Published online Jun 25, 2025. doi: 10.5501/wjv.v14.i2.100001
Revised: December 30, 2024
Accepted: January 23, 2025
Published online: June 25, 2025
Processing time: 322 Days and 1 Hours
The evolutionary mutational changes of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) since its emergence in Chhattisgarh, India in 2020 have warranted the need for the characterization of every lineage/sublineage that has evolved until February 2024.
To unravel the evolutionary pathway of SARS-CoV-2 in Chhattisgarh from 2020 to February 2024.
A total of 635 coronavirus disease 2019 cases obtained between 2020 and February 2024 were investigated by whole genome sequencing.
Whole genome sequencing analysis identified the evolution of SARS-CoV-2 into seventeen lineages from 2020 to 2024. SARS-CoV-2 initially emerged in Chha
SARS-CoV-2 from 2020 to 2024 has evolved into 17 lineages/sublineages in Chhattisgarh. The presently circulating JN.1 harbored 40 mutations, especially E554K, A570V, P621S, and P1143 L, capacitating the virus with features of host cell entry, stability, replication, rapid transmissibility, and crucial immune evasion. Therefore, earlier immunity from either vaccination or prior infection may not protect against the current lineage and increases the possibility of future outbreaks. Thus, the periodical genomic surveillance of SARS-CoV-2 is essential for the genomic blueprint of the circulating virus, which may help in updating the vaccine strain and various basic research for developing appropriate therapeutics and diagnostics.
Core Tip: Chhattisgarh, a central state of India is still being affected by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causing coronavirus disease 2019 infection in humans, even in 2024 after four years from the time of its first emergence in April, 2020. No specific report giving details of each and every variant with their mutational pattern evolved over these four years is available. This study has provided insight of evolutionary journey underwent by the virus to unravel SARS-CoV-2 Lineages circulating predominantly in Chhattisgarh at different time period along with genomic pattern of crucial mutations and their effect. The details of circulating lineages with their prevalence rate and mutational pattern would help the researchers and policy makers to update the vaccine strain for persisting its effectivity against new variants or providing baseline data for developing new therapeutics.
