INTRODUCTION
Cell culture and animal models are the accepted evaluative methodology in all types of preclinical studies, including oncology research. These models have contributed a lot to the overall understanding of the pathological mechanisms of several diseases including different types of cancers, however, their value in predicting the effectiveness of treatment options and strategies in clinical trials have remained doubtful[1,2]. Apart from the ethical controversies; lead by the animal activist, the main problems with animal models lays in the species differences when compared with human. These species differences often causes misleading interpretation[3]. In fact, clinical trials are mandatory because preclinical studies on cell culture and animal models do not envisage with sufficient confidence the likely outcomes in human studies.
In oncology research, due to the ethical concerns associated with human experimentation, animal models and cell culture studies have become an important source of information. However, the average rate of successful clinical translation from animal models to clinical trials are not very encouraging; at present not more than 8%[4]. Animal models have the restricted ability to mimic the complex process of human cell proliferation and pathophysiology conditions. In oncology research, studies on cell culture and animal models are critical instruments in determining the efficacy, pharmacodynamics and mechanism of action of novel anti-cancer drugs. It should be remembered that heterogeneity of the tumour cells leads to the huge diversity with a high degree of genetic instability and phenotypic variation.
Prior to plunge into the trial of a promising anticancer drug, pharmaceutical companies and institutional investigators conduct wide pre-clinical experimental studies. In vitro and in vivo studies preliminary covers safety, efficacy, toxicity and pharmacokinetic profiles of the candidate molecules. Early in vivo testing aims specifically to provide initial safety and efficacy data to supports investigators claims about compound under investigation. To justify further development, preclinical experiments add sufficient confidence to the research data. This is important because as per the Food and Drug Administration guidelines, successful animal need/preclinical testing have to be completed before humans are exposed to the potential therapeutic entity[5].
Apart from possible misleading in vitro results, relating to inaccuracies in potency, efficacy, toxicity, genotoxicity and carcinogenicity, the financial cost of clinical research also plays a decisive role in the development and establishment of the successful therapeutics. Given that three-dimensional (3D) bio-printed structures could produce better models of the in vivo microenvironment, there is the significant potential for cost reductions in pre-clinical research. The 3D bio-printed tissues and organs have the capacity to provide viable substitutes to cell cultures and animal models. The 3D printing of solid objects is already guiding major innovations in diverse areas, such as education, manufacturing, engineering, art, pharmaceuticals and medicine[6]. Recent innovation in 3D printing and material science have enabled construction of complex 3D functional living constructs (tissues and organs)[6]. Without worrying about the rejection, 3D bio-printing has already been used for the generation and transplantation of several important tissues including, bones, skin, heart tissue, etc. Other lucrative applications include developing more reliable 3D bio-printed tissue models for pharmaceutical and drug discovery research. Accurate reproduction of the tissue or an organ is a significant feature of the 3D bio-printing which ultimately could lead to the standardisation of therapeutic testing[7]. This is possible to achieve by reproducing all the functional components of the tissues and organs, such as mimicking the exact branching patterns of the tinniest capillary in a complex organ like the heart, kidney, liver and lungs, or manufacturing the biomaterials to take care of the natural physiology.
PRECLINICAL IN VITRO MODELS AND THERAPEUTIC DEVELOPMENT
New drug development programmes generally take about 12 years to get an experimental lead compound to the patient bedside. The average cost involved in this process can be as high as exceeding $1.2 billion dollars[8,9]. The drug development process is highly risky in terms of economic gain; evident by an overall average attrition rate of approximately 90%, which means that only 10% of clinical trial compounds could finally reach to the market[10]. Consequently, scientists are now putting greater efforts in reducing the cost of the drug development process. Computer aided drug design[11], in silico pharmacokinetics[12] and toxicity testing[13] are few of the newer methodologies available, which could reduce the initial cost of the drug development process.
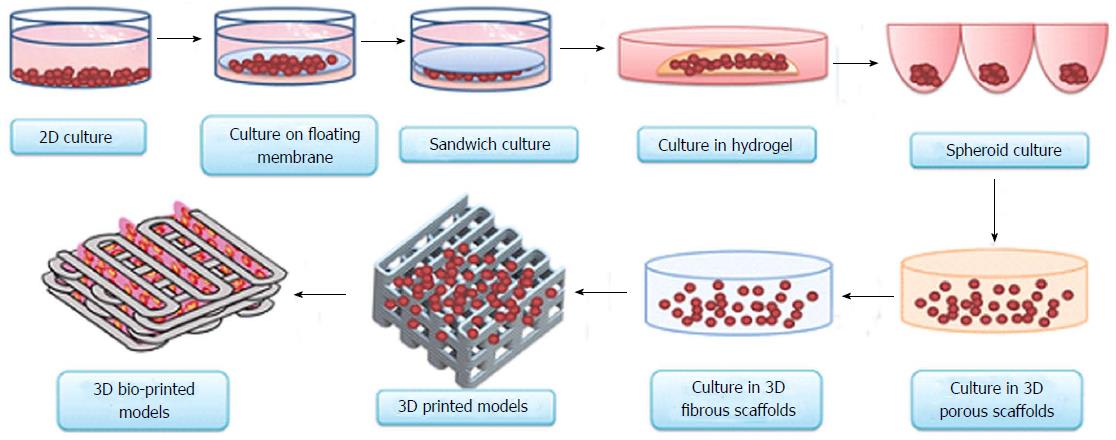
Accurate preclinical determination of efficacy and toxicity would lower the failure rate of new molecules during the important stage of clinical evaluation. Drug testing on 3D bio-printed human organs could eliminate the possibility of drawing uncertain conclusions from preclinical animal and cell culture studies. Conflicting conclusions from preclinical animal models and human experiments usually surface during the final stage of the clinical trials, when most of the resources have already been invested in the research and development process. Several promising lead candidates have faced failures in clinical trials after successful animal testing[14-19]. Preventing these problems in the first place would improve the cost and time involved in bringing a new drug to the market. To accurately predict the unwanted parameters of the drug candidates in clinical trials, various classical, existing and emerging technologies (models) are available. This comprehensive list includes traditional 2D tissue culture[20], classical whole rodent models[21], humanised mouse models[22], 3D culture models[23], co-culture systems[24] and 3D tissue models[25] (Figure 1).
