©The Author(s) 2021.
World J Virol. Nov 25, 2021; 10(6): 288-300
Published online Nov 25, 2021. doi: 10.5501/wjv.v10.i6.288
Published online Nov 25, 2021. doi: 10.5501/wjv.v10.i6.288
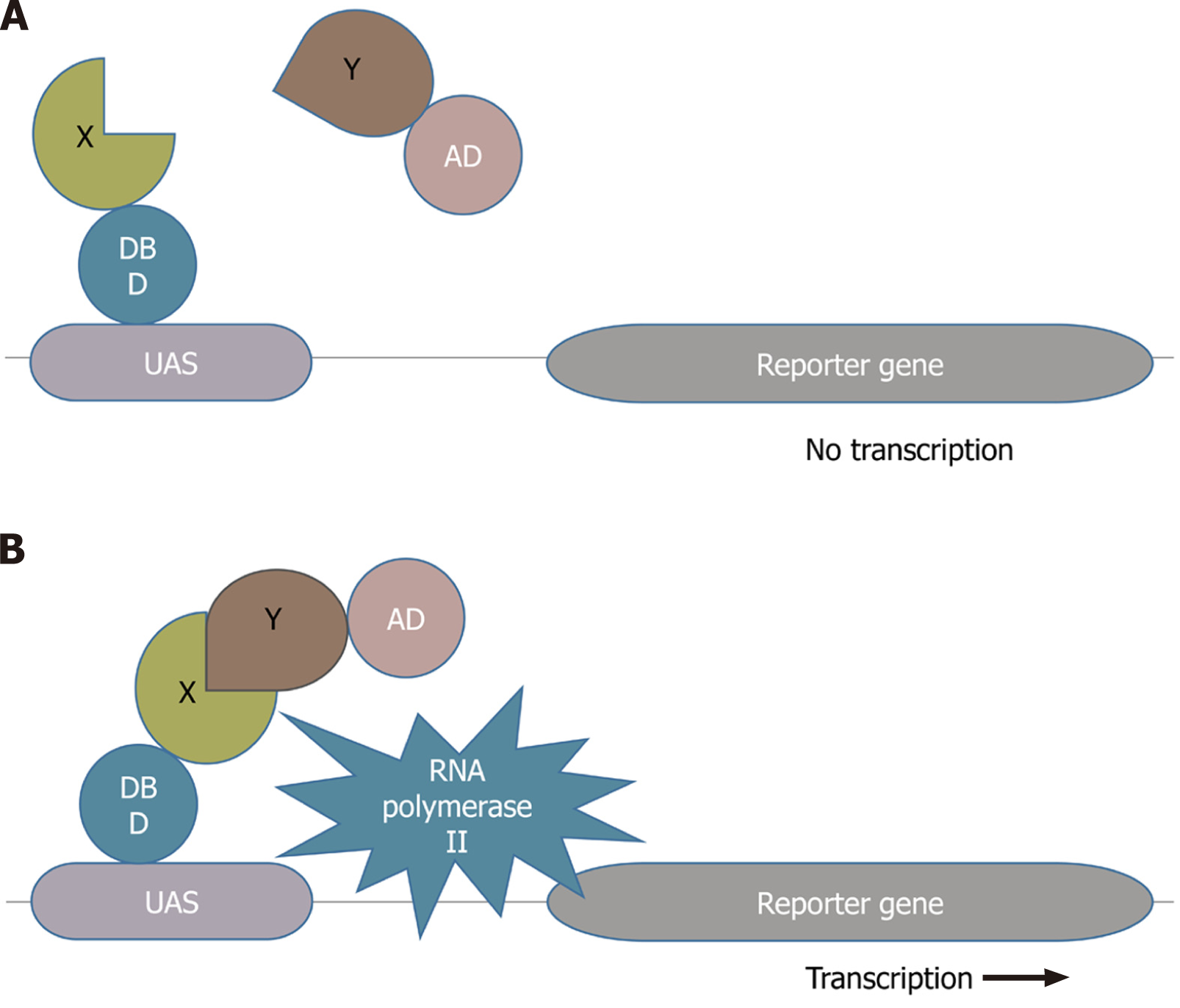
Figure 1 Yeast two-hybrid technique.
A: There is no transcription of the reporter gene because the transcriptional factor is broken down into two halves; B: The reporter gene is being transcribed because the two halves of the transcription factor are brought together by the interaction between bait (X) and prey (Y) proteins[10]. DBD: DNA binding domain; AD: Activation domain; UAS: Upstream activation domain.
