©The Author(s) 2016.
World J Virol. Nov 12, 2016; 5(4): 161-169
Published online Nov 12, 2016. doi: 10.5501/wjv.v5.i4.161
Published online Nov 12, 2016. doi: 10.5501/wjv.v5.i4.161
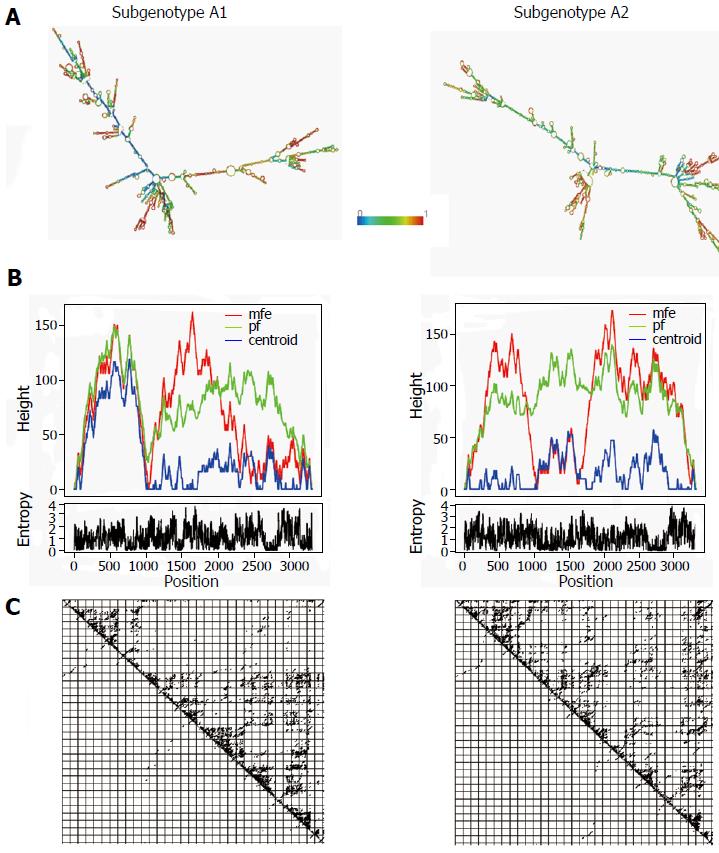
Figure 1 Comparative diagram showing the differences between different aspects of the predicted secondary structures of the pgRNA, specific for HBV subgenotype A1 and subgenotype A2.
A: Predicted minimum free energy (mfe) structures, coloured by base-pairing probabilities (according to the rainbow scale shown in the middle, denoting base pair probabilities from 0 to 1). Colour of the unpaired regions denotes the probability of being unpaired; B: Mountain plot representing the mfe structure (red line), the thermodynamic ensemble of RNA structures (green line), and the centroid structure (blue line). Positional entropy for each position is presented below the mountain plot; C: Dot-plot showing the base-pairing probabilities of the two predictions.
