNA 21 expressions in HCV study group and control group were 1.37 ± 1.69 and 0.26 ± 0.16. These values clearly depict the upregulation of miR-21. Median and IQR values of cases and controls are stated in Table 2. Also, Fold of change value i.e. NER for miR-21 was found to be 6.85 ± 11.21 and it showed significant up regulation relative to control group.
The HCV cases were divided into four groups on basis of the degree of fibrosis (Table 3). The routine and special investigations (mean ± SD) with respect to fibrosis severity i.e. no fibrosis, mild, moderate and severe fibrosis is shown in Table 3. A notable distinction was noted across all groups for AST, ALT, ALP, Platelet count, and all the miRNAs (P ≤ 0.5).
| No fibrosis (n = 25) | Mild (n = 24) | Moderate (n = 25) | Severe (n = 26) | P value | |||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||
| Hb | 12.79 | 2.33 | 12.73 | 2.48 | 12.36 | 2.37 | 12.06 | 2.41 | 0.674 |
| INR | 1.11 | 0.18 | 1.21 | 0.23 | 1.15 | 0.21 | 1.15 | 0.21 | 0.478 |
| TSP | 6.77 | 0.65 | 6.89 | 0.41 | 6.91 | 0.64 | 6.95 | 0.50 | 0.683 |
| Albumin | 3.95 | 0.61 | 3.80 | 0.63 | 3.87 | 0.59 | 3.84 | 0.56 | 0.849 |
| Bilirubin | 0.63 | 0.45 | 0.71 | 0.27 | 0.68 | 0.32 | 0.78 | 0.37 | 0.488 |
| AST | 48.92 | 22.21 | 72.96 | 52.12 | 99.92 | 48.72 | 137.88 | 100.97 | 0.000 |
| ALT | 59.40 | 34.72 | 85.43 | 84.73 | 112.56 | 71.68 | 127.69 | 73.42 | 0.003 |
| ALP | 85.28 | 11.89 | 83.00 | 8.52 | 112.16 | 22.44 | 145.19 | 31.32 | 0.000 |
| TLC | 8584.00 | 1182.05 | 7987.50 | 1375.66 | 7304.00 | 1347.37 | 7203.85 | 1076.10 | 0.000 |
| Plt. Cnt. (X1000MM3) | 226.56 | 92.48 | 194.08 | 72.86 | 166.44 | 61.72 | 137.27 | 45.06 | 0.000 |