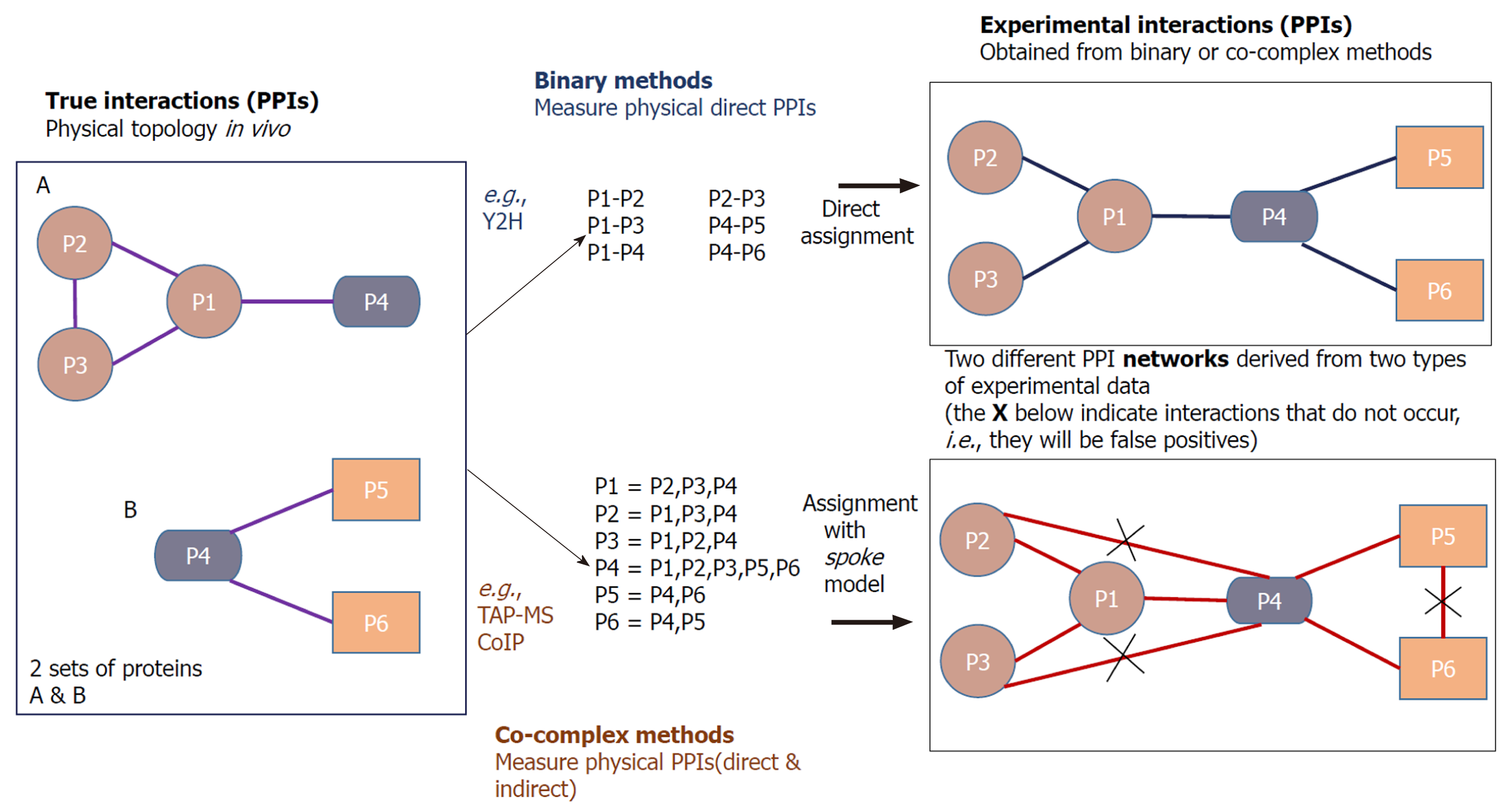
Figure 2 Binary and co-complex methods to determine protein-protein interactions.
Yeast two-hybrid (Y2H) and tandem affinity purification mass spectrometry are the two most extensively used approaches for detecting protein-protein interactions. Given here are the two sets of proteins (4 proteins in set A while 3 proteins in set B) in the left panel and the connections show the genuine interactions between them. The right side shows the experimentally determined interaction network among the six proteins. The network in the upper right shows the interactions derived from Y2H, and the network in the lower right shows the interactions got from co-complex method, in which three of the interactions inferred do not exist[4]. PPI: Protein-protein interaction; TAP-MS: Tandem affinity purification mass spectrometry; CoIP: Co-immunoprecipitation.