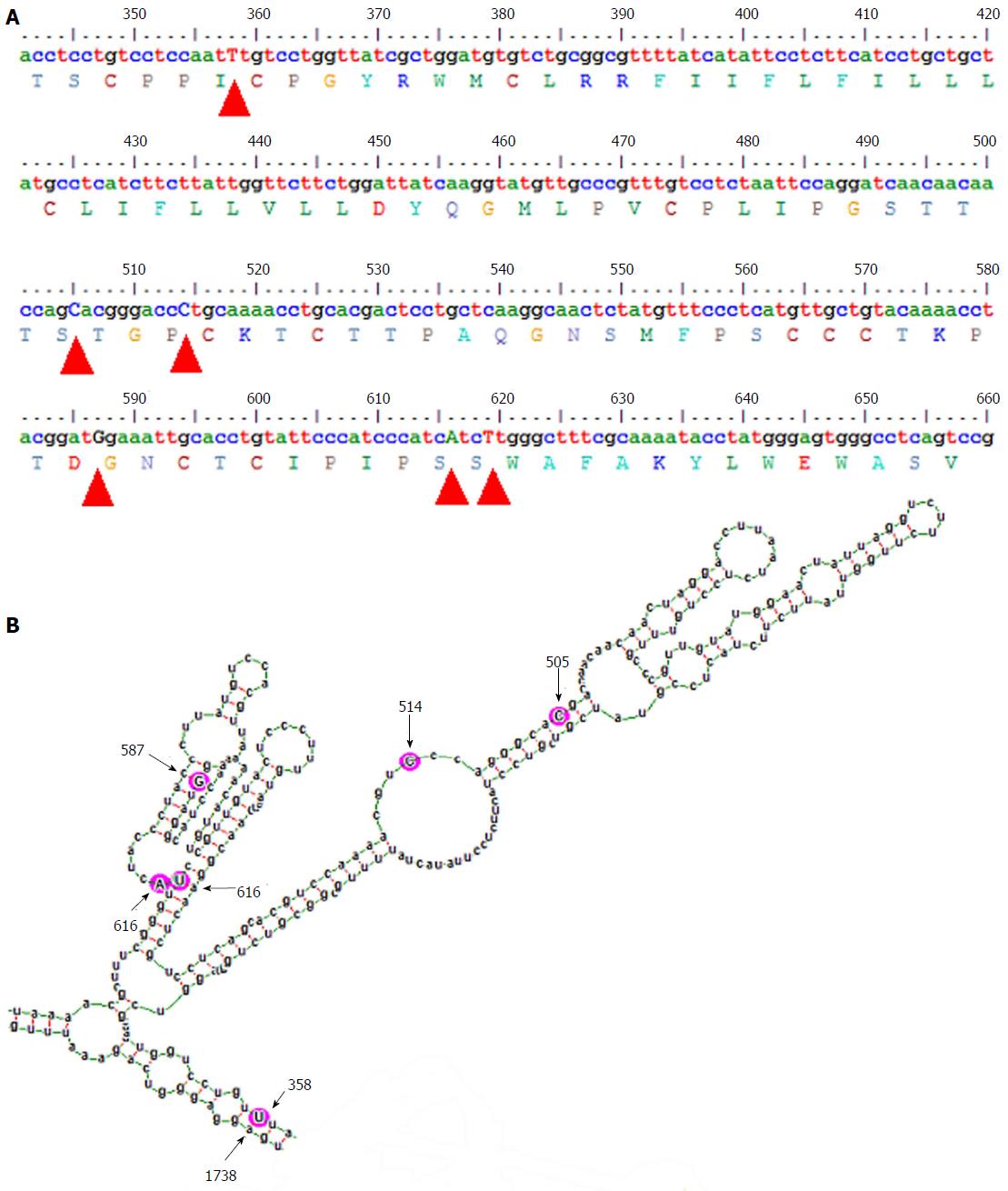
Figure 2 Diagram showing the genetic variability and the part of the predicted mfe structure, corresponding to the genetic region encoding the major hydrophilic loop of subgenotype A1.
A: Consensus nucleotide sequence (corresponding to positions 341 to 660 of the HBV genome) and predicted amino acid sequence (corresponding to residues 63 to 168 of the HBsAg). Subgenotype A1 specific nucleotides (505C, 514C, 616A and 619T) and two variable nucleotides (358T and 587G) are indicated by arrowheads; B: Detailed base pairing pattern of the mfe pgRNA structure, specific for subgenotype A1. Aforementioned variable sites are encircled by pink circles and indicated by arrows and numbers correspond to their nucleotide position in the HBV genome. HBV: Hepatitis B virus; HBsAg: Hepatitis B surface antigen.