| miR122 | 1.10 | 0.97 | 2.32 | 1.97 | 6.90 | 5.66 | 15.65 | 13.81 | 0.000 |
| miR155 | 1.34 | 1.17 | 3.75 | 3.22 | 10.09 | 7.56 | 7.92 | 7.44 | 0.000 |
| miR199a | 1.53 | 1.73 | 0.36 | 0.37 | 0.07 | 0.07 | 0.02 | 0.02 | 0.000 |
| miR21 | 0.93 | 0.92 | 2.40 | 1.77 | 6.52 | 4.24 | 16.97 | 17.77 | 0.000 |
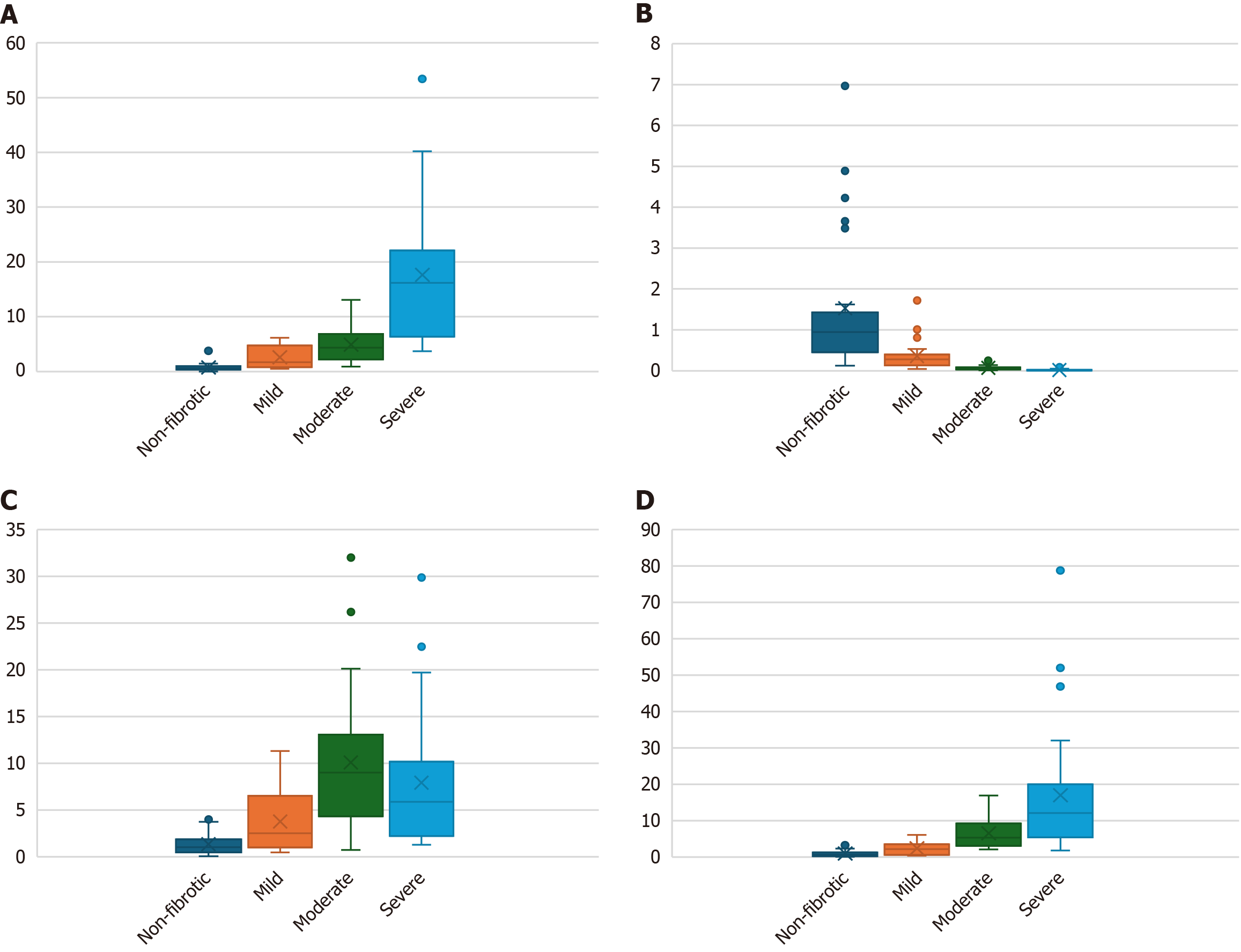
miR-122: Circulating miR-122 was estimated in all the four groups in HCV cases (Table 3). As the staging progresses, the circulating miR-122 expression was also increased in a significant pattern (P ≤ 0.05) as depicted in Figure 3A.
miR-199a: It was estimated in all the four stages in the HCV cases (Table 3). We observed that there was a decrease in miR-199a expression with fibrosis staging as depicted in Figure 3B.
miR-155: MiRNA 155 expression was calculated for all the groups (Table 3). It was observed that up to moderate fibrosis miR-155 was increased. But there was a decrease in severe fibrosis cases relative to the moderate fibrotic group (P ≤ 0.05) as shown in Figure 3C.
miR-21: The expression for miRNA-21 was significantly higher in severe cases (Table 3). As the severity progresses, the miR-21 expression was also elevated (P ≤ 0.05) as shown in Figure 3D.
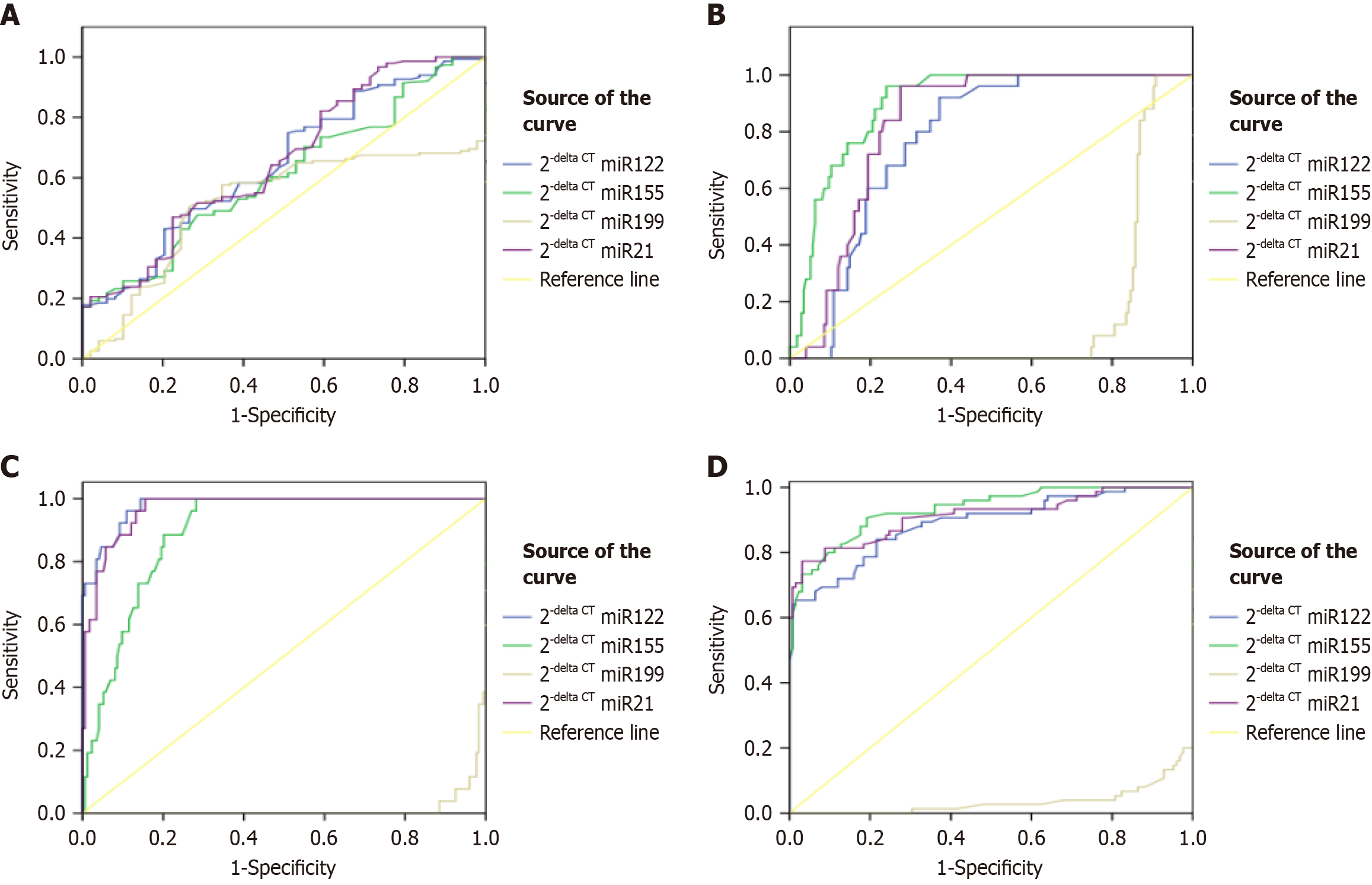
We also revealed the potential of all the miRNAs to discriminate between HCV fibrotic cases. Figure 4 shows the ROC for mild, moderate, severe, and overall HCV cases, respectively. Table 4 depict the area under the receiver operating characteristic curve (AUROC) plot for special investigations (four miRNAs) conducted in the present