Figure 1 Evolution of cell-culture models from simple two-dimensional to complex three-dimensional bio-printed models.
Currently, 3D bio-printing is the most sophisticated technique used to make tissue/organ constructs[65]. 3D: Three-dimensional.
Traditional 2D cell culture systems which employ cell lines in a single layer, themselves contain abundant genetic mutations. 2D cell culture systems also lack the important natural microenvironment present in the tissues and organ from which they were originally seeded[26]. Traditional culture performed with primary cells do not offer 3D microenvironmental characters similar to that of its origin[27]. Classical cell culture systems not only lack the influential tissue microenvironment and gradient but may also include the rapid loss of important proteins and its functions and gene expression profiles. To get a better representation of tissue complexity, microenvironment and whole-body physiological impact, studies on the animal model have become the backbone of preclinical studies. However, as discussed earlier, basic molecular, physiological and pathophysiological differences between the species lead to the likelihood of erroneous conclusions being drawn about an under trial candidate. Erroneous conclusions are the leading cause of failures in clinical trials.
Co-culture systems, 3D culture models, 3D tissue models and humanised mouse models which could mimic the host microenvironment are available for preclinical studies. To some extent, these methodologies allow drug testing in human-like systems, eliminating the species differences and, thereby, increasing acceptability in clinical trials. Developing pharmacological assays based on configuring multiple cells into a 3D-orientated structure could provide more realistic data. The 3D cell culture and models could mimics native tissue architecture more closely and hence could address drug development concerns in a more actual ambience than traditional 2D culture models.
Humanised mice model is another approach to testing drugs in more human-like conditions. This type of the animal models include mice bearing tumours derived from humans, known as xenografts or mice in which the endogenous liver has been compromised and repopulated with human liver cells[22]. Xenografts are important and proving useful in anticancer drug development. Xenografts often enable the assessment of drug efficacy, safety and toxicity in the context of tumour phenotypic and genotypic heterogeneity. Similarly, mice with humanised liver offer the ability to assess drug pharmacokinetics and metabolism preclinically in vivo. Humanised liver is an important tool to understand drug excretion and toxicity[28]. One important thing to remember about all humanised models is their chimeric nature. They are a single human tissue or cell type planted within the animal body, which may lead them to behave differently from their native environment. This may propagate false interpretation due to inter-species variations. For example, the stromal and vascular components of xenograft models largely come from an animal in origin[29]. Similarly, mice with humanised livers contain human hepatocytes, however, the other cell types found in the liver and all of the interrelated organ systems are of mouse[30] which ultimately could affect the liver functions. Hence, such models cannot be considered as the perfect model for human systems modelling. However, as stated earlier, humanised mouse models are a popular model in the study of human cancer. They provide an understanding of factors involved in pathology, physiology, metastasis and invasion.
In xenograft models, human tumour cells are transplanted into a different species, either into the organ type in which the tumour originated or under the skin. Human tumour cells are commonly transplanted into the mice which are severely immunocompromised. The weak immune system of such mice accepts foreign human cells readily. For example, the xenograft (foreign cells or organ) will be readily accepted by athymic nude mice (lacking T cells producing thymus), severe combined immunodeficiency mice strains, or other immunocompromised mice[31,32]. Therapeutic agents can be studied in these immunocompromised mice as it readily allows the growth of human tumour within itself. The size of the tumour is generally depends on upon the number of cells originally transplanted, however, growth occurs over 1 to 8 wk to give more natural humanised environment. Genetically engineered mouse (GEM) model is another type of widely used animal model used for studying human cancer.
GEM mouse model allows the investigator to study the genes which are speculated to be the reason of the malignancy. Such genes are deleted, silenced or sometimes overexpressed and the animal is observed for the molecular and phenotypical changes over the period of time to study the therapeutic response. GEM provides an opportunity to study the therapeutic response in vivo. Xenograft models and immunocompromised athymic nude mice have been in used for several decades to increase our understanding of pathophysiological and genetic factors involved in uncontrolled cell proliferation and metastasis. Recent information about the role of the microenvironment on the tumour progression, growth and resistance towards the drugs has made GEM and primary human xenografts in humanised mouse models a primary choice for the experiments. However, because of the species difference, xenografts of human cell lines in mice to test drug responses do not always necessarily correlate with the actual pathophysiological condition in patients[29].
The importance of the tumour microenvironment on tumour growth not only leads to the general acceptance of the humanised mouse models and GEM for the development of the cancer therapeutics but also paved ways for the development of 3D printed tissues and organs in oncology research. The 3D culture and co-culture systems already exist and recent refinement increases their availability for therapeutic research. Certain drawbacks, such as low cell density, and use of artificial matrices and scaffolds add a non-human or non-native aspect to the system, which could affect the final outcome. However, more recent approaches that generate 3D culture systems, such as 3D bio-printing, could help nullify the non-human aspect.