- Citation: Singh P, Khare R, Sharma K, Bhargava A, Negi SS. Alpha to JN.1 variants: SARS-CoV-2 genomic analysis unfolding its various lineages/sublineages evolved in Chhattisgarh, India from 2020 to 2024. World J Virol 2025; 14(2): 100001
- URL: https://www.wjgnet.com/2220-3249/full/v14/i2/100001.htm
- DOI: https://dx.doi.org/10.5501/wjv.v14.i2.100001
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), since its emergence at the end of 2019, has exhibited remarkable, continuous genomic mutational changes to evolve into several variants of interest (VOI), variants of concern (VOC), and others. The World Health Organization (WHO) has described these variants and has disseminated information about their transmission to cause coronavirus disease 2019 (COVID-19) pandemic[1,2]. The persistent emergence of these variants has been associated with higher transmissibility, virulence, and resistance to antibody-mediated immune response developed either due to prior infection or vaccination, capacitating SARS-CoV-2 to infect more people[2]. Ineffective/inadequate immune response has in fact provided a conducive milieu for virus evolution, leading to its further diversification.
India has witnessed three catastrophic waves of the COVID-19 pandemic, starting first in 2020 (D614G SARS-CoV-2 variant), followed by the second in 2021 (the deadly Delta variant wave), and the third in 2022 (the extremely transmissible Omicron variant). These variants were reported in all the states of India. Chhattisgarh, a state located in central India, also underwent these waves and reported 1188403 cases infected with COVID-19 till February 2024, along with 14196 deaths[3]. It is pertinent to decipher different variant forms of the virus since its emergence in Chhattisgarh to better characterize mutational events and their effect[4]. Accordingly, this study was undertaken to unravel the progressive emergence of various lineages/sublineages of SARS-CoV-2 that evolved in Chhattisgarh from 2020 to February 2024, along with their mutational changes and their possible effects.