study. We conducted statistical analysis for all four miRNA and computed the area under the curve (AUC). The statistical results concerning microRNAs for considered cases were compared on the basis of LSM. To compare patients with liver fibrosis with respect to LSM, the AUROC of miRNAs were determined. AUC for depicting severe fibrosis by miR-122, miR-199a, miR-155, and miR-21 was calculated as 0.979, 0.013, 0.896, 0.969, respectively (P ≤ 0.05). Similarly, The AUC for depicting whole fibrotic group in HCV patients by miR-122, miR-199a, miR-155, and miR-21, when compared to LSM values was calculated as 0.889, 0.035, 0.933, and 0.912 respectively. The results depicted that miRNAs’ expressions were highly significant in discriminating between fibrosis patients.
| HCV groups | Micro RNAs | AUC | Sensitivity | Specificity | Youden Index |
| Mild | miR-122 | 0.646 | 79.5% | 40.8% | 0.203 |
| miR-155 | 0.607 | 70.2% | 44.9% | 0.151 | |
| miR-21 | 0.518 | 98% | 24.5% | 0.225 | |
| Moderate | miR-122 | 0.774 | 92% | 62.9% | 0.549 |
| miR-155 | 0.898 | 96% | 76% | 0.720 | |
| miR-21 | 0.826 | 96% | 72.6% | 0.686 | |
| Severe | miR-122 | 0.979 | 96.2% | 89.1% | 0.852 |
| miR-155 | 0.896 | 88.5% | 79.9% | 0.683 | |
| miR-21 | 0.969 | 92.3% | 87.9% | 0.802 |
Correlation coefficient of routine investigations with respect to LSM values: We determined Pearson's correlation coefficient and investigated the importance of liver function tests; Platelet count and TLC. The correlation coefficient (r) for these parameters i.e. AST, ALT, ALP, TLC and Platelet count was 0.428, 0.30, 0.747, -0.391 and -0.473 respectively (P ≤ 0.05).
