©The Author(s) 2025.
World J Virol. Jun 25, 2025; 14(2): 100001
Published online Jun 25, 2025. doi: 10.5501/wjv.v14.i2.100001
Published online Jun 25, 2025. doi: 10.5501/wjv.v14.i2.100001
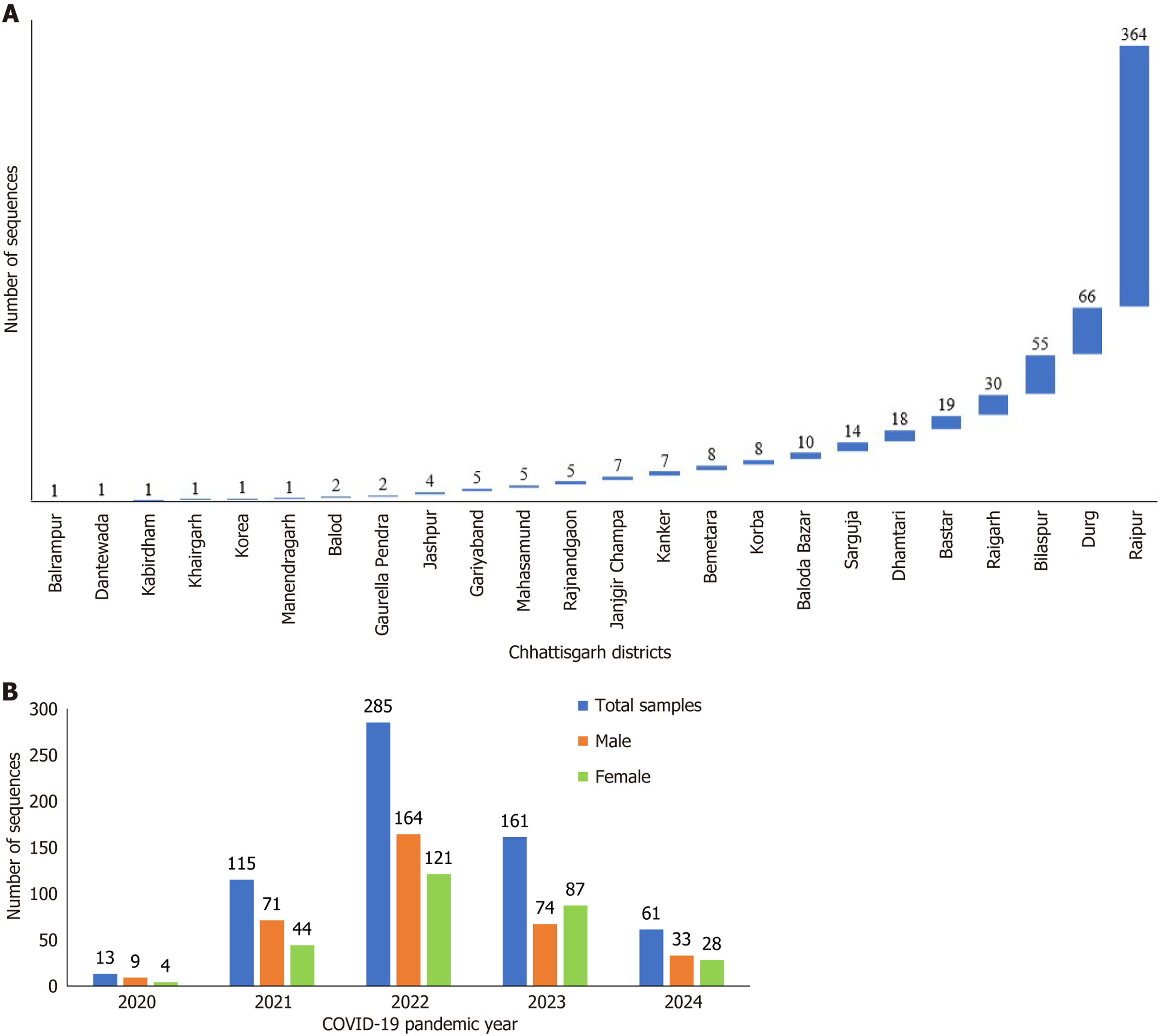
Figure 1 Results of 635 whole genome sequencing samples.