Being a member of the Indian SARS-CoV-2 Genomics Consortium, Government of India, the genomic surveillance of SARS-CoV-2 routinely performed at the state-level Viral Research and Diagnostic Laboratory (VRDL), All India Institute of Medical Sciences, Raipur, Chhattisgarh.
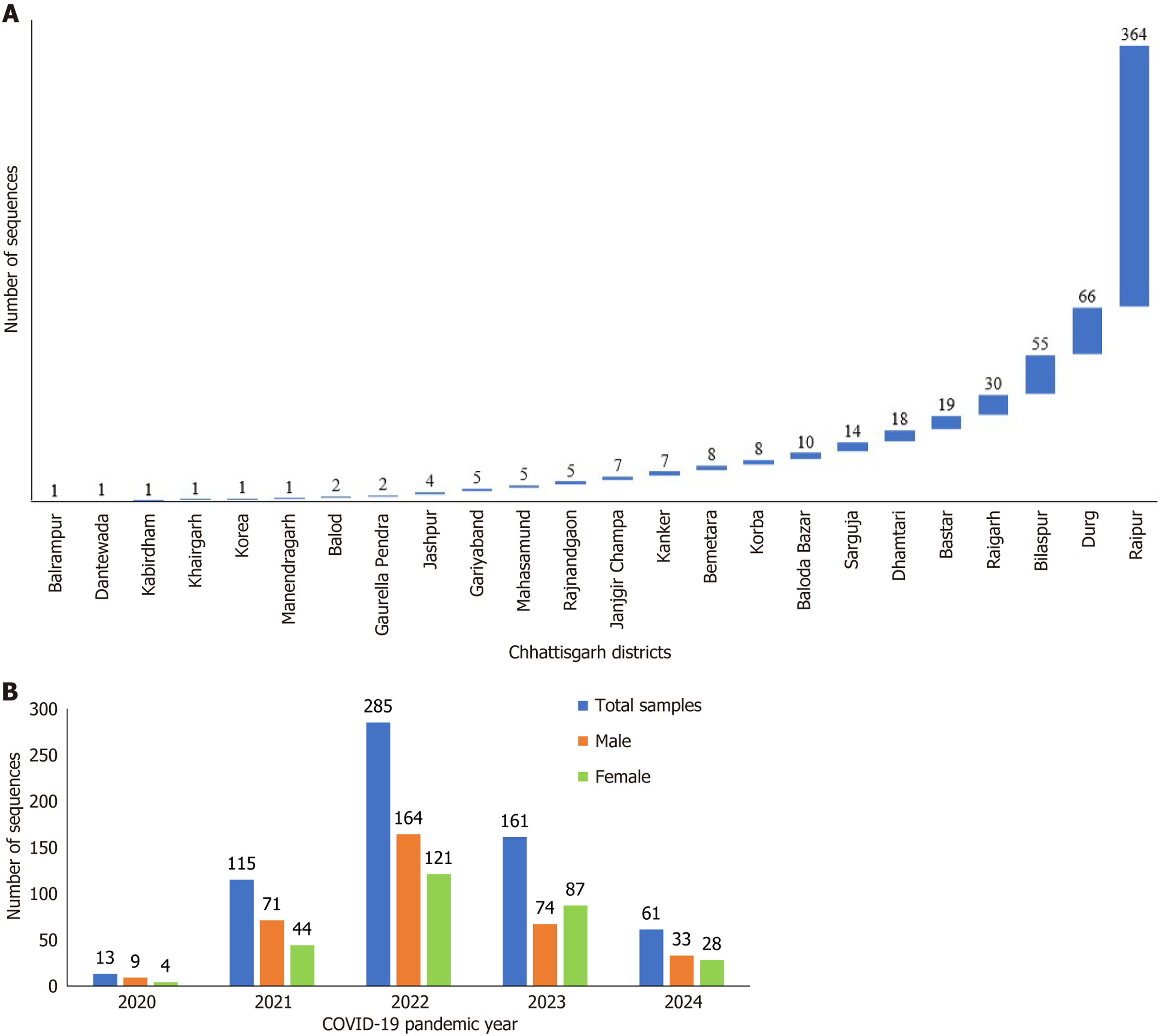
A total of 635 cases of whole genome sequencing (WGS) of SARS-CoV-2 were included in the study. Five hundred and fifty-eight cases of WGS of SARS-CoV-2 were reported by the Viral Research and Diagnostic Laboratory from an equal number of patients who were positive for COVID-19 during the earlier three waves of the pandemic between 2020 and 2023 and were retrospectively analyzed in this study. The year-wise details of these sequences comprised 13 in 2020, 115 in 2021, 285 in 2022, and 145 in 2023 until the month of October. From November 2023 to February 2024, an upsurge in COVID-19 cases was reported, and the laboratory received a total of 149 SARS-CoV-2-positive combined samples of naso- and oropharyngeal swabs from different districts of Chhattisgarh. Out of 149 samples, only 77 samples showing Ct value ≤ 25 giving evidential confirmation of optimal RNA levels for WGS were included in the study and further processed for sequencing[5]. The institutional ethical approval was received via letter number IEC/AIIMS/RPR/1453/2021.
Briefly, processing of clinical samples included RNA isolation, cDNA preparation, DNA amplification, enrichment, tagmentation, and adapter ligation as described earlier[6,7]. The SARS-CoV-2 WGS was performed on the Illumina MiniSeq sequencer using the Mid Output Reagent Cartridge (300 cycles) in 150 × 2 PE read and Fast Q mode using the MiniSeq local run manager Illumina BaseSpace (https://basespace.illumina.com) DRAGEN COVID Lineage (version: 3.5.10) bioinformatics was used for data QC, FASTQ generation, genome assembly, and SARS-CoV-2 variant detection and to generate a single consensus FASTA file[6,7]. The sequences were successfully submitted to GISAID and assigned the accession number EPI_SET_240227op.
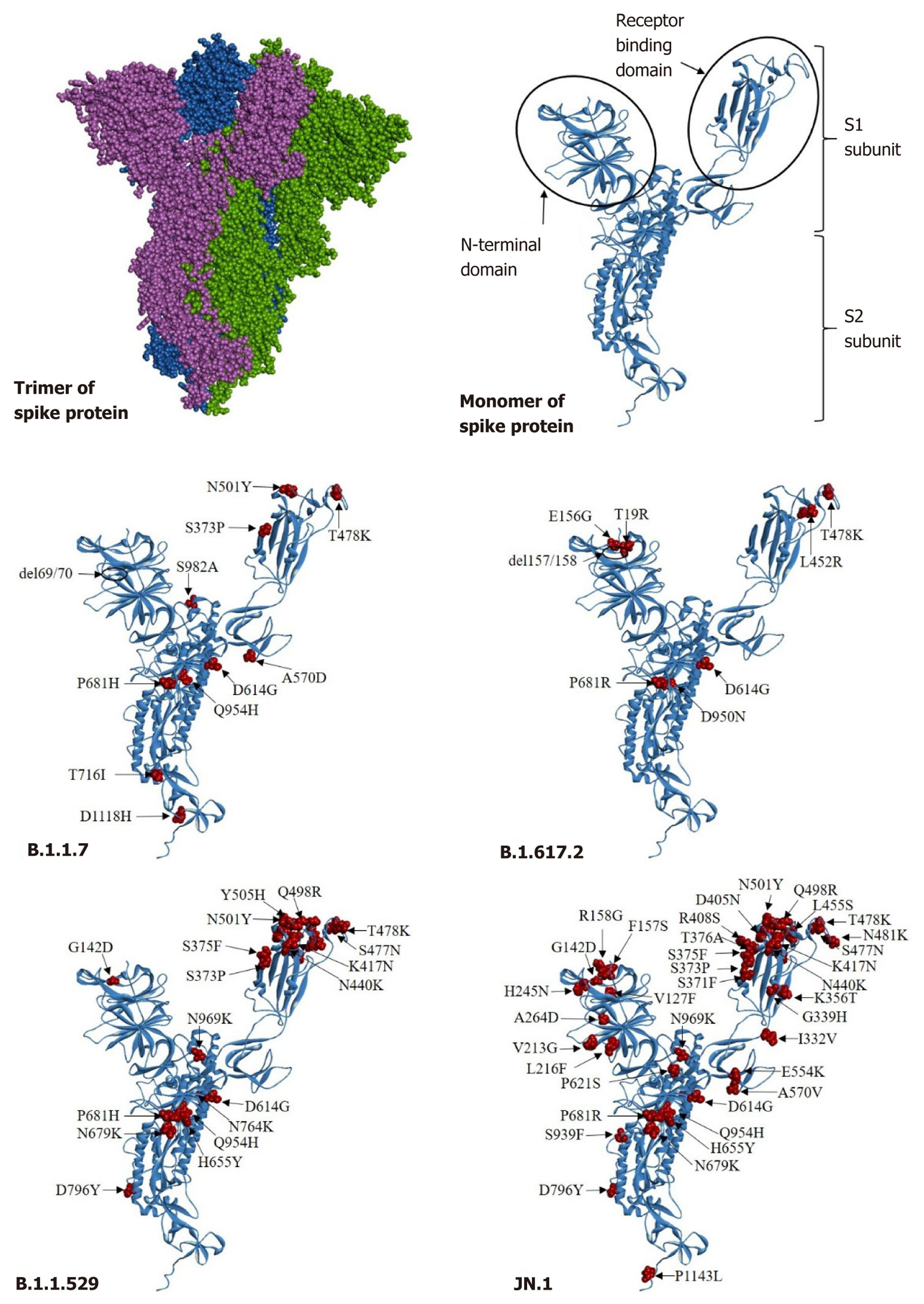