Correlation coefficient of MiRNAs with respect to LSM values: Table 5 shows the correlation coefficient for all the four miRNAs against LSM values, that were highly significant (P ≤ 0.05).
| miR-122 | miR-155 | miR-199a | mir-21 | ||
| Liver stiffness measurement | Correlation Coefficient | 0.687 | 0.546 | -0.864 | 0.793 |
| P value | 0.000 | 0.000 | 0.000 | 0.000 |
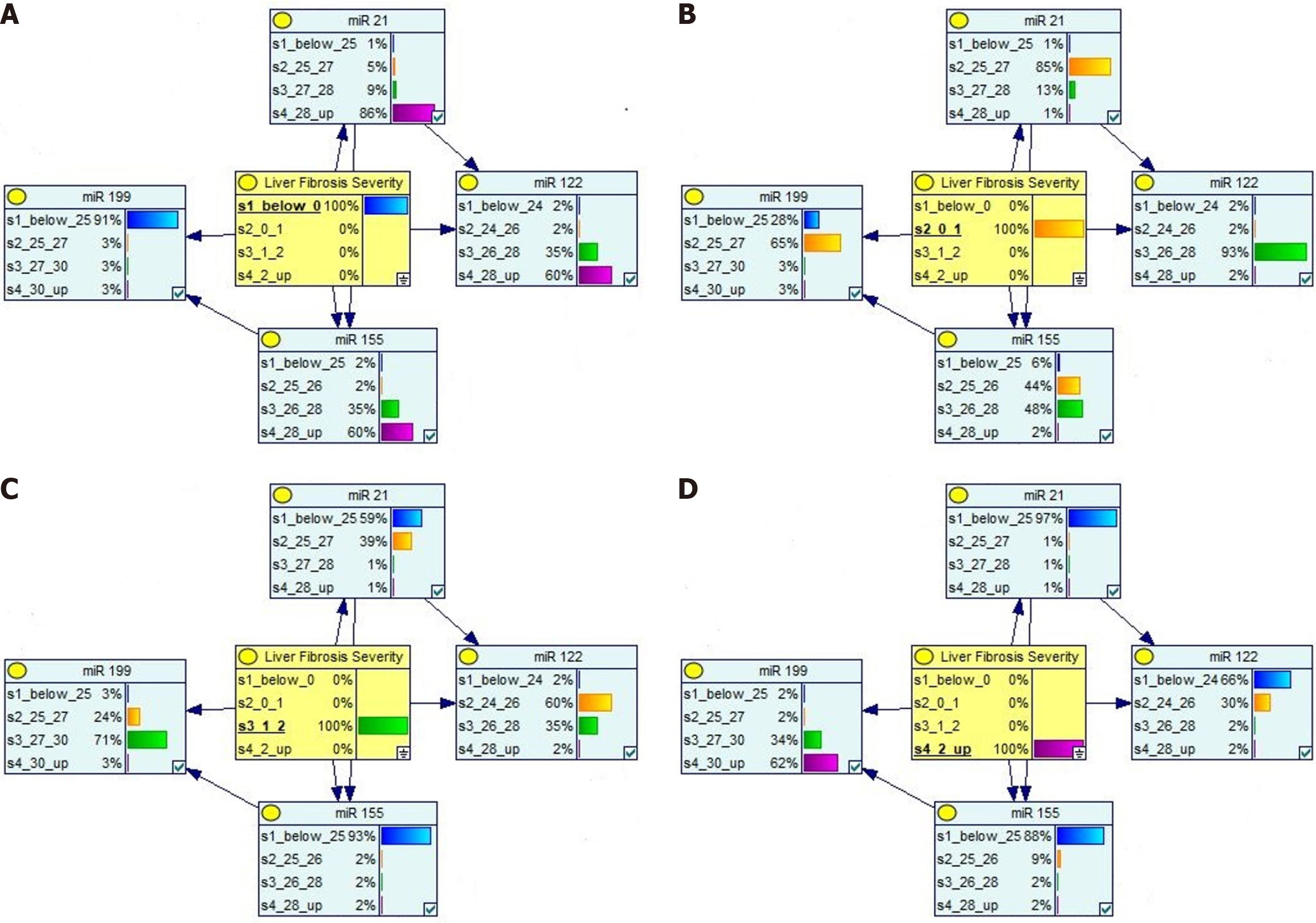
We conducted Bayesian analysis for analyzing the relationship of liver fibrosis severity with respect to conducted investigations. The results clearly depict a strong dependence of all four miRNAs and other routine investigations with respect to an increase in the severity of liver fibrosis. Figure 5 depicts the outcome for Bayesian analysis for four fibrotic stages. These figures clearly show reverse change for miR-199a values with respect to change in miR-21, miR-122, and miR-155 CT values, thereby suggesting a clear change with respect to increase in severity stage.