A: District detail of 635 whole genome sequencing cases; B: Year-wise male and female whole genome sequencing cases from 2020 to 2024. COVID-19: Coronavirus disease 2019.
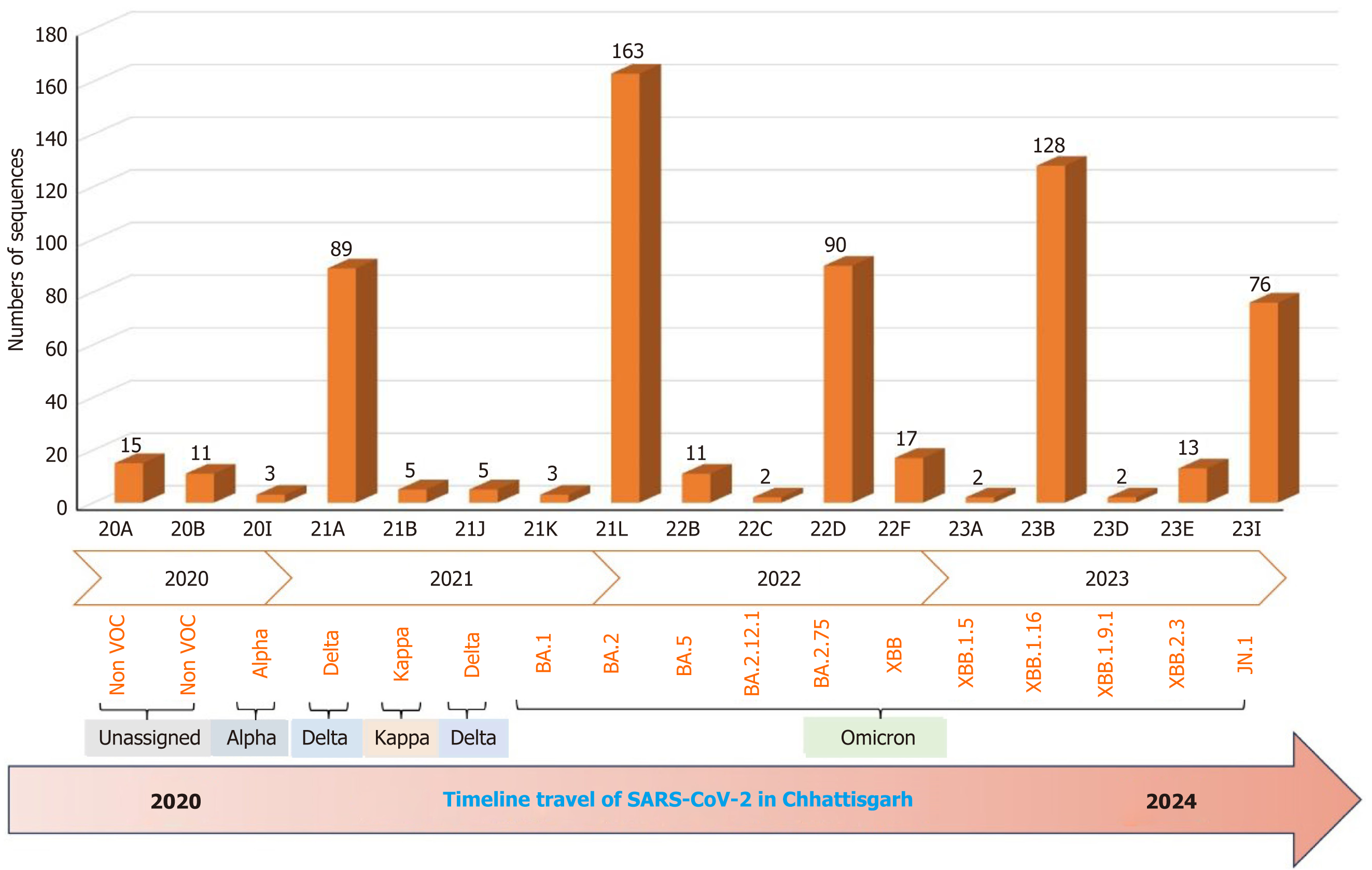
Figure 2 Severe acute respiratory syndrome coronavirus 2 variants identified in Chhattisgarh between 2020 to 2024.