3D BIO-PRINTING
The 3D bio-printed tissues and organs could be designed to mimic the exact cellular density of target tissues and organs, with proper consideration given for cellular component, extracellular matrix and three-dimensional spatial components. Since complex tissues are not constructed exclusively from a single cell type, 2D mono-cell culture models are of debatable value[33]. However, 3D bio-printers deposit more than one cell type, co-culturing them in one single spatial arrangement making them a closer match for the natural architectural arrangement. With the recent advancement in bio-printing, it is now feasible to combine the most important elements of spatial patterning to generate 3D in vitro tissue/organ systems that could mimic the key cellular and extracellular functional machinery, including innervation[33]. The 3D printers use various types of cells in the form of bio-inks, which technically have enhanced the speed of 3D printing of organs and tissues. The 3D organ scaffold generated with the help of computed tomography or another imaging technology and the solid surface made up of the biocompatible materials is used as the substrate to generate the 3D tissues and organs. Bio-inks are made up of cells suspended in a biocompatible gel-like material then deposited on the substrate using 3D printers which work on the principal like mechanical extrusion[33]. During and after deposition on the substrate the bio-ink is gelled by polymeric inter-linking with the help of photo or thermal activation. Because of the involvement of the high energy, care is always taken to leave the cells intact and functional. Hydrogels not only play an important role in physically restraining the suspended cells and in the maintenance of the cell viability but also can be personalised and tailored according to the biocompatible material or dimensions[33].
The development of aqueous-based systems enabled direct printing of bio-inks into 3D scaffolds[34]. Sequential deposition of the living cells, biocompatible extracellular and materials with spatial control over the 3D architectural parameters is the heart of the 3D bio-printing and 3D bio-printed organs. The 3D bio-printing works on the several established principals based on bio-mimicry, autonomous self-assembly and mini-tissue building blocks[6].
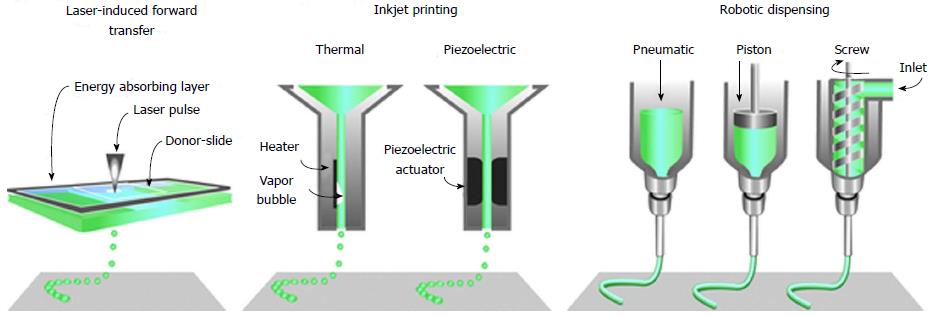
Technological advancement in imaging and digital design technology has positively impacted the 3D bio-printing by reproducing and visualising the very complex, heterogeneous architecture of complex tissues and organs. Non-invasive imaging techniques, like computed tomography, magnetic resonance imaging, computer-aided design and computer-aided manufacturing tools and mathematical modelling, are used to collect and digitise the complex tomographic and architectural information of the tissues/organs. The 3D digital images of complex organs are then used to print tissues and organs using techniques like inkjet[35-38], micro-extrusion[39-41] and laser-assisted printing (Figure 2)[42-44].
Figure 2 The common approaches currently used to bio-print tissue, are laser-assisted, inkjet-based and extrusion-based robotic dispensing techniques[110].
The 3D printing technologies first became prominent in non-biological applications, such as the deposition of ceramics, metals and thermoplastic polymers in heavy and light industries. Organic solvents, high temperatures and cross-linking agents (e.g., photo-activation) used in 3D printing poses immediate compatibility problems for delicate living cells, thermal liable biological (e.g., proteins) and biocompatible materials[6,45,46]. Among several, one of the main and dare challenges in the 3D bio-printing of tissues and organs is to develop the compatible materials that not only should go well with the several other biological materials and the harsh printing process but should also provide the required mechanical and functional properties to the 3D bio-printed constructs. Materials currently used in the field of regenerative medicine are based on either natural polymers (e.g., alginate, gelatin, collagen, chitosan, fibrin and hyaluronic acid etc.) or synthetic molecules (e.g., polyethylene glycol). Some of the major advantages of the natural polymers in 3D bio-printing are its similarity to the human extracellular matrix, non-toxic nature and inherent bioactivity. Whereas the typical advantage of the synthetic polymers is that they can be personalised and tailored to the specific application and can also be obtained in the most purified form. But like other synthetic molecules, synthetic polymers not only possess the risk of the poor bio-acceptability but could also lead to the toxicity because of the toxic degradation. Other challenges could be the loss of the mechanical strength over the period of time and immunogenicity. Despite this, synthetic hydrogels polymers owing to its hydrophilic, absorbent and manageable physical and chemical properties are an attractive alternative in 3D bio-printing. The correct functioning of the 3D fabricated tissue or organ does not only depend on upon the accurate deposition of the cells but the choice of the cells is also crucial. Other criteria need to be satisfied is that the cell chosen for 3D bio-printing should have the capability to proliferate of its own. Precise control of cell proliferation (in vitro and in vivo) ensures the functionality of the construct. In addition to the primary cell of interest (e.g., hepatocytes in liver construct), most tissues also contain other cell types that are involved in supportive, structural or barrier functions (selective transport) (e.g., liver also contains sinusoidal endothelial cells and phagocytic Kupffer cells) and may also be involved in vascularization or may play role in stem cell maintenance and differentiation.