HCV is transferred via exposure to contaminated blood, typically through the sharing of needles or injection equipment, getting a blood transfusion or organ transplant, or being born to an HCV-positive mother. Liver disease must be detected in the initial stages so that appropriate treatment may be given. But unfortunately, there is currently limited validation of the non-invasive biomarkers to detect liver disease in its initial stages for varied populations. miRNA-based biomarkers possess the potential for clinical application in evaluating liver disease, hence eliminating the necessity for invasive liver biopsy. The adoption of non-invasive miRNAs for identifying liver fibrosis in HCV patients improves patient care by reducing the need for invasive procedures, enhancing diagnostic precision and disease staging, and enabling more efficient and prompt treatment. Thus, the role of miRNA-based biomarkers to regulate HCV via host gene expression modification has been recently popular in clinical studies[30,31].
In this investigation, total of 100 HCV person within Malwa belt of Indian Punjab were considered for investigation alongside healthy controls. The HCV cases grouped into non-fibrotic, mild, moderate and severe showcased significant increase in ALT, AST, ALP and platelet count when compared with each fibrotic group individually. In our study, miR-122 exhibited elevated expression and upregulation in HCV patients relative to the control group (P ≤ 0.05). Also, circulating miR-122 depicted a significantly increase alongside severity of fibrosis (P ≤ 0.05). MiR-122 was recently established as the dominant miRNA in normal liver tissues, representing approximately seventy percent of the miRNAs found inside hepatocytes[32]. Our findings were in line with McMahon et al[33] 2017, which stated that miR-122 increase was observed in early inflammatory phase of fibrosis. Also, it was mentioned in another study that as the cirrhosis progress, the miR-122 release is decreased due to less parenchymal tissue. As the scarring of hepatocytes will increase, the miR-122 release will slow down in cirrhosis phase. Moreover, the miRNA-122 expression in controls vs HCV patients was significant (P ≤ 0.05). The correlation coefficient of miR-122 with respect to LSM was significant (r = 0.687, P = 0.01). These results agree to study by Bihrer et al[18], which stated that ALT and necro inflammatory activities being associated with the circulatory miR-122 high levels in blood. It is highly probable that miR-122 has been liberated from compromised hepatocytes. This pertains to the research conducted by Bala et al[34], which identified elevated levels of miR-122 in the supernatant of JFH-1 infected cells.
The levels of expression of miR-155 increased considerably in all groups among HCV infected. It was significantly higher in HCV patients when contrasted with controls. The potential for growth may arise from these miRNAs being released non-specifically by immune cells or specifically by other cell types in response to HCV infection. A study by Bala et al[34] has established that each of immune cells as well as hepatocytes plays a role to the elevation of miR-155 in circulation. These findings agree with Zhang et al[35] 2012 which stated that up regulation of miR-155 is HCV patients was seen, and it further promote hepatocyte proliferation alongside tumorigenesis through activation of Wnt signaling pathway. Also, it has been mentioned in the study that miR-155 overexpression tends to stimulate β-catenin and further it leads to increase in cyclin D1 which promote cell proliferation. It was concluded by Bala et al[34] that HCV core, Nonstructural 3 and NS5 proteins can mediate upregulated manifestations of miR-155 within HCV population.
miR-21 was also increased in all the groups in HCV cases. Moreover, it was increased in HCV cases relative to the healthy controls. It was actually already stated that miR-21 promotes fibrosis by targeting mothers against decapentaplegic homolog 7 (SMAD7) and activating transforming growth factor-β signaling. MiR-21 can provoke fibrosis by activating HSC, subsequently leading to collagen synthesis. The overexpression of miR-21 facilitates oxidation, thereby enhancing collagen formation, which in turn activates angiotensin. MiR-21 can influence the expression of several proteins by interacting with the 3’-UTR of particular mRNAs. This leads to a complicated interaction network due to the downstream consequences of the signaling pathways[36]. MiR-21 has been demonstrated to serve as a survival factor during hepatic damage and the progression of HCC. Khairy et al[37] identified four miRNAs (miRNA 21, 199, 448, and 181c) and shown that miR-21 is elevated in instances of HCV. Our research pertains to Nasser et al[38].
Expression of circulating miR-199a was depicted a decrease in HCV severity groups. Moreover, it was found to be downregulated when compared to the controls. El-Guendy et al[39] discovered the fact that miR-199a was down-regulated in cases with HCV in contrast to controls. In similar lines, El-Abd et al[40] indicated that serum levels of miR-199a were reduced in chronic HCV patients relative to the control group. As previously mentioned, miR-199a possesses a target sequence within domain II of the IRES region of the HCV 5' UTR[41] and is essential for viral replication. The introduction of miR-199a into replicon cells resulted in an enhancement of viral replication. MiR-199a had an inhibitory effect on HCV replication when mutagenesis analysis was performed[42]. In[43], miR-199a has been reported to inhibit HCV replication through directly targeting stem-loop II region of 5’UTR of HCV genomes when over expressed while its silencing enhances the HCV replication.