VOC: Variants of concern; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2.
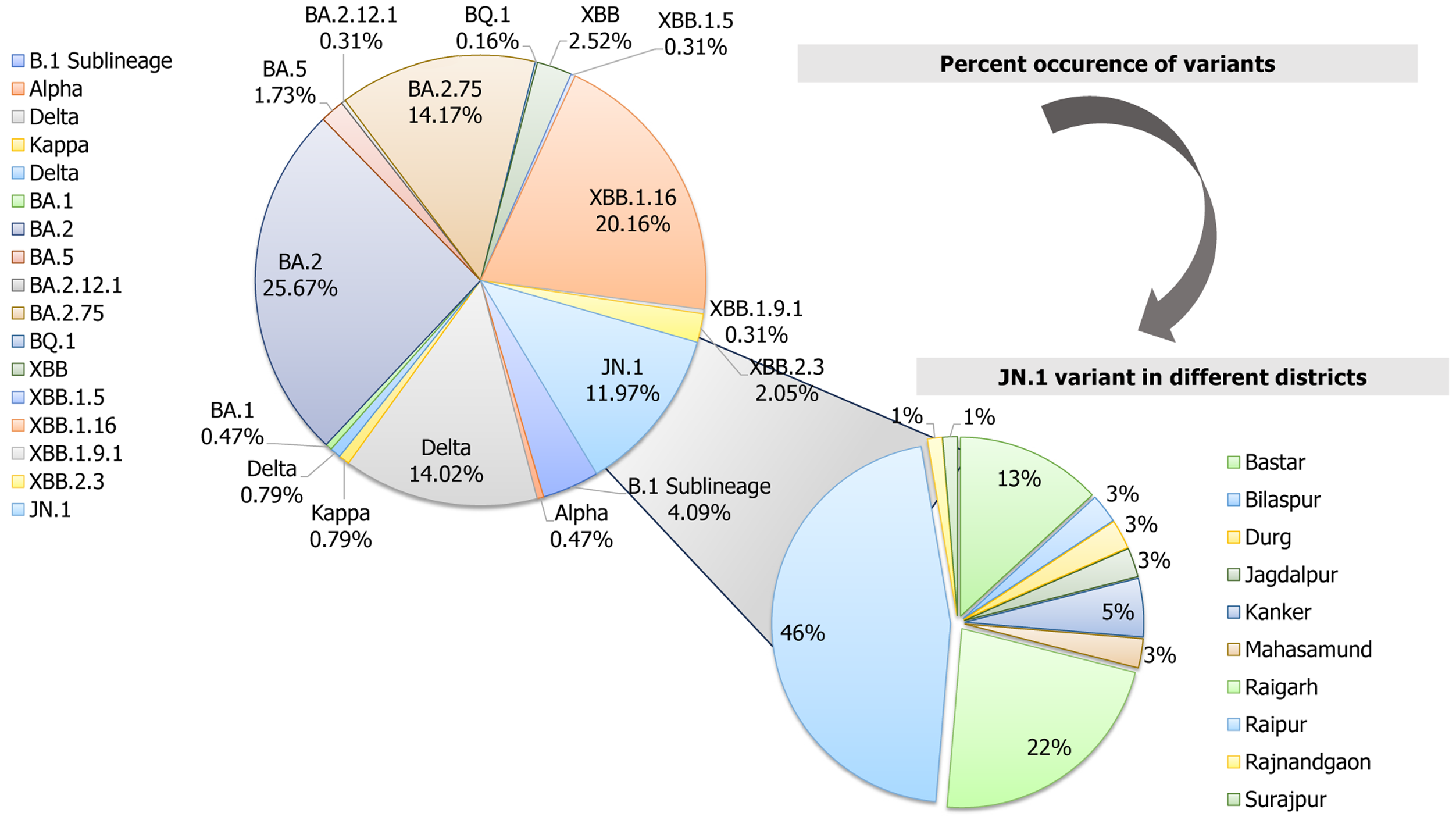
Figure 3 Circulation of different variants of severe acute respiratory syndrome coronavirus 2 in Chhattisgarh with detailed demographic appearance of JN.