The consensus FASTA files identified using the online Phylogenetic Assignment of Named Global Epidemic Lineages v4.3 (https://pangolin.cog-uk.io) and Nextclade v3.2.0 (https://clades.nextstrain.org/) were used for depicting a phylogenetic tree using the neighbor-joining method and the Kimura 2-Parameter model of MEGA-X software[8]. A bootstrap resampling process with 1000 replications was performed to analyze the robustness of the individual nodes. The annotation was performed using Interactive Tree of Life version 6 (iTOLv6)[9]. The reference sequences of SARS-CoV/2003 were used to create outgroup. The mutations accumulated at the spike protein in monomeric and trimeric structures were compared among the co-circulating variants.
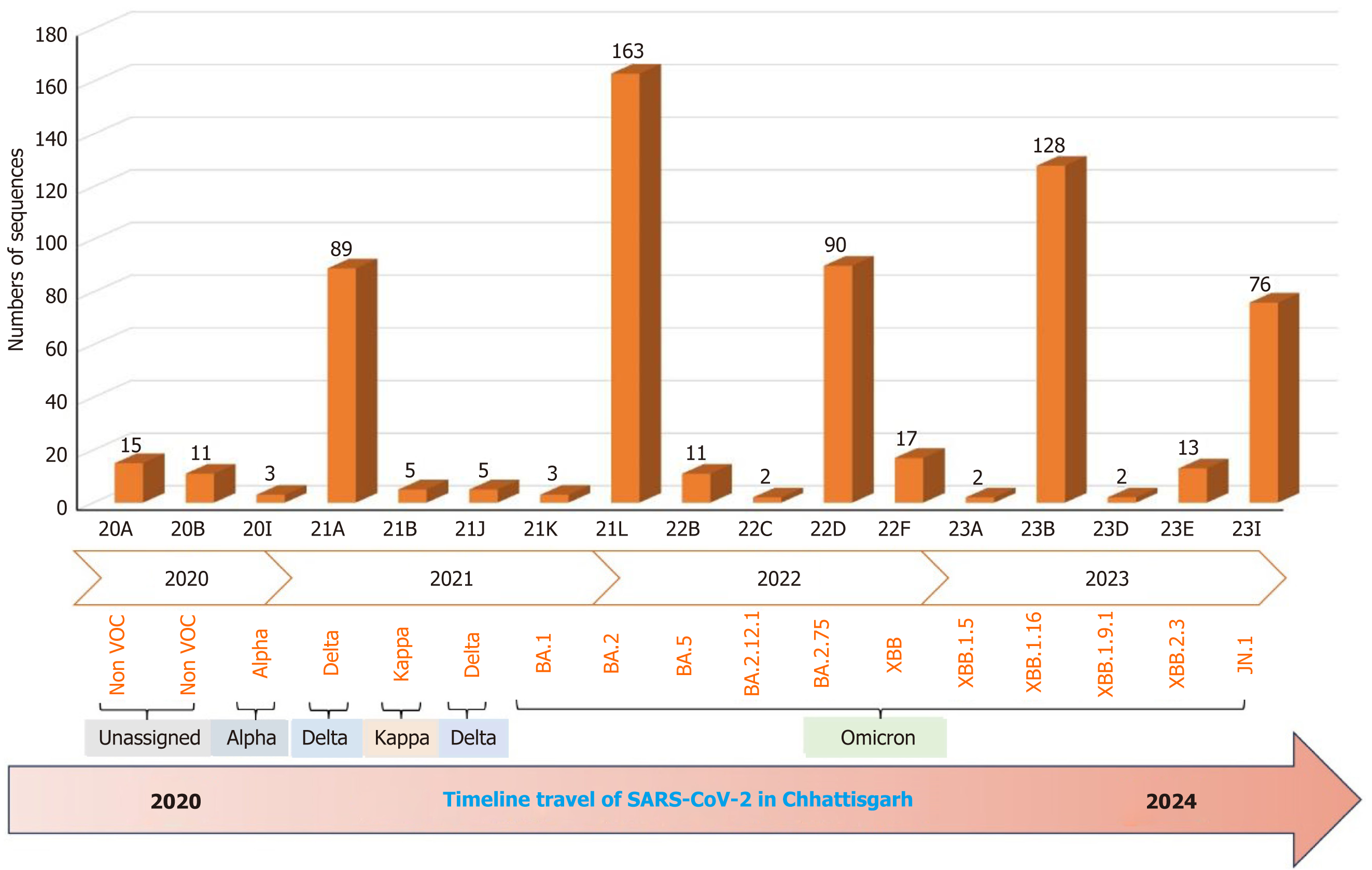
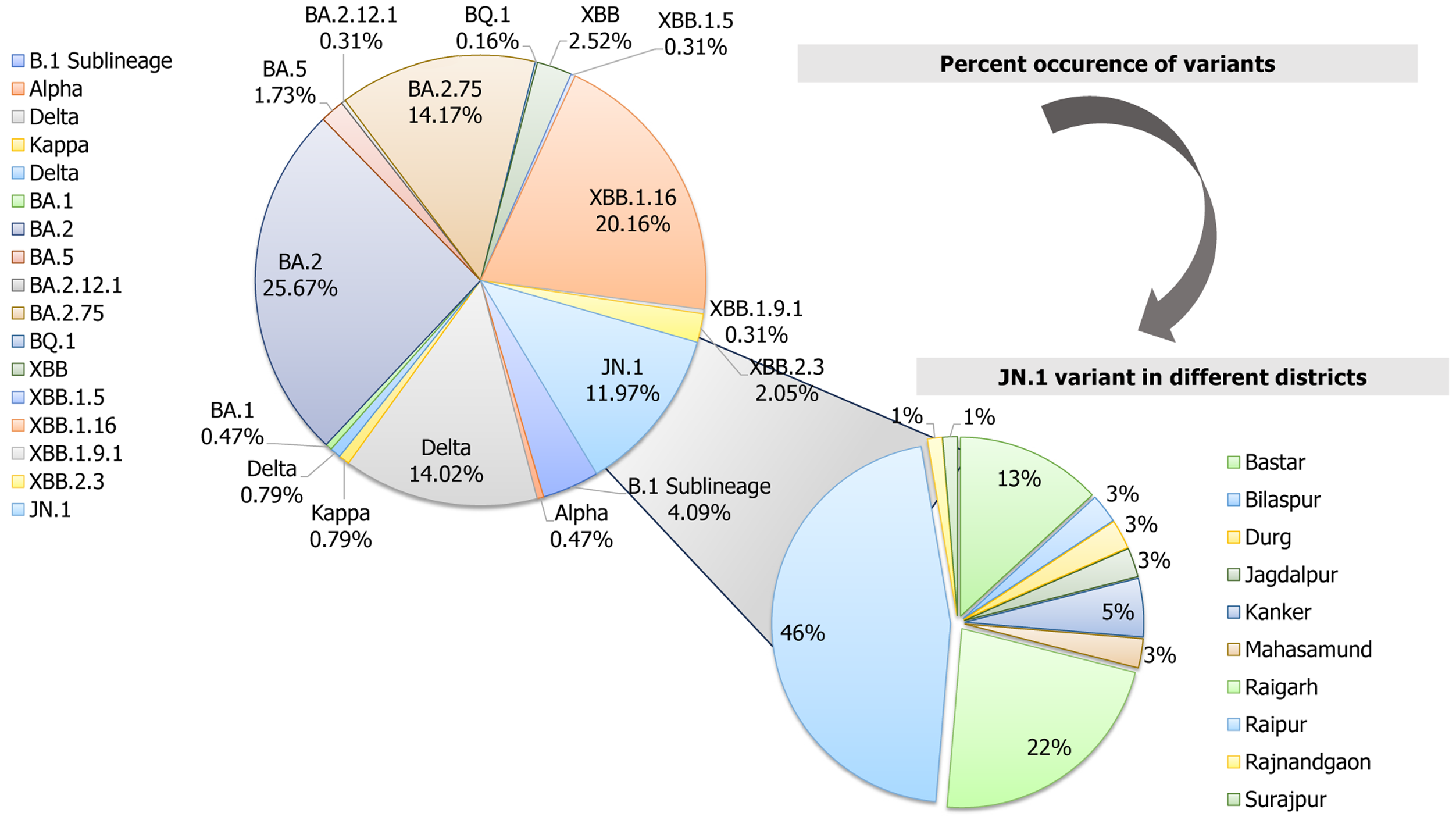
WGS analysis unraveled the dramatic evolution of SARS-CoV-2 over the last 4 years into 17 various lineage/clade variants (Figure 1). The major four lineages observed were BA.1.1.7 (Alpha) in 2020, B.1.617.2 (Delta) in the period between 2021 and mid-2022, B.1.1.529 (Omicron) in late 2022-2023, and Omicron-JN.1 in early 2024 (Figure 2). WGS analysis revealed that the JN.1 variant is presently circulating in Chhattisgarh with its evidential prevalence in 80.51% (n = 62/77) of cases, followed by its sublineage JN.1.1 in 18.18% (n = 14/77) of cases and XBB.2.3 in 1 case with a prevalence of 1.29% (n = 1/77) during the period between November 2023 and February 2024. The predominance of JN.1 was also found to be statistically significant (P < 0.05, χ2 = 120.67). The JN.1 was observed predominantly in Raipur (46.10%), followed by Raigarh (22.40%), Bastar (13.20%), and Kanker (5.30%), with other districts contributing minor proportions (Figure 3).
Over the past 4 years, BA.2 was the predominant variant found in 25.67% of sequences, followed by recombinant Omicron lineage XBB.1.16 (20.16%), BA.2.75 (14.17%), Delta (14.02% for clade 21A and 0.78% for clade 21J), and JN.1 (11.97%) (Figure 3). These variants were all observed to be dominantly circulating in Chhattisgarh during the different specific time periods of the COVID-19 pandemic (Figure 3). This analysis revealed the highest number of sequences, i.e. 163 (25.66%) belonged to 21L, followed by 128 (20.15%) sequences to 23B, 90 (14.17%) to 22D, 89 (14.07%) to 21A, and 76 (11.96%) to 23I, apart from the other thirteen variants (20A, 20B, 20I, 21B, 21J, 21K, 22B, 22C, 22E, 22F, 23A, 23D, and 23E), which were found in co-circulation with less than 5% frequency.