Presently, 3D bio-printing involves the deposition of multiple primary cell types into patterns that accurately represent the native tissue. In the case of the auto-rearrangement and self-assembly to the 3D construct, printing involves the bio-ink of the stem cells that can proliferate and differentiate into the required cell types. Maintenance and exact mimicking of the physiological function of cells in 3D construct are important and hence the criteria applied for selecting the cells plays the decisive role in proper functioning[47].
Rejection by the host immune system is the challenge in the tissue and organ transplant. This issue can be sort out by using the autologous cells for 3D bio-printing of organs and tissues. Autologous cell source involves biopsies, generation and differentiation of autologous stem cells or induced pluripotent stem cells. Although autologous cells are the very reliable source, it’s of no use in case if the patient is already ill, cells are infected or have metabolic or hereditary disorders. In such cases, especially in the case of genetic disorder, 3D construct is not useful for the transplant but could be useful in case of therapeutic development (e.g., genetic mutation in cancer cells will be useful to construct 3D bio-printed tumour model). In the case of the metabolic disorder, autologous cells may not be able to produce the normally desired function in bio-printed organs.
Prolong functionality of any 3D bio-printed tissues and organs are the key to the success. However, cells types like heart, liver and immune cells are not only difficult to isolate from the source but is also difficult to culture in a lab because of their limited lifespan[48]. Self-renovating, ability to differentiate into any cell type and capability to generate multi-functional tissue-specific cell phenotypes is the solution for such problems. Embryonic stem cells and induced pluripotent stem cells have all these characters and hence are the promising cell types for 3D bio-printed organs and tissues[49]. The 3D bio-printed organs require the self-renovating or self-replenishing character to maintain the functionality, in this regard pluripotent stem cells ability to multiply several times highlight its potential in 3D bio-fabricated construct. Other types of stem cells, such as stem cells from bone marrow[50-52] and fat[53] or perinatal stem cells from amniotic fluid[54] or placenta[55], have limited multi-potent differentiation ability. These cell types but are considered safer for 3D bio-printed construct. These cells also satisfy the criteria of the autologous cell types and hence have the potential application in regenerative medicine. Mesenchymal stromal cells (MSC) are also a good cell source but its Isolation is difficult. However, the establishment of the new protocols for isolation, expansion and differentiation now make them the reliable and promising source for bio-fabricated constructs. Clinically required amount of MSC has been effectively generated in vitro and have found application in clinical trials and regenerative medicine[50-52]. Future development in biotechnology and cell-culture techniques is likely to be useful to exploit other stem cell populations for bio-printing and regenerative medicine; this is not just a hypothesis but a potential possibility.
3D PRINTING IN PRE-CLINICAL TESTING AND THERAPEUTIC DEVELOPMENT OF ANTI-CANCER DRUGS
Therapeutic drug development and therapy optimisation experiments in genetically modified mouse, 2D cell culture, 3D co-culture and xenografts of human tumour cells into nude mice are the important tool and have immensely contributed in the oncology research[31,56,57]. Physiologically, tumour microenvironment is extremely complex in which genetically mutant and phenotypically proliferative cancerous cells not only interact with each other but also reciprocally interact with the stromal and immune system microenvironment[58]. Modelling the heterogeneous complexity of a typical tumour using 3D bio-printed tissues and organs for preclinical testing could be an innovative and novel approach for the pre-clinical testing and therapeutic development of anti-cancer drugs.
Determination of the efficacy, toxicity, pharmacodynamics, pharmacokinetics and mechanism of action are the critical studies towards the development of efficient anti-cancer therapeutics. Cell culture and animal studies have played important roles in this process. Tumour cells and host microenvironment interaction leads to the recruitment of the components essential for the inflammatory and immune signalling. This recruitment of the signalling components is preceded by the fibroblasts and endothelial cells activation. The microenvironment of the host tumour is modified to select and adapts the genetic and phenotypic characters of the tumour cells. In fact, the modified microenvironment of the host organ in cancer pathology ultimately helps in the growth of the tumour cells. This reciprocal interaction between tumour cells and the microenvironment is actually essential for tricking the immune system, proliferation and metastasis[59]. Host microenvironment not only subjected to the different environmental stimuli but if looked from the population perspective it is genetically and phenotypically so diverse that the same tumour will grow and behave differently in different physiological condition (different patient). Simulation of such huge diversity (thousands of genes) in 2D cell culture and in animal models to test the toxicity and efficacy of drug candidate is the mammoth task. Essentially, it is impossible to extrapolate the results obtained from single or two test models to the numerous tumour variants in a broad genetically heterogeneous population.
Cell cultures derived from the human tumour cell line only offers the advantage of the biology to the primary tumour but it cannot simulate or mimic the complexity involved in the interaction between the proliferating tumour cells and microenvironment. Xenografts in immunocompromised mice interact with the surrounding cell types which are different from the native cell types and hence grafted tumour cells could behave differently in mice. Overall, the xenografts mice models have added limited value to the 2D cell culture. Similarly, lack of working the immune system and insufficient interactions between the human tumour cells and human stromal cells do not essentially represent the human tumour microenvironment.
Organovo is now an early-stage but established medical research company, which designs and develops functional 3D human tissues and organs for medical and pharmaceutical research and therapeutic development. The main focus of this innovative company is to speed up the preclinical and clinical drug testing by bio-printing human tissues and organs which mimics the human organ in vitro. The 3D bio-printed constructs enable the researcher to develop treatments and therapeutics faster, at very low cost and without risk to the living subjects. To assist the drug development process, Organovo now associated itself with biopharmaceutical and pharmaceutical companies and renounced academic medical research centres to design, build, standardised and validate more human-like in vitro tissues for disease simulation and drug, efficacy and toxicity testing.