1 variant in various districts.
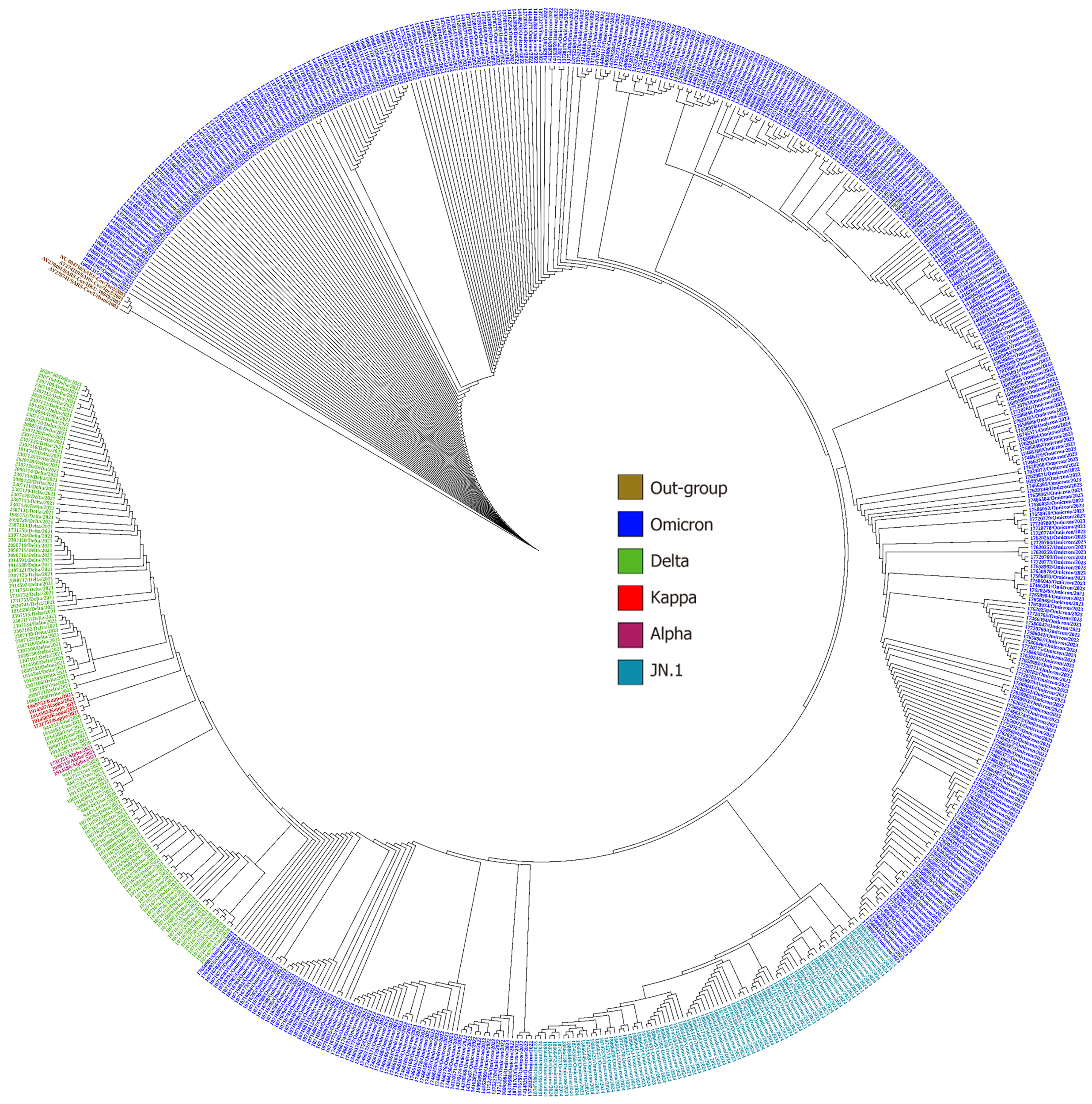
Figure 4 Circular phylogenetic tree representing whole genome sequencing of 635 strains from Chhattisgarh.