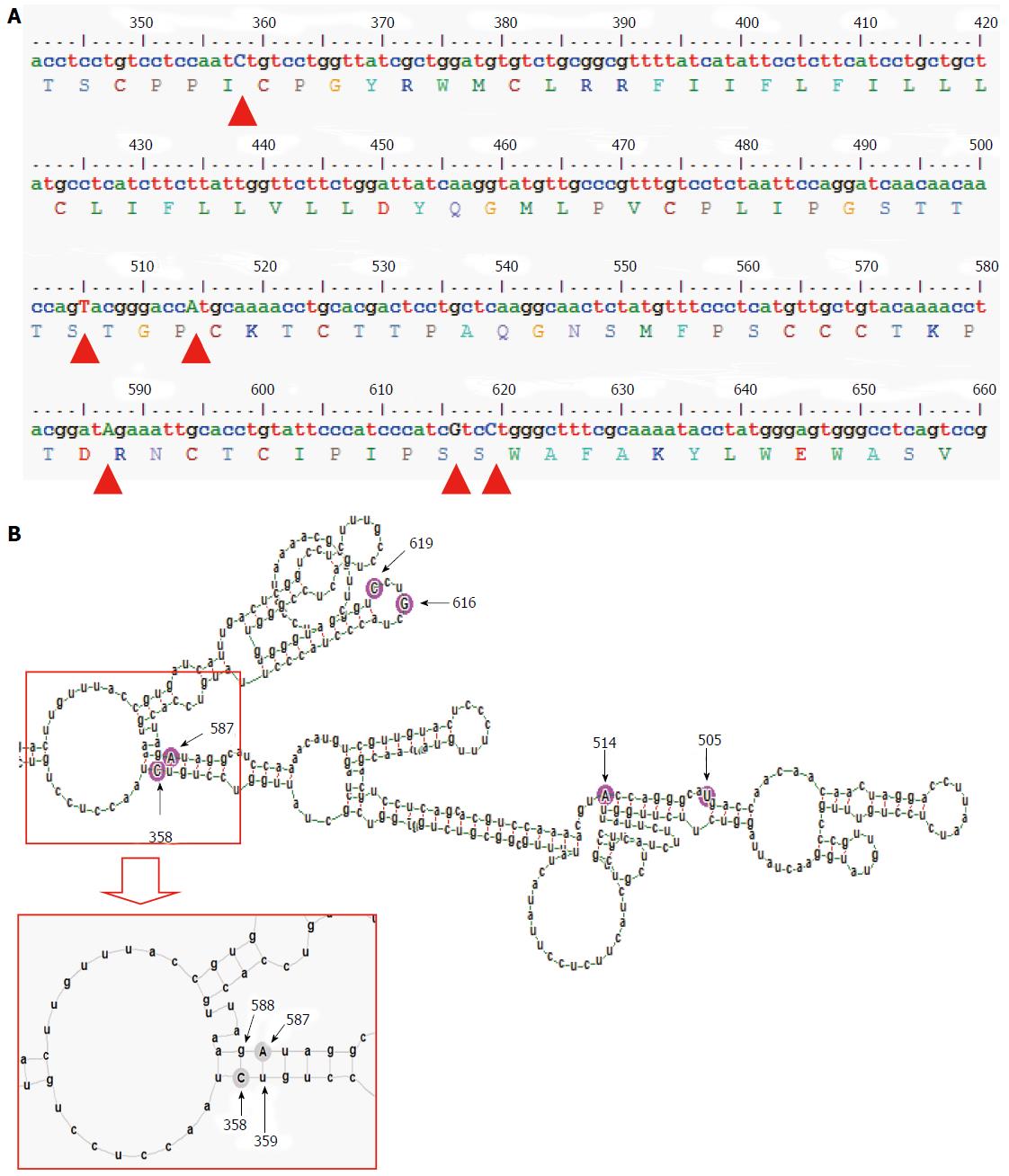
Figure 3 Diagram showing the genetic variability and the part of the predicted mfe structure, corresponding to the genetic region encoding the major hydrophilic loop of subgenotype A2.
A: Consensus nucleotide sequence (corresponding to positions 341 to 660 of the HBV genome) and predicted amino acid sequence (corresponding to residues 63 to 168 of the HBsAg). Subgenotype A2 specific nucleotides (505T, 514A, 616G and 619C) and two co-evolving nucleotides (358C and 587A) are indicated by arrowheads; B: Detailed base pairing pattern of the mfe pgRNA structure, specific for subgenotype A2. Aforementioned variable sites are encircled by pink circles and indicated by arrows and numbers correspond to their nucleotide position in the HBV genome. Part of the mfe structure is amplified in the inset for better visualization of the base pairing. HBV: Hepatitis B virus; HBsAg: Hepatitis B surface antigen.
- Citation: Datta S, Chakravarty R. Role of RNA secondary structure in emergence of compartment specific hepatitis B virus immune escape variants. World J Virol 2016; 5(4): 161-169
- URL: https://www.wjgnet.com/2220-3249/full/v5/i4/161.htm
- DOI: https://dx.doi.org/10.5501/wjv.v5.i4.161