In line with the above finding, the trends visible from Bayesian analysis agree to the change with respect to change is severity levels. For example, the CT value for miR-21 varies from > 28 for non-fibrotic stage to < 25 for severe fibrosis stage. The CT value for miR-199 shows opposite trend, with < 25 value for non-fibrotic stage and > 30 for severe fibrosis. This trend agrees with statistical findings, making it strong to conclude the impact of miRNAs in detecting liver disease as well as its severity stages.
We have highlighted several studies in introduction and discussions that have considered different population and regions, but they have also observed similar findings. This suggests that the observed associations between miRNA expression and fibrosis stages are consistent across different populations. Although the specific region under consideration is one of the HCV hotspots, but we suggest further validations on different population and regions. Based on the analysis of existing studies, we considered four miRNAs in the current study, however, we anticipate the need to expand this study to include additional miRNAs. This study highlights the role of miRNAs in assessing the liver fibrosis progression. In the future, this study can be adopted to determine the approximate range that clinicians and doctors can utilize for additional research or to identify the phases of fibrosis in HCV patients. Furthermore, a large-scale study should be conducted on varied and diverse population base to validate the outcomes. Nowadays, anti-miRNAs[44] and miRNA mimics are also considered to have potential to suppress the targeted miRNA in a specific disease. So, we aim to perform a follow-up study considering anti-miRNAs action and lifestyle/diet changes to slow the fibrosis progression in HCV patients.
In conclusion, the four miRNAs are found to be reliable markers in comparison to LSM values. They play an essential part in HCV replication and are substantially expressed in HCV groups juxtaposed with controls. Also, these parameters are showing significance with increase in degree of fibrosis severity in our study. We recommend further studies on large scale population.
We would like to acknowledge Dr Satnam Singh, Senior Entomologist, Punjab Agriculture University, Regional Research Station Faridkot, India for his support and advice during the study.
| 1. | Wan H, Adams RL, Lindenbach BD, Pyle AM. The In Vivo and In Vitro Architecture of the Hepatitis C Virus RNA Genome Uncovers Functional RNA Secondary and Tertiary Structures. J Virol. 2022;96:e0194621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 2. | Chevaliez S, Pawlotsky JM. HCV Genome and Life Cycle. Hepatitis C Viruses: Genomes and Molecular Biology. Norfolk, United Kingdom: Horizon Bioscience, 2006.. |
| 3. | Viral Hepatitis Surveillance-United States. Numbers and rates of deaths with hepatitis C listed as a cause of death† among residents, by demographic characteristics-United States, 2018-2022. Available from: https://www.cdc.gov/hepatitis/statistics/2022surveillance/hepatitis-c/table-3.8.htm. |
| 4. | Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61:77-87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1077] [Cited by in RCA: 1160] [Article Influence: 105.5] [Reference Citation Analysis (0)] |
| 5. | Yang J, Qi JL, Wang XX, Li XH, Jin R, Liu BY, Liu HX, Rao HY. The burden of hepatitis C virus in the world, China, India, and the United States from 1990 to 2019. Front Public Health. 2023;11:1041201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 35] [Reference Citation Analysis (0)] |
| 6. | Puri P, Anand AC, Saraswat VA, Acharya SK, Dhiman RK, Aggarwal R, Singh SP, Amarapurkar D, Arora A, Chhabra M, Chetri K, Choudhuri G, Dixit VK, Duseja A, Jain AK, Kapoorz D, Kar P, Koshy A, Kumar A, Madan K, Misra SP, Prasad MV, Nagral A, Puri AS, Jeyamani R, Saigal S, Sarin SK, Shah S, Sharma PK, Sood A, Thareja S, Wadhawan M. Consensus Statement of HCV Task Force of the Indian National Association for Study of the Liver (INASL). Part I: Status Report of HCV Infection in India. J Clin Exp Hepatol. 2014;4:106-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 7. | Dhiman RK, Satsangi S, Grover GS, Puri P. Tackling the Hepatitis C Disease Burden in Punjab, India. J Clin Exp Hepatol. 2016;6:224-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 8. | Woo J, Choi Y. Biomarkers in Detection of Hepatitis C Virus Infection. Pathogens. 2024;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 9. | Suk KT, Kim DJ. Staging of liver fibrosis or cirrhosis: The role of hepatic venous pressure gradient measurement. World J Hepatol. 2015;7:607-615. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 61] [Article Influence: 5.5] [Reference Citation Analysis (2)] |