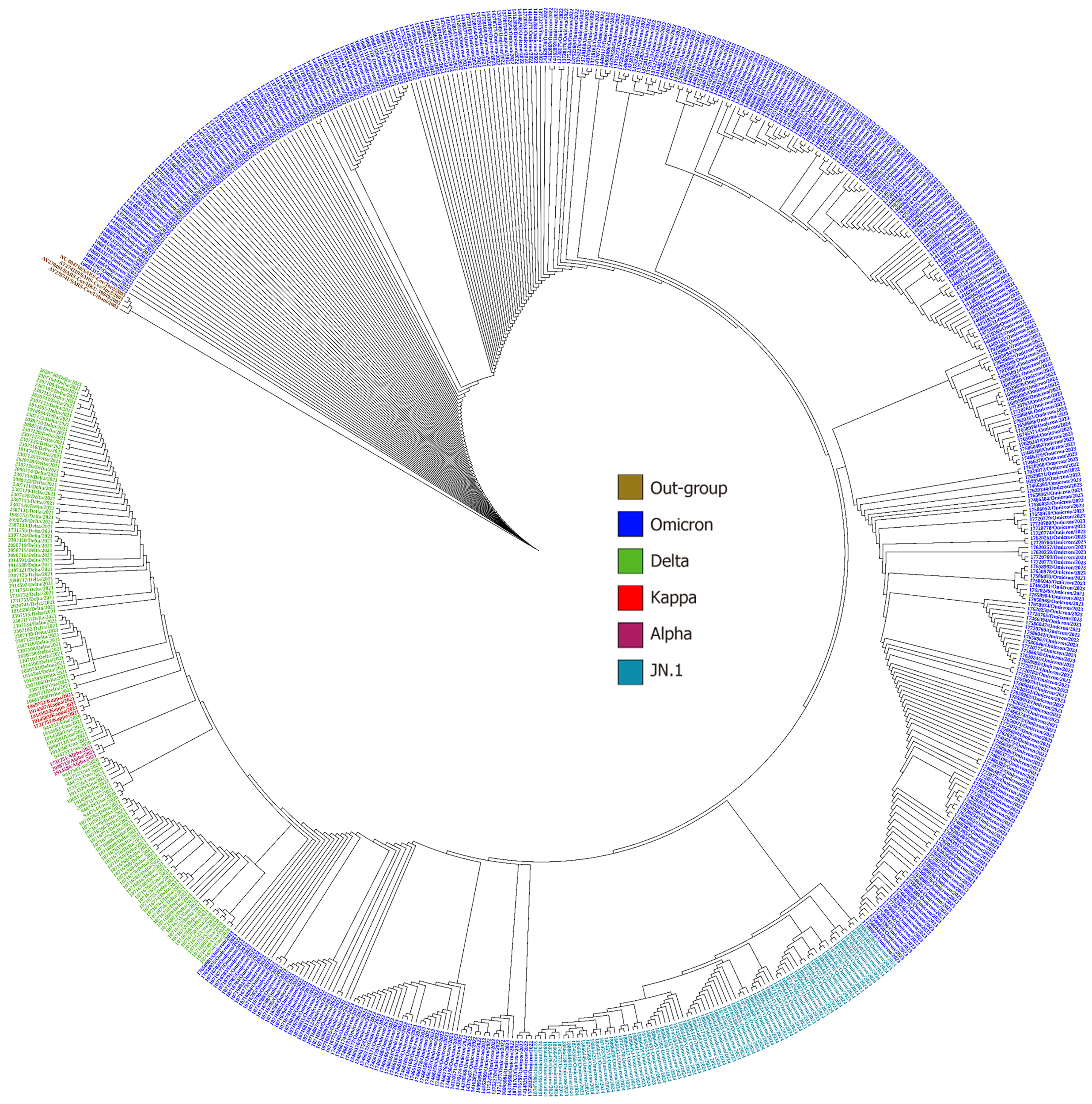
The analysis of the circular phylogenetic tree represented the spike protein as the site of interest to read various mutations detected in all 635 sequences (Figure 4). A total of 0.47% of sequences belonged to the Alpha and Kappa variants, 14.80% of cases belonged to Delta variant, 67.7% of cases belonged to the Omicron variant, and 11.96% of cases belonged to the JN.1 variant. The phylogenetic tree comprised sequences of four lineages: Alpha (B.1.1.7) denoted by pink color (3 male cases only); Delta (B.1.617.2) sequences denoted by green color (55 male and 39 female); Omicron (B.1.1.529) sequences denoted by purple color (226 male and 205 female patients); and JN.1 sequences denoted by brown color (45 male and 31 female). The prominent co-circulating lineage B.1.1.7 in 2020 had DEL69/70, S373P, T478K, N501Y, A570D, D614G, P681H, T716I, Q954H, S982A, and D1118H as a total of 11 signature mutations in the spike protein (Figure 5). This lineage was replaced by B.1.617.2 between 2021 and mid-2022 and contained eight signature mutations in the spike protein: T19R; T478K; D614G; P681R; E156G; DEL157/158; L452R; and D950N. The circulating prevalence was 95%.
Soon the Omicron (B.1.1.529) variant had evolved in late 2022-mid 2023 with 18 signature mutations: G142D; S373P; S375F; K417N; N440K; S477N; T478K; Q498R; N501Y; Y505H; D614G; H655Y; N679K; P681H; N764K; D796Y; Q954H; and N969K. The Omicron lineage was found to be persistently evolving into various sublineages from BA.1 to JN.1 in the period between the end of 2021 and February 2024. These sublineages were characterized by specific inherent numbers of mutations in the spike protein, especially in the receptor binding domain. Between the end of 2023 and February 2024, JN.1 has been the most predominant lineage evolved in Chhattisgarh, harboring the maximum 40 synonymous and non-synonymous mutations with a 95% prevalence viz., V127F, G142D, F157S, R158G, V213G, L216F, H245N, A264D, I332V, G339H, K356T, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L455S, N460K, S477N, T478K, N481K, Q498R, N501Y, Y505H, E554K, A570V, D614G, P621S, H655Y, N679K, P681R, N764K, D796Y, S939F, Q954H, N969K, and P1143 L. The pan mutation D614G in the spike protein was observed to be omnipresent in all four major lineages described above.