The 3D bio-printed tissues and organs printed form human/autologous cells theoretically provides similar microenvironment as that of tissues and organs inside the body. Individual cells of the 3D construct experience the similar microenvironment as that of the tissues of the body. This provides an opportunity to the researcher to carry out the drug testing experiments in vitro in living tissues and organs. This also eliminates the possibility of the testing of drugs in living human subject; thereby bridging the gap between preclinical experiments and clinical trials.
Organovo’s bio-printed tissues are created from human cells. Bio-printed construct recreates various biological aspects in vitro, e.g., microenvironment and biology, reciprocal interactions between cells and micro-environmental factors and simulation of original tissue extracellular matrix including extracellular electrolytes. Organovo’s exVive3DTM bio-printed human tissues may reduce the failure risks and costs involved in the drug and therapeutic development process. Drug testing experiments in vitro 3D printed human tissues enable to secure human tissue-specific data prior to initiating the clinical trials in humans.
The liver is the primary site for the metabolism of many endogenous (e.g., hormones) and exogenous (e.g., xenobiotics) substances. Organovo’s exVive3D liver is a bio-printed human liver model composed primary of hepatocytes, hepatic stellate cells and endothelial cells. Organovo’s exVive3D liver tissue secretes important proteins like fibrinogen, albumin and transferrin proportional to levels in whole liver. Levels of ATP and lactate dehydrogenase secreted are also in the normal range when compared with the whole liver. This liver model could be a very important tool to study the route of metabolism of various exogenous and endogenous substances.
The realistic implications of 3D printing technology in drug discovery and development process involves the optimisation of the preclinical and clinical research methodologies. The research gap present between the lead molecule optimisation, preclinical studies and clinical research could be filled by the 3D construct of human tissues. Moreover, 3D constructs can reduce the failure risk and cost associated with the final stages of the drug discovery and development process. The 3D bio-printed models, unlike traditional cell culture models, could be standardised and validate for answering the complex questions related to the human cancer biology at molecular and tissue levels.
Today’s 3D bio-printed human research data is not sufficient enough to replace the classical cell culture and animal models. However, the recent pragmatic shift towards the 3D bio-printed tissues and organs may be sufficient enough to generate enough evidence to prove its usefulness in drug discovery process. Sooner or later the researcher will be confident enough to make a call with a high level of confidence. The Early conclusion at the preclinical stage could be possible with the advancement in the 3D bio-printed technology; thereby reducing the risk associated with final-stage clinical trials.
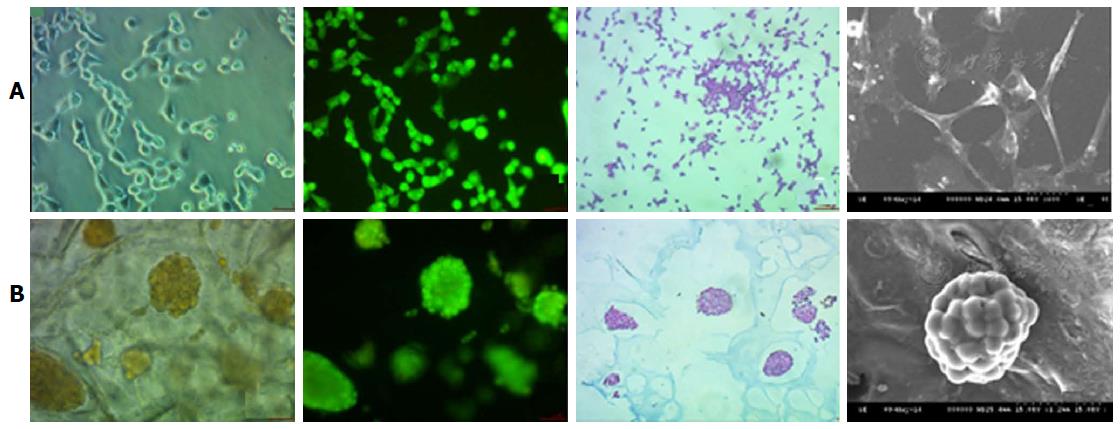
Early prediction of the risk associated with the drug discovery process could be reduced with the help of 3D printed tissues, e.g., Mou et al[60] used non-small cell lung cancer 95D cells to co-culture with a 3D bio-printed scaffold to construct a lung cancer model in vitro. This study of Mou et al[60] was focused on the relative comparison of the biological functions of lung cancer cells under the 2D and 3D environmental conditions. The 3D scaffold was constructed using the natural products like agarose and alginate and 3D printing technique was utilised to deposit the cell cultures on the scaffold. 95D cells types were used to co-cultured with this scaffold. The most important observation of this research tells us about the spindle and polygonal morphology of the cell cultured in 2D wells, whereas those cells which were grown in the 3D culture aggregated into spheroids and was able to migrate and invade the surrounding area of the scaffold (Figure 3).
Figure 3 Nonsmall-cell lung cancer 95D cell morphology under two-dimensional and three-dimensional culture conditions.
The 2D cultured cells (A) are tiled, polygonal, of long spindle shape and display more pseudopodia. In contrast, 3D morphology culture groups (B) are a combination of round and oval shapes, display intercellular tight aggregation and adhesion. Furthermore, there is evidence of multiple sizes of cells distributed in different scaffold pores[60]. 2D: Two-dimensional; 3D: Three-dimensional.