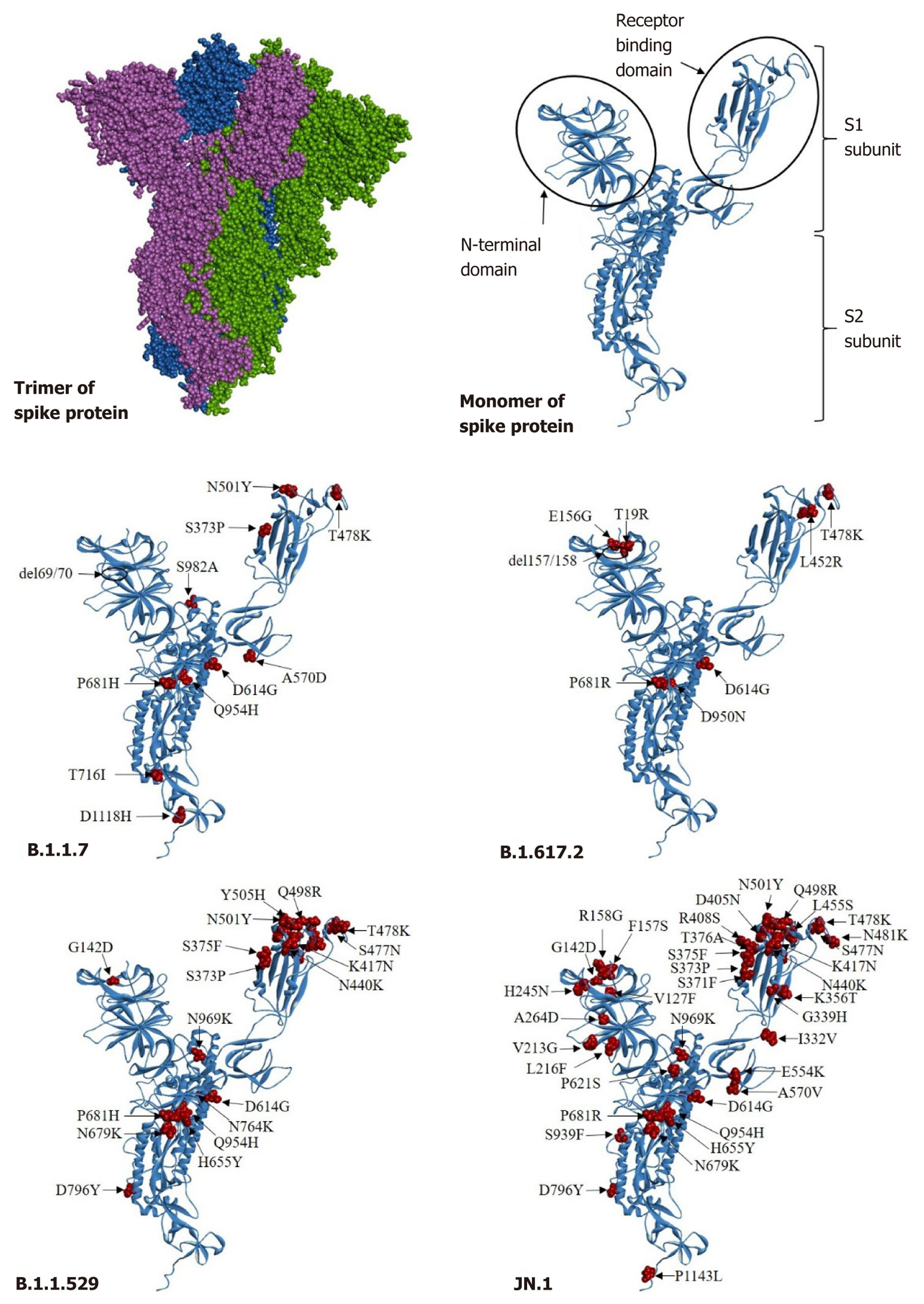
Figure 5 The three-dimensional structure and mutations in the spike protein of severe acute respiratory syndrome coronavirus 2 detected in different lineages.