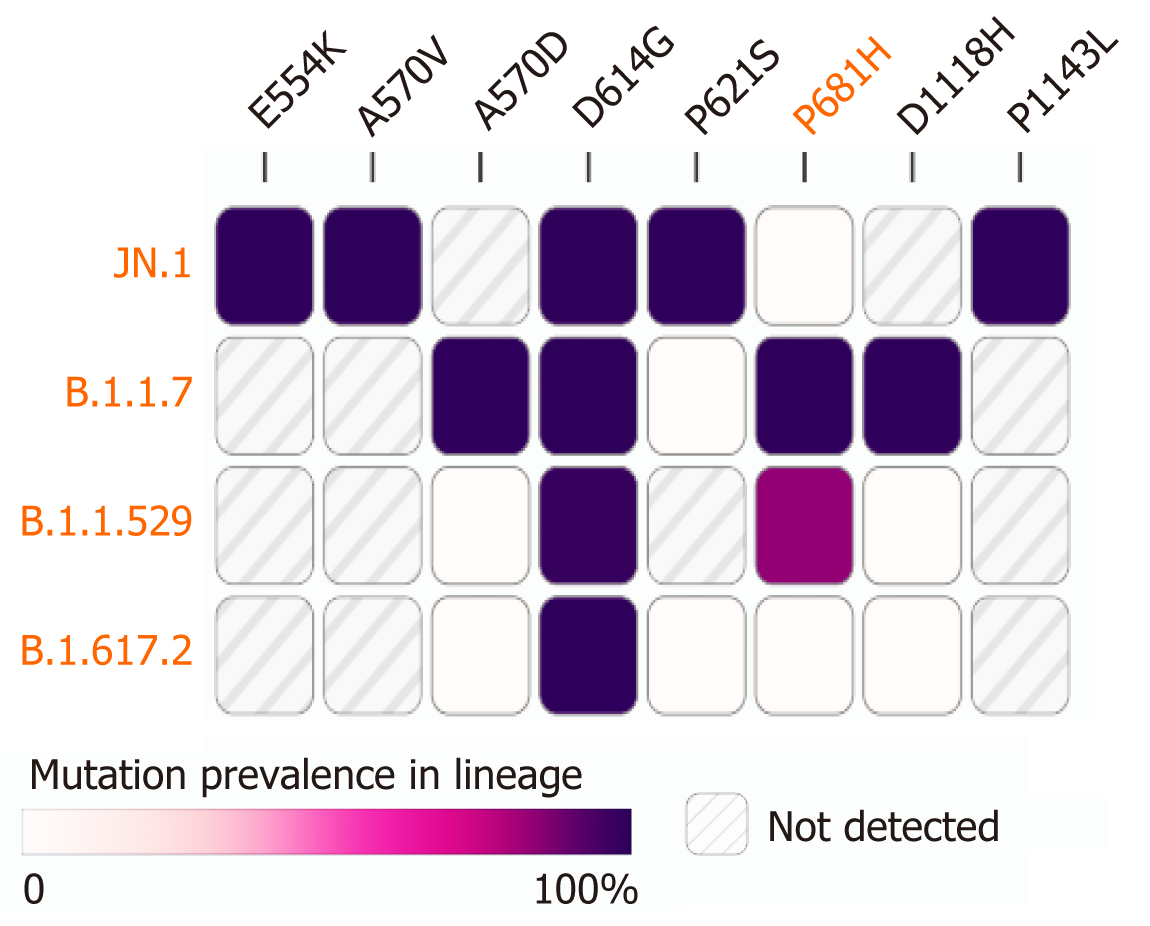
Among the four predominantly circulating lineages in Chhattisgarh during different time periods, the JN.1 lineage has been observed expressing a maximum of five mutations (E554K, A570V, D614G, P621S, and P1143 L) with 99% prevalence, followed by the B.1.1.7 lineage with A570D, D614G, P681H, and D1118H as the major five mutations. The B.1.1.529 lineage had two mutations (namely D614G and P681H) while the B.1.617.2 lineage has only one mutation (D614G) in the spike protein with 99% prevalence (Figure 6). In comparison to the three lineages (B.1.1.7, B.1.617.2, and B.1.1.529), four mutations in the spike protein (E554K, A570V, P621S, and P1143 L) were exclusively detected in the JN.1 lineage.
Among these four mutations, the E554K in the spike protein was reported to have a cumulative prevalence of 98% (710/727) in India. In the past 2 months, Karnataka, a state in South India, had the highest E554K prevalence (99%; 292 out of 294 cases). Other than Karnataka, an identical pattern of prevalence (96%) was observed in three more states, namely Chhattisgarh (26/27 cases), Gujarat (102/106), and Maharashtra (264/274). The presence of this mutation, albeit with lesser prevalence, was also reported from five more states of India: Assam; Haryana; Tamil Nadu; Telangana; and West Bengal.
The other mutation, A570V, has a 97% (707/727 cases) cumulative prevalence in India, with the highest in Karnataka (99%; 291/294 cases), followed by Chhattisgarh (96%, 26/27 cases), Maharashtra (96%, 264/274 cases), and Gujarat (95%, 101/106 cases). The P621S mutation had a 96% prevalence (697/727 cases) nationwide, including 97% in Karnataka (285/294 cases), followed by Chhattisgarh (96%; 26/27 cases), Maharashtra (95%; 261/274 cases), and Gujarat (94%; 100/106 cases).
The fourth mutation, P1143 L, was circulating among the Indian patients with COVID-19 with a 98% (709/727 cases) prevalence. The prevalence was highest in Karnataka (99%; 292/294 cases), followed by Maharashtra (98%; 269/274 cases), Chhattisgarh (96%; 26/27 cases), and Gujarat (93%; 99/106 cases). In Chhattisgarh, 96.29% of isolates of the JN.1 lineage harbored these four mutations.
The mutations A570D and D1118H were detected in the B.1.1.7 lineage with 99% and 99.4% prevalence, respectively, since 2020. However, in the prior 2 months, they were not found in circulation in any of the states in India (Figure 6). The P681H mutation, designated as a mutation of interest, was detected in lineages B.1.1.529 and B.1.1.7 with 83.4% and 99.4% prevalence, respectively. In India, it has been found to be in circulation in the prior 2 months with a 4% prevalence in 26/727 cases. It was highest in Gujarat with a 6% prevalence (6/106 cases), followed by 5% in Maharashtra (15/274 cases), 4% in Chhattisgarh (1/27%), and 1% in Karnataka (3/274 cases). The cases reporting this mutation from Chhattisgarh were detected exclusively in the Raipur district. The D614G mutation was the exclusive single mutation found in all four lineages: B.1.1.7 with 99.7%; B.1.617.2 with 99.5%; B.1.1.529 with 97.6% and JN.1 with 99.5% prevalence (Figure 6).