Cell metabolic activity assay showed that the multiplication rates of the 3D-cultured cells for 2-6 d were significantly lower when compared with the 2D-cultured cells. On the other hand, those cells which were cultured for a longer time (8-9 d) were significantly higher than that of the 2D-cultured cells, demonstrating the proliferative activity of the cancer cells grown in 2D cultures for 8-9 d was inhibited. It is also observed that the cells grown on 3D scaffolds maintained a high rate of proliferation over the longer period of time. At the end, it was concluded that not only the cell morphology and proliferation rate was different but also the associated protein expression was different. The growth of the lung cancer cells in 3D culture was also found to be different from the 2D cultured cells. We can also conclude that the agarose-alginate 3D scaffold can better simulate the microenvironment of lung cancer in vivo and in future this 3D construct may be established as a promising model for research in lung cancer.
Bone were constructed using human mesenchymal stem cells which were co-printing with acrylated peptides and acrylated poly (ethylene glycol). Inkjet bio-printing technique was used to make this construct[61]. Bone marrow stem cells with hydrogels like alginate, agarose, Matrigel®, and Lutrol® F127 were dispensed together using 3D bio-printer[62]. The printed bone marrow stem cells in combination with hydrogels were found to be functional and viable in the 3D construct. A mechanically stronger 3D bio-printed construct containing two different cell types has also been fabricated for osteochondral tissue regeneration[63].
Adipose-derived stem cells have the versatile ability to differentiate along with multiple lineage pathways. These cells could be isolated from human adipose tissue and could play the crucial role in regenerative medicine. Yao et al[64] used adipose-derived stem cells along with hydrogel (gelatin-alginate) to bio-print 3D construct in cubical shape. This work has significantly contributed to the idea of 3D construct of adipose tissue with functional vessels for efficient blood flow. Development of blood vessels inside the 3D printed adipose tissue means the better simulation to study complex biological phenomenon’s in vitro, e.g., differentiation of stem cell, cell signalling and interaction etc. One important finding of this study is that adipose stem cells not only proliferated of its own but were also found to differentiate within the 3D construct. When basic fibroblast growth factor was added, cells present in the 3D scaffold converted into endothelial cells and the cells rooted in the hydrogel separated into adipose-like cells. The constructs were found to remained intact for around 60 d[65].
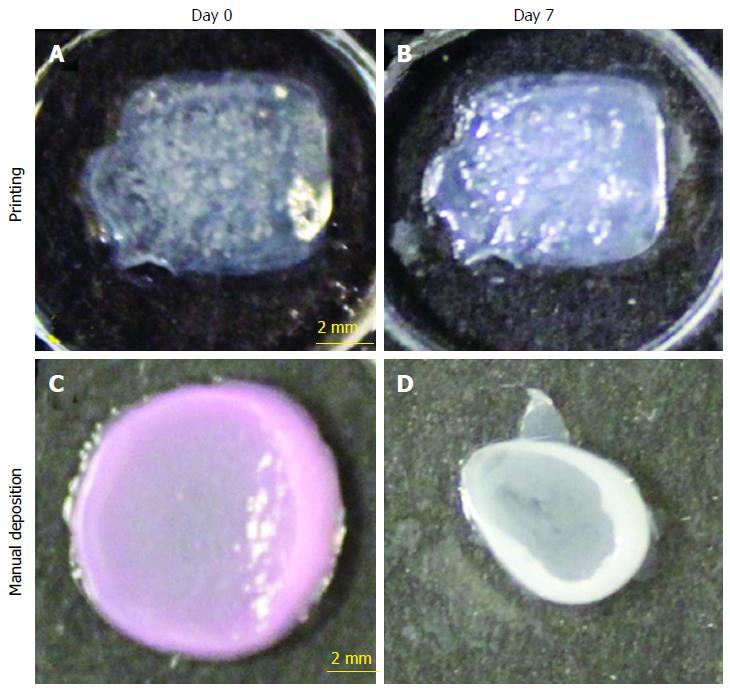
Lee et al[66] used cells like keratinocytes, fibroblasts and collagen to develop the skin construct in vitro. Keratinocytes represented and converted to epidermis layer, fibroblasts into dermis layer and collagen epitomised the extracellular matrix of the skin (Figure 4). Histological, biochemical, light and fluorescence microscopic examinations have proved that the 3D printed skin was not only morphologically but was also found to be biologically similar to the natural skin[66,67]. Koch et al[68] on the other hand utilised laser-induced forward transfer (LIFT) for the development of 3D skin. Koch et al[68] used skin cells like fibroblasts and keratinocytes to represent the cells of dermis and epidermis layers of skin respectively and also used human mesenchymal stem cells for differentiation into other useful cells. All these cells were used in the form of bio-ink and were then deposited using laser-induced forward transfer method.
Figure 4 Shape and form of printed skin tissue.
A comparison of skin tissues fabricated via 3D bio-printing and manual deposition indicates that printed skin samples (A, B) retain their form (dimensions) and shape, whereas manually deposited structures (C, D) shrink and form concave shapes (buckle) under submerged culture condition after 7 d.
Vascular system transports oxygen, nutrients and toxic residue to-and-fro from the cell and hence considered as the very important component of the complex organ system. In regenerative medicine, development of the in vitro vascular structures could help us to bio-print the bigger and hugely complex organ[69]. Skardal et al[70] was the first to cross-linked tetrahedral polyethylene glycol tetracrylates with hyaluronan hydrogels to generate the 3D bio-constructed vascular system. Skardal et al[70] utilised bio-printers which work in the principle of extrusion (Figure 5). Recently Kolesky et al[71] also developed the complex vascular scaffold using gel-based cellular suspensions, sacrificial and fugitive gel and casting cavity filled with a GelMA gel.
Figure 5 Cross-sectional images of three-dimensional bio-printed tissue (NIH 3T3 cells) containing an encapsulated fluorescent HA-BODIPY tracer for increased visualisation.