| 10. | Rockey DC, Caldwell SH, Goodman ZD, Nelson RC, Smith AD; American Association for the Study of Liver Diseases. Liver biopsy. Hepatology. 2009;49:1017-1044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1449] [Cited by in RCA: 1638] [Article Influence: 96.4] [Reference Citation Analysis (2)] |
| 11. | Thanapirom K, Suksawatamnuay S, Tanpowpong N, Chaopathomkul B, Sriphoosanaphan S, Thaimai P, Srisoonthorn N, Treeprasertsuk S, Komolmit P. Non-invasive tests for liver fibrosis assessment in patients with chronic liver diseases: a prospective study. Sci Rep. 2022;12:4913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (1)] |
| 12. | Bedossa P, Dargère D, Paradis V. Sampling variability of liver fibrosis in chronic hepatitis C. Hepatology. 2003;38:1449-1457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1193] [Cited by in RCA: 1409] [Article Influence: 61.3] [Reference Citation Analysis (0)] |
| 13. | Yamauchi M, Nakajima H, Ohata M, Hirakawa J, Mizuhara Y, Nakahara M, Kimura K, Fujisawa K, Kameda H. Detection of fibronectin receptor in sera: its clinical significance as a parameter of hepatic fibrosis. Hepatology. 1991;14:244-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 14. | Keskin S, Babaoglu O, Keskin Z. An investigation of the efficacy of shear wave elastography in the characterization of benign and malignant liver lesions. Pol J Radiol. 2022;87:e462-e468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 15. | Suda T, Okawa O, Masaoka R, Gyotoku Y, Tokutomi N, Katayama Y, Tamano M. Shear wave elastography in hepatitis C patients before and after antiviral therapy. World J Hepatol. 2017;9:64-68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 16. | Inglot M, Pozowski P, Misiak P, Fleischer-Stępniewska K, Lewandowski Ł, Bilski M, Zaleska-Dorobisz U. Evaluation of liver fibrosis in HCV-infected patients using two-dimensional shear-wave elastography (2D-SWE) before and after antiviral treatment. J Ultrason. 2024;24:1-10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 17. | Zayadeen AR, Hijazeen S, Smadi M, Fayyad L, Halasa M, Alqusous S, Alrabadi O, Hijazeen R, Ajlouni Y, Tulenko K, Malik P. Comparing shear wave elastography with liver biopsy in the assessment of liver fibrosis at King Hussein Medical Center. Egypt Liver J. 2022;12:24. [RCA] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 18. | Bihrer V, Friedrich-Rust M, Kronenberger B, Forestier N, Haupenthal J, Shi Y, Peveling-Oberhag J, Radeke HH, Sarrazin C, Herrmann E, Zeuzem S, Waidmann O, Piiper A. Serum miR-122 as a biomarker of necroinflammation in patients with chronic hepatitis C virus infection. Am J Gastroenterol. 2011;106:1663-1669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 161] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 19. | Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 423] [Cited by in RCA: 476] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 20. | Ullah A, Yu X, Odenthal M, Meemboor S, Ahmad B, Rehman IU, Ahmad J, Ali Q, Nadeem T. Circulating microRNA-122 in HCV cirrhotic patients with high frequency of genotype 3. PLoS One. 2022;17:e0268526. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 21. | Kitano M, Bloomston PM. Hepatic Stellate Cells and microRNAs in Pathogenesis of Liver Fibrosis. J Clin Med. 2016;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 99] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 22. | Shrivastava S, Steele R, Ray R, Ray RB. MicroRNAs: Role in Hepatitis C Virus pathogenesis. Genes Dis. 2015;2:35-45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 72] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 23. | El-Ahwany E, Nagy F, Zoheiry M, Shemis M, Nosseir M, Taleb HA, El Ghannam M, Atta R, Zada S. Circulating miRNAs as Predictor Markers for Activation of Hepatic Stellate Cells and Progression of HCV-Induced Liver Fibrosis. Electron Physician. 2016;8:1804-1810. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 24. | Lemoinne S, Thabut D, Housset C, Moreau R, Valla D, Boulanger CM, Rautou PE. The emerging roles of microvesicles in liver diseases. Nat Rev Gastroenterol Hepatol. 2014;11:350-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 140] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 25. | Shaker OG, Senousy MA. Serum microRNAs as predictors for liver fibrosis staging in hepatitis C virus-associated chronic liver disease patients. J Viral Hepat. 2017;24:636-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 26. | Osman AM, El Shimy A, Abd El Aziz MM. 2D shear wave elastography (SWE) performance versus vibration-controlled transient elastography (VCTE/fibroscan) in the assessment of liver stiffness in chronic hepatitis. Insights Imaging. 2020;11:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 27. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149116] [Cited by in RCA: 139369] [Article Influence: 5574.8] [Reference Citation Analysis (3)] |