The genomic surveillance of SARS-CoV-2 in the past 4 years in Chhattisgarh revealed the Alpha variant predominance in 2020, followed by the Delta variant predominance in 2021, and Omicron variant and its sub-lineages in 2022 and 2023[4,7]. Thereafter, from November 2023 to February 2024, a sudden upsurge in COVID-19 cases was again observed due to the evolution of a new variant, JN.1.The WGS analysis performed at our laboratory revealed the emergence of predominantly JN.1 (80.51%), followed by JN.1.1 (18.18%) and XBB.2.3 (1.29%) lineages that are currently circulating in Chhattisgarh. JN.1 and its subvariant JN.1.1 were analyzed, replacing the earlier dominant lineage of XBB.1.16 in Chhattisgarh[4].
The genomic data of 4 years revealed the presence of 17 different types of lineages and sublineages, namely non-variants of concern (20A, 20B), BA.1.1.7 (Alpha) in 2020, B.1.617.2 (Delta) between 2021 and mid-2022, B.1.617.1 (Kappa) in 2021, Omicron (B.1.1.529, BA.1, BA.2, BA.5, BA.2.12.1, BA.2.75, XBB.1, XBB.1.5, XBB.1.16, XBB.1.19.1, XBB.2.3) from late 2022 and 2023, and Omicron-JN.1 from November 2023 to February 2024. The JN.1 variant was encompassed new mutations as well as earlier reported mutations from three lineages, B.1.17 (Alpha), B.1.617.2 (Delta), and B.1.1.529 (Omicron). The JN.1 variant is reported to be a descendent lineage of BA.2.86 with an additional mutation, L455S[10]. BA.2.86 has been reported with more than 30 mutations over its parent BA.2 lineage in the spike protein[11]. The distinct L455S mutation in the receptor binding domain that binds to human angiotensin-converting enzyme 2 receptors in the JN.1 variant provides resistance against class 1, 2, and 3 antibodies, leaving sera unprotected post-vaccination and inflicting an enhanced transmissibility rate. The effective reproductive number of JN.1 exceeds BA.2.86.1 and HK.3 in the sequences obtained from France, the United Kingdom, and Spain[12]. The JN.1 variant has also exhibited a relative growth advantage of 57% (confidence interval: 56%-57%)[11]. An earlier report demonstrated that JN.1 is superior in evading an immune response to its predecessor BA.2.86 by the neutralization assay[12]. Taking cognizance of these characteristics, the WHO has shifted JN.1 from the earlier category of variant under monitoring to VOI on December 18, 2023. Globally, JN.1 is the most reported VOI (now reported by 130 countries), accounting for 54.3% of SARS-CoV-2 sequences submitted globally as of May 17, 2024[13].
In India, JN.1 has been reported by various states. It was first identified in Kerala on October 6, 2023. Thereafter, its presence was also reported in other states; the highest cases being reported from Maharashtra (40.03%, 1342/3352), followed by Karnataka (9.81%, 329/3352), Kerala (9.42%, 316/3352) Andhra Pradesh (9.39%, 315/3352), Gujarat (7.48%, 251/3352), West Bengal (4.50%, 151/3352), Goa (3.57%, 120/3352), Tamil Nadu (2.77%, 93/3352), Telangana (2.41%, 81/3352), and Chhattisgarh (2.26%, 76/3352) as of May 20, 2024[14].
The risk evaluation of causing symptomatic and severe disease of the JN.1 variant appeared minimal as per the WHO report[15]. The probable reason could be the presence of cross-reactive immunity generated by the previous dominant lineage of XBB.1.16 and vaccination. Thus, even the continued spread of JN.1 alone will be unlikely to increase the burden on national public health systems compared to other Omicron sublineages. It is further confirmed in Maharashtra, where mild infections were reported by JN.1 with a hospitalization rate of 8.03% and oxygen support requirement in 3.11% of positive cases.
In mutation analysis, the role of predominant mutations present in 99% of sequences was analyzed (Figure 6). The JN.1 variant exhibited five such mutations, namely E554K, A570V, D614G, P621S, and P1143 L, with 99% prevalence in sequences obtained from Chhattisgarh. The E554K mutation, also reported in the parent BA.2.86 lineage, is attributed to the acquisition of resistance towards antibodies, resulting in an efficient immune escape mechanism[16-18]. The D614G mutation is said to be responsible for increasing infectivity, as well as the stability of the viral genome[16,17]. This mutation was present in all four lineages. The mutation A570V increases the stability of the spike protein by balancing the hydrophobic interactions to provide conformational stability to the viral genome[18]. In serological studies post-vaccination against the BA.2.86 lineage, it was shown that the P621S mutation in SARS-CoV-2 led to immune escape[19].
The JN.1 lineage constituted 40 mutations accumulated in the spike protein region, out of which P681R was declared as a mutation of interest. This mutation was also expressed earlier at the furin cleavage site of the spike protein in B.1.617.2 lineage of the Delta variant. The furin cleavage site plays an essential role in the entry of the virus into the host epithelial cells as well as in viral replication[20]. It was hypothesized that the P681R mutation at the furin cleavage site has altered the cleavage efficiency, resulting in smooth entry of the virus into the host epithelial cells. It can be considered one of the significant mutations responsible for the higher infection rate of JN.1 worldwide.
This study had the limitation of not being able to evaluate the clinical spectrum of the patients infected with COVID-19 by the JN.1 variant. Nonetheless, being the nodal center in Chhattisgarh, we aimed to unravel the evolutionary trajectory of SARS-CoV-2 from the time of its emergence in 2020 to February 2024. An earlier study from Maharashtra reported that JN.1 predominantly causes mild symptoms with cold (67.03%), fever (55.91%), cough (40.86%), and headache (15.64%) and requires hospitalization in only 13.26% of cases, while the rest recovered at home[11]. Overall, a 98.57% recovery rate with 1.43% mortality is also evidently reiterated in our observation of the high infectivity but less mortality of JN.1.
All mutations found in JN.1 with 99% prevalence were responsible for major events, viz., host cell entry, stability, replication, resistance against antibodies, and increased transmission that eventually led to the rise in COVID-19 cases recently in Chhattisgarh and other states of India and worldwide. Although most of the cases of JN.1 were clinically mild with no intensive care unit admissions, the transmissibility was high, and many were breakthrough infections. Therefore, it is important, both clinically and from a public health perspective, to identify various mutations inherited by the virus. This will help to initiate appropriate measures to manage any future outbreak, thereby preventing morbidity and mortality in the long run.