Cross-sectional views of the bio-printed vascular constructs were taken (A) immediately after printing; (B) at 14 d; and (C) at 28 d of culture using LIVE/DEAD staining to highlight viable and dead cells. Green fluorescence indicates calcein AM-stained live cells[70].
Miller et al[72] first time used bio-printed complex vascular structure using carbohydrate glass. Carbohydrate glass was used as a sacrificial substrate/template for the cell adhesion. The sacrifice of the carbohydrate glass after cell deposition lead to the formation of the cylindrical vessels. Carbohydrate glass wall was lined with endothelial cells and the blood was forced through it under high pulsated pressure. After sacrifice of the carbohydrate glass wall, the hollow channel network left behind was populated with human umbilical vein endothelial cells to attach themselves to the wall of hollow channels. As compared with the other methods discussed earlier, Miller et al[72] approach is not only simple and gives greater control over the network geometry but is also well-suited with the different types of natural and synthetic extracellular materials, different variety of cells and various cross-linking methods. Miller et al[72] also proved that the vascular system was able to tolerate the metabolic function of rat hepatocytes in 3D engineered constructs[72]. Norotte et al[73] on the other hand, developed a method for preparation of the scaffold-free vascular tissue l. Norotte et al[73] utilised fibroblasts and smooth muscle cells with agarose as the supporting gel.
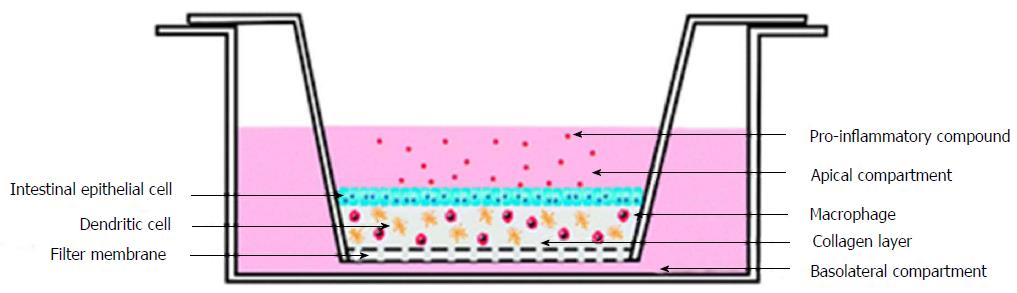
To study the inflammation in the intestinal mucosa Leonard et al[74] developed a complex in vitro model. Leonard et al[74] have utilised enterocyte cell line, immunocompetent macrophages and dendritic cells to construct 3D-fabricated intestinal mucosa model. This 3D printed intestinal mucosa model was then stimulated with the help of lipopolysaccharides from Escherichia coli and Salmonella typhimurium, interleukin-1β, and interferon-γ. Stimulation helped to develop the natural pathophysiological changes which occur in the intestine during inflammation. Different cell lines like Caco-2, HT-29 and T84, were used to develop the 3D constructs and were stimulated with the same pro-inflammatory molecules. It was observed that the Caco-2 cells were highly responsive towards the pro-inflammatory interleukin-1β molecules (Figure 6).
Figure 6 Experimental setup of three-dimensional co-culture comprising of intestinal epithelial cells, macrophages and dendritic cells[74].
The above-mentioned examples of 3D bio-printed tissues and organs could fasten the therapeutics development process and would facilitate the in vitro study of cancer pathophysiology. Recent advancement in the stem cell technology (Induced pluripotent stem cell) will hugely supplement the research in 3D bio-printing. Induced pluripotent stem cell has the unique character of dedifferentiated and then redifferentiated into tissues of choice[75]. Induced pluripotent stem cell technology has the very important role to play in 3D bioprinting and in solid organ transplantation. In the future, patient specific 3D tumour model also has the ability to revolutionised the field of personalised treatment.
ADVANTAGES OF 3D PRINTED TUMOUR MODELS - A COMPARISON WITH 2D PLANAR MONO-CULTURE AND 3D CO-CULTURE MODELS
The most efficient way of learning about the tumour progression and anticancer drug evaluation is by regulated and structured clinical trials on humans. However, direct evaluation of pathophysiological process in cancer development and anticancer activity of drugs is highly unethical because of the safety concerns. To overcome ethical challenges, preclinical studies on tumour models are highly appreciated. Several preclinical tumour models like cell culture, xenograft, mouse model and 3D tissue culture are developed which are thought to resemble with the natural tumours in terms of pathophysiological processes involved[76-78]. Evidence are now available which proves that the tumour microenvironment is the key regulator of the several stages involved in the pathophysiology of cancer progression. Tumour microenvironment is particularly very important in terms of the development of resistance, inventions of the distance organs and escape from the immune surveillance
This recent development not only challenged the past concept which mostly focused on the tumour cells but also impacted the research strategies of future. In future, the medical interventions in clinical oncology will also involve the therapeutics targeting the microenvironments. A systematic and methodological study of the tumour microenvironment, with the help of 3D bio-printed tumour models, would promote evaluation and selection of candidate agents from preclinical trials[79]. This would not only fasten the drug development process but would also save the resources.
A factor that plays an important role in the advanced malignancies is inappropriate activation of the supportive tissue called stroma. In most of the malignancy cases, stroma loses its connective and structural role. The various types of stromal cells are pericytes, smooth muscle cells, adipocytes immune cells, endothelial cells, fibroblasts, etc. Tumour microenvironment also found to contain various growth factors, many hormones, several structural and functional proteins, enzymes, cytokines and small cytokines of which most works as a primary and secondary signalling molecules and ligands for the receptors. The presence of all these functionalities in microenvironment could widely affect not only the pharmacokinetics but also the pharmacodynamics of the anticancer drugs. Thus the therapeutic outcome is widely regulated by the normal or abnormal expression of these extracellular proteins. It is now well recognised that protein and gene function varies strangely when studied them in vivo and in vitro. Studying the effect of these genetic alterations on drug response in either original or damaged neoplastic microenvironment is very critical for the fruitful drug development, translational anticancer regimes, and optimisation of therapies. These and several other factors are vital for the development of malignancies and are very difficult to re-orchestrate in 2D and co-culture models[80,81].