| 28. | Ma SX, Dhanaliwala AH, Rudie JD, Rauschecker AM, Roberts-Wolfe D, Haddawy P, Kahn CE Jr. Bayesian Networks in Radiology. Radiol Artif Intell. 2023;5:e210187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 29. | Campbell G, Irony T, Pennello G, Thompson L. Bayesian Statistics for Medical Devices: Progress Since 2010. Ther Innov Regul Sci. 2023;57:453-463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 30. | Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577-1581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1993] [Cited by in RCA: 2001] [Article Influence: 95.3] [Reference Citation Analysis (0)] |
| 31. | Schult P, Roth H, Adams RL, Mas C, Imbert L, Orlik C, Ruggieri A, Pyle AM, Lohmann V. microRNA-122 amplifies hepatitis C virus translation by shaping the structure of the internal ribosomal entry site. Nat Commun. 2018;9:2613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 104] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 32. | Demerdash HM, Hussien HM, Hassouna E, Arida EA. Detection of MicroRNA in Hepatic Cirrhosis and Hepatocellular Carcinoma in Hepatitis C Genotype-4 in Egyptian Patients. Biomed Res Int. 2017;2017:1806069. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 33. | McMahon BJ, Bruden D, Townshend-Bulson L, Simons B, Spradling P, Livingston S, Gove J, Hewitt A, Plotnik J, Homan C, Espera H, Negus S, Snowball M, Barbour Y, Bruce M, Gounder P. Infection With Hepatitis C Virus Genotype 3 Is an Independent Risk Factor for End-Stage Liver Disease, Hepatocellular Carcinoma, and Liver-Related Death. Clin Gastroenterol Hepatol. 2017;15:431-437.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 34. | Bala S, Tilahun Y, Taha O, Alao H, Kodys K, Catalano D, Szabo G. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J Transl Med. 2012;10:151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 131] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 35. | Zhang Y, Wei W, Cheng N, Wang K, Li B, Jiang X, Sun S. Hepatitis C virus-induced up-regulation of microRNA-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology. 2012;56:1631-1640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 260] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 36. | Liu RH, Ning B, Ma XE, Gong WM, Jia TH. Regulatory roles of microRNA-21 in fibrosis through interaction with diverse pathways (Review). Mol Med Rep. 2016;13:2359-2366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 37. | Khairy RMM, Hmmad SS, Sayed M, Ahmed HA, Esmail MAM. Serum MicroRNAs as predictors for fibrosis progression and response to direct-acting antivirals treatment in hepatitis C virus genotype-4 Egyptian patients. Int J Clin Pract. 2021;75:e13954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 38. | Nasser MZ, Zayed NA, Mohamed AM, Attia D, Esmat G, Khairy A. Circulating microRNAs (miR-21, miR-223, miR-885-5p) along the clinical spectrum of HCV-related chronic liver disease in Egyptian patients. Arab J Gastroenterol. 2019;20:198-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 39. | El-Guendy NM, Helwa R, El-Halawany MS, Abdel Rahman Ali S, Tantawy Aly M, Hasan Alieldin N, Fouad SA, Saeid H, Abdel-Wahab AH. The Liver MicroRNA Expression Profiles Associated With Chronic Hepatitis C Virus (HCV) Genotype-4 Infection: A Preliminary Study. Hepat Mon. 2016;16:e33881. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 40. | El-Abd NE, Fawzy NA, El-Sheikh SM, Soliman ME. Circulating miRNA-122, miRNA-199a, and miRNA-16 as Biomarkers for Early Detection of Hepatocellular Carcinoma in Egyptian Patients with Chronic Hepatitis C Virus Infection. Mol Diagn Ther. 2015;19:213-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 41. | Honda A, Arai Y, Hirota N, Sato T, Ikegaki J, Koizumi T, Hatano M, Kohara M, Moriyama T, Imawari M, Shimotohno K, Tokuhisa T. Hepatitis C virus structural proteins induce liver cell injury in transgenic mice. J Med Virol. 1999;59:281-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 42. | Pietschmann T. Regulation of hepatitis C virus replication by microRNAs. J Hepatol. 2009;50:441-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 43. | Murakami Y, Aly HH, Tajima A, Inoue I, Shimotohno K. Regulation of the hepatitis C virus genome replication by miR-199a. J Hepatol. 2009;50:453-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 171] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 44. | de Paz A, Cerro E, Barquero N. AntimiRs: A New Frontier in the Treatment of Complex Diseases. Available from: https://www.farmabiotec.com/digital-versions/magazines/pdf/13/60/index.html. |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/