We are grateful for the support provided by the All India Institute of Medical Sciences, Raipur, the Indian Council of Medical Research New Delhi, and the Department of Health Research New Delhi. We are thankful to Md. Rafiqullah Khan, laboratory technician, and Ms. Khushbu Khwase, laboratory technician, for their technical support.
| 1. | World Health Organization. WHO Director-General's opening remarks at the media briefing on COVID-19 - 11 March 2020. Mar 11, 2020. [cited 30 December 2024]. Available from: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020. |
| 2. | World Health Organization. Tracking SARS-CoV-2 variants. [cited 26 February 2024]. Available from: https://www.who.int/activities/tracking-SARS-CoV-2-variants. |
| 3. | Government of India. COVID-19 state wise data. [cited 28 February, 2024] Available from: https://www.mygov.in/covid-19-archive. |
| 4. | Negi SS, Sharma K, Bhargava A, Singh P. A comprehensive profile of SARS-CoV-2 variants spreading during the COVID-19 pandemic: a genomic characterization study from Chhattisgarh State, India. Arch Microbiol. 2024;206:68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 5. | Sharma K, Aggarwala P, Gandhi D, Mathias A, Singh P, Sharma S, Negi SS, Bhargava A, Das P, Gaikwad U, Wankhede A, Behra A, Nagarkar NM. Comparative analysis of various clinical specimens in detection of SARS-CoV-2 using rRT-PCR in new and follow up cases of COVID-19 infection: Quest for the best choice. PLoS One. 2021;16:e0249408. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 6. | Singh P, Sharma K, Singh P, Bhargava A, Negi SS, Sharma P, Bhise M, Tripathi MK, Jindal A, Nagarkar NM. Genomic characterization unravelling the causative role of SARS-CoV-2 Delta variant of lineage B.1.617.2 in 2nd wave of COVID-19 pandemic in Chhattisgarh, India. Microb Pathog. 2022;164:105404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (1)] |
| 7. | Singh P, Sharma K, Shaw D, Bhargava A, Negi SS. Mutational characterization of Omicron SARS-CoV-2 lineages circulating in Chhattisgarh, a central state of India. Front Med (Lausanne). 2022;9:1082846. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 8. | Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35:1547-1549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16161] [Cited by in RCA: 23254] [Article Influence: 3322.0] [Reference Citation Analysis (0)] |
| 9. | Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293-W296. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1256] [Cited by in RCA: 7034] [Article Influence: 1406.8] [Reference Citation Analysis (0)] |
| 10. | World Health Organization. Updated Risk Evaluation of JN.1. Jan 09, 2023. [cited 30 December 2024]. Available from: https://www.who.int/docs/default-source/coronaviruse/09022024_jn.1_ure.pdf?sfvrsn=a153518c_3.2023. |
| 11. | Karyakarte RP, Das R, Rajmane MV, Dudhate S, Agarasen J, Pillai P, Chandankhede PM, Labhshetwar RS, Gadiyal Y, Kulkarni PP, Nizarudeen S, Yanamandra S, Taji N, Joshi S, Potdar V. Appearance and Prevalence of JN.1 SARS-CoV-2 Variant in India and Its Clinical Profile in the State of Maharashtra. Cureus. 2024;16:e56718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 12. | Kaku Y, Okumura K, Padilla-Blanco M, Kosugi Y, Uriu K, Hinay AA Jr, Chen L, Plianchaisuk A, Kobiyama K, Ishii KJ; Genotype to Phenotype Japan (G2P-Japan) Consortium, Zahradnik J, Ito J, Sato K. Virological characteristics of the SARS-CoV-2 JN.1 variant. Lancet Infect Dis. 2024;24:e82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 203] [Article Influence: 101.5] [Reference Citation Analysis (0)] |
| 13. | World Health Organization. COVID-19 epidemiological update – 17 May 2024. May 17, 2024. [cited 30 December 2024]. Available from: https://www.who.int/publications/m/item/covid-19-epidemiological-update-edition-167. |
| 14. | India Biological Data Centre. Submission track for INSACOG member labs. [cited 30 December 2024]. Available from: https://inda.rcb.ac.in/insacog/indexpage.2024. |
| 15. | World Health Organization. Updated Risk Evaluation of JN.1. April 15, 2024. [cited 30 December 2024]. Available from: https://www.who.int/docs/default-source/coronaviruse/15042024_jn1_ure.pdf?sfvrsn=8bd19a5c_7. |
| 16. | Zhang L, Jackson CB, Mou H, Ojha A, Peng H, Quinlan BD, Rangarajan ES, Pan A, Vanderheiden A, Suthar MS, Li W, Izard T, Rader C, Farzan M, Choe H. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun. 2020;11:6013. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 704] [Cited by in RCA: 707] [Article Influence: 117.8] [Reference Citation Analysis (0)] |
| 17. | Zhang L, Jackson CB, Mou H, Ojha A, Rangarajan ES, Izard T, Farzan M, Choe H. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 365] [Cited by in RCA: 249] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 18. | Qu P, Xu K, Faraone JN, Goodarzi N, Zheng YM, Carlin C, Bednash JS, Horowitz JC, Mallampalli RK, Saif LJ, Oltz EM, Jones D, Gumina RJ, Liu SL. Immune Evasion, Infectivity, and Fusogenicity of SARS-CoV-2 Omicron BA.2.86 and FLip Variants. bioRxiv. 2023;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 19. | Bdeir N, Lüddecke T, Maaß H, Schmelz S, Rand U, Jacobsen H, Metzdorf K, Kulkarni U, Cossmann A, Stankov MV, Hoffmann M, Pöhlmann S, Blankenfeldt W, Dopfer-jablonka A, Behrens GM, Čičin-šain L. Reverse mutational scanning of spike BA.2.86 identifies epitopes contributing to immune escape from polyclonal sera. 2024 Preprint. [DOI] [Full Text] |
| 20. | Liu Y, Liu J, Johnson BA, Xia H, Ku Z, Schindewolf C, Widen SG, An Z, Weaver SC, Menachery VD, Xie X, Shi PY. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. bioRxiv. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 98] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/