The genetically activated stroma of sarcomas and carcinomas is not only composed of cancer associated fibroblasts and myofibroblasts but can be identified due to altered matrix components, change in the proteins synthesis associated with repair machinery and reprogrammed breakdown process[80,81]. Except for the supportive function, stromal cells also play the important role in the physical and biological protection of microenvironment protection. This functionality actually limits the effective delivery of the therapeutic drugs to the cancer cells. Altered components of the tumour microenvironment, including the synthesis of the proteins involved in the repair mechanism, allows the unrestricted growth of the tumour cells. Tumour cells in favourable environment successfully evade the apoptosis signals triggered by cytotoxicity and develop various resistance strategies to select the malignant phenotypes.
Correlation of the survival rate and capability of stroma to overpower the carcinogenesis is already established[82]. However, once distorted to a tumour-associated neighbour because of the stimuli like inflammation, infection, mutation, etc., the stromal protective function can be altered to stimulate the proliferation[83-85]. Under the altered condition, stromal cells start to evolve with the cancer cells and begin synthesis of growth factors, cytokines, chemokines, etc., which fast-track the disease progression[86]. In addition to this, many in vitro studies have proved the complex role of the tumour microenvironment in cancer development. Experiments with genetically modified stroma proved the importance of the tumour microenvironment in disease progression[87,88].
Infection, immune-associated signalling and inflammation have been found to be associated with several cancer types. For example, liver carcinoma which is the leading cause of death in patients with liver cirrhosis and increased the risk of colorectal cancer in the patients with increased inflammation is credited to unresolved inflammatory signalling[89]. Similarly viruses, bacteria and parasites are also the leading cause of the variety of cancers. A higher incident of multiple cancers like gastrointestinal tract, lung, reproductive and skin cancers has been found in female immunosuppressed organ transplant recipients[90]. Retrospective analysis revealed a higher incidence of AIDS-associated cancers (e.g., Kaposi’s sarcoma, Cervical cancer, Non-Hodgkin lymphoma), and non-AIDS-related cancers (e.g., tongue, skin, lung, CNS and multiple myelomas) in HIV-infected patients[91]. Various enzymatic proteins, like matrix metalloproteinase, in particular, matrix metalloproteinase-2 and matrix metalloproteinase-9 have a role in the tumour progression. For example, matrix metalloproteinase-2 and matrix metalloproteinase-9 allow cancer cells to breach through the extracellular matrix of the tumour microenvironment and are closely related to cancer metastasis. The activity of the various matrix metalloproteinase is found to increase with the development of cervical cancer[92] and can be studied efficiently in 3D bio-printed tumour models[93].
Development of the resistance towards the therapeutic intervention is the foremost challenge in clinical oncology. In addition to fuelling the tumour growth, the altered tumour microenvironment modifies treatment responses by affecting cell sensitivity towards anticancer agents. Decreased cell sensitivity towards anticancer drugs gives rise to the drug resistance. The drug resistance facilitated by the alteration tumour microenvironment is not limited to classical agents like chemotherapies. Instead, it covers various therapeutic materials, including targeted agents and targeted drug delivery systems[94]. The role of tumour microenvironment in the protection of acute myeloid leukaemia or chronic lymphocytic leukaemia cells from pharmaceutical agents like anthracyclines, alkylating agents, imatinib and nucleoside analogues has been recently evaluated. The defending role of tumour microenvironment is detected in the protection of the mutant Janus kinase 2 cells from Janus kinase inhibitors. Tumour microenvironment role is also observed in protecting solid tumours from erlotinib and cetuximab. Similarly, recent findings described the protection of melanoma against RAF inhibitors, like vemurafenib[95-97]. Tumour microenvironment assisted resistance is found to be directed through several cell lineages and alteration in the stromal components (e.g., fibroblasts, endothelial cells, etc.)[94,98].
Tumour microenvironment assisted protection of tumour cells applies to multiple therapeutic strategies and varies with the inter-individual differences. For example, in the treatment of melanoma by mitogen-activated protein kinase pathway inhibitors, tumour-associated macrophages multiplies and release cytokine-like tumour necrosis factor-α as a crucial growth factor that provides resistance to the targeted therapy through the microphthalmia transcription factor[99]. Similarly, certain cancer endothelial cells secrets interleukin-6 and tissue inhibitor of metalloproteinases-1 as the survival factors. Both of the factors were found to be significantly involved in the resistance of lymphoma when the Eμ-Myc mice model of Burkitt’s lymphoma treated with anticancer antibiotic doxorubicin. This could be reversed or good chemotherapeutic efficacy could be achieved by the inhibition of these survival factors or by stimulating the p38 mitogen-activated protein kinase pathway[100]. Another noted example of tumour microenvironment-exerted protection of cancer cells is the chemoresistance caused by the amplification of the CXCL1/2-S100A8/9 loop by antineoplastic agents used in breast cancer treatment[101].
The examples illustrated above demonstrate various pathways by which therapies or targeted agents can be affected by the changes in the tumour microenvironment. Tumour microenvironment not only contains the tumour cells but also contains the several other cells, e.g., immune cells, lymphatics cells fibroblasts, pericytes, etc. This composition