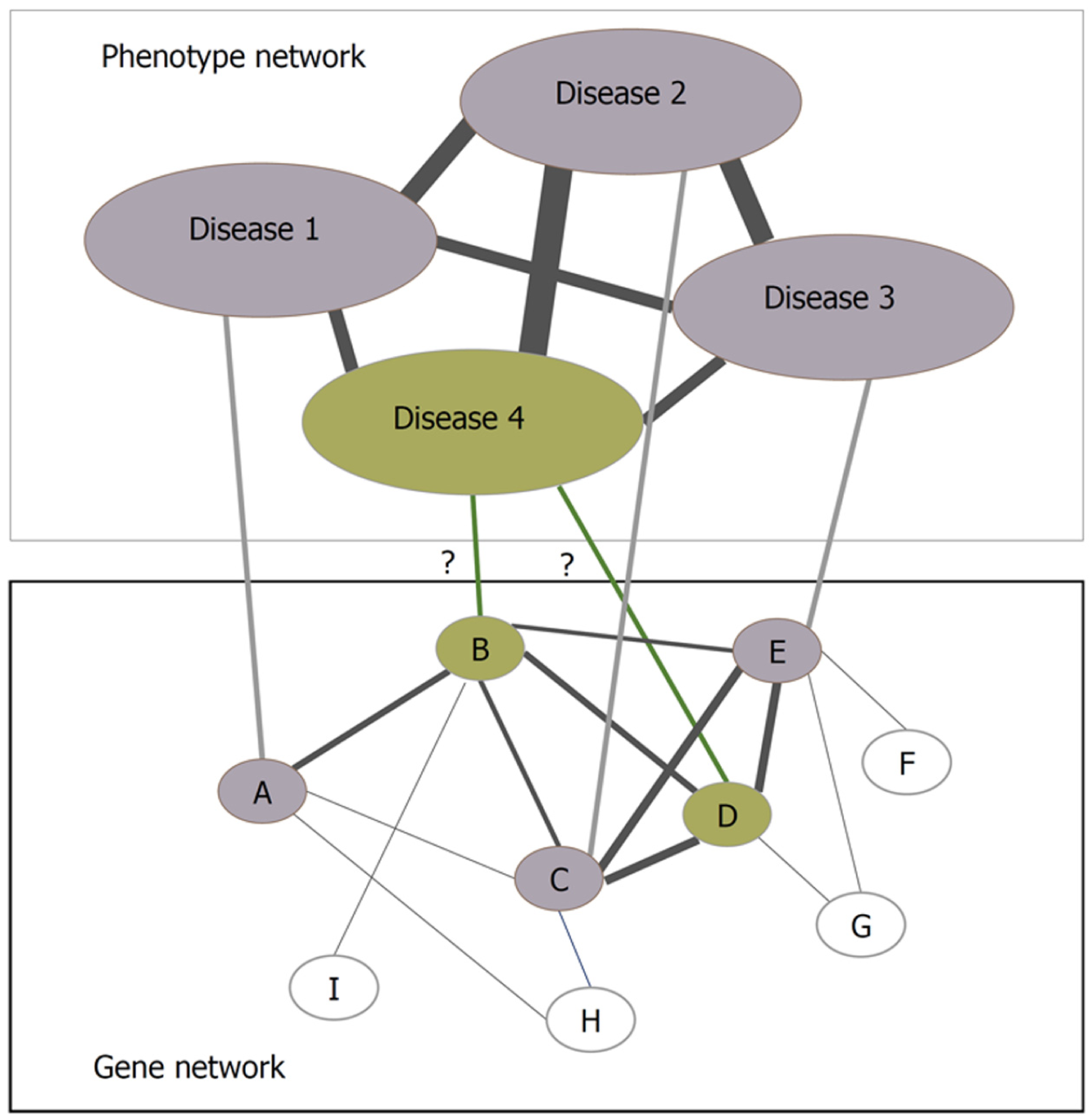
Figure 3 Protein interaction networks can help find new disease-related genes.
The concept depicts that diseases 1, 2, and 3 are caused subsequently by genes A, C, and E, and the genes causing disease 4 are unknown but disease 4 is phenotypically associated with diseases 1, 2, and 3. If the known genes, i.e., A, C, and E are closely associated functionally, it can be hypothesized that genes B and D are the cause of disease 4[86].