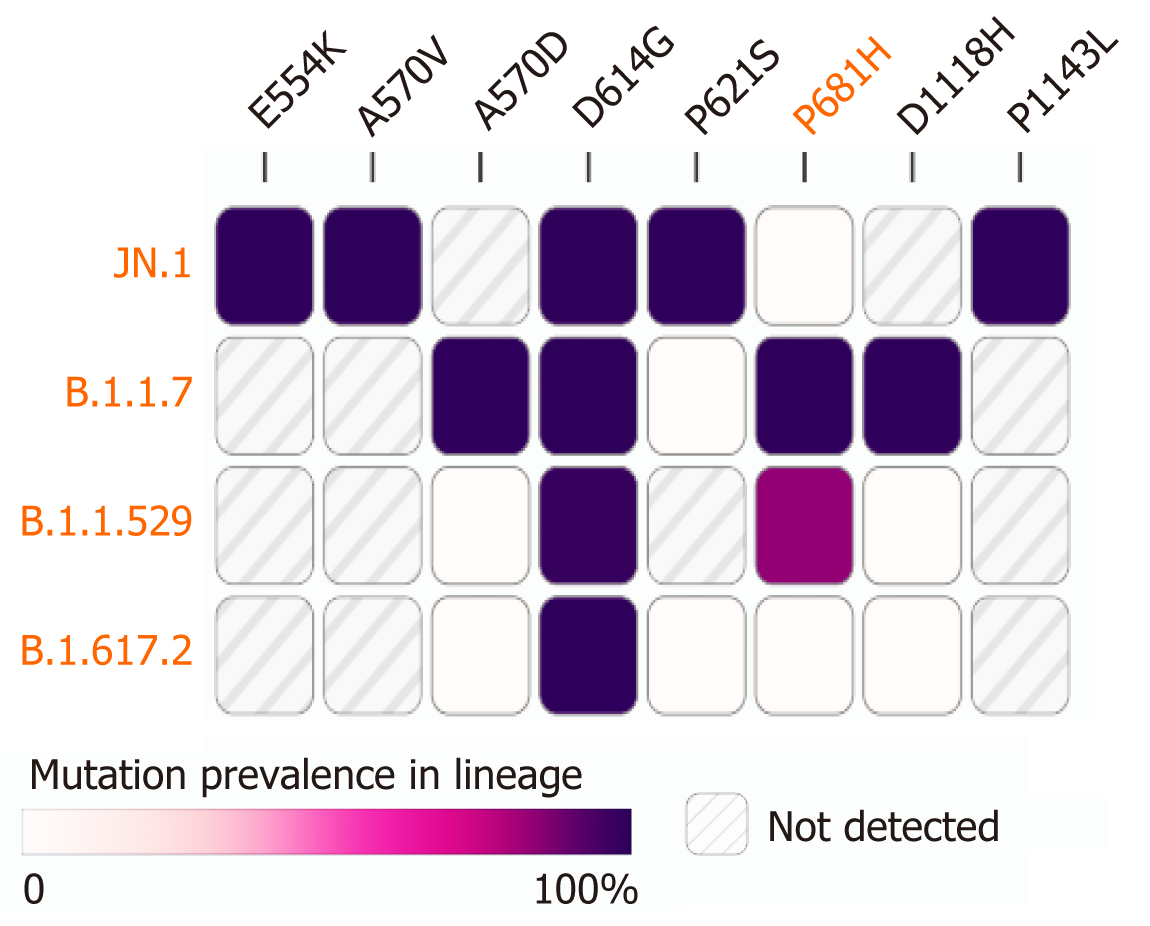
Figure 6 Mutations of > 90% prevalence across four lineages.
- Citation: Singh P, Khare R, Sharma K, Bhargava A, Negi SS. Alpha to JN.1 variants: SARS-CoV-2 genomic analysis unfolding its various lineages/sublineages evolved in Chhattisgarh, India from 2020 to 2024. World J Virol 2025; 14(2): 100001
- URL: https://www.wjgnet.com/2220-3249/full/v14/i2/100001.htm
- DOI: https://dx.doi.org/10.5501/wjv.v14.i2.100001