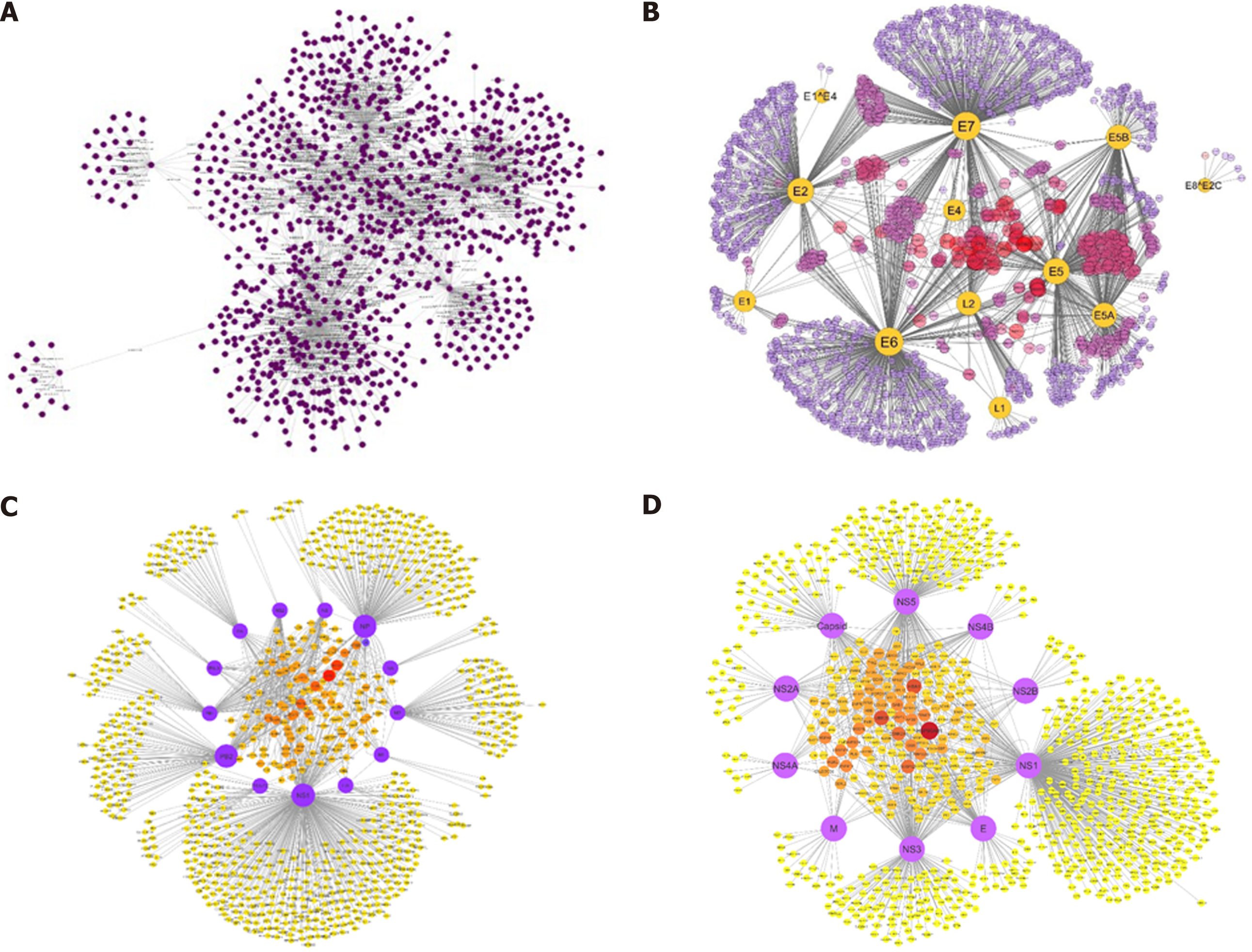
Figure 4 Comprehensive protein interaction networks of hepatitis C virus, human papillomavirus, influenza A virus, and dengue virus with host Homo sapiens constructed in Cytoscape by literature curated experimentally verified and computationally predicted protein-protein interactions.
The network explains virus-host relationship between the infectious agents and host factors which contribute to disease pathways in human body. A: Hepatitis C virus; B: Human papillomavirus; C: Influenza A virus; D: Dengue virus.
- Citation: Farooq QUA, Shaukat Z, Aiman S, Li CH. Protein-protein interactions: Methods, databases, and applications in virus-host study. World J Virol 2021; 10(6): 288-300
- URL: https://www.wjgnet.com/2220-3249/full/v10/i6/288.htm
- DOI: https://dx.doi.org/10.5501/wjv.v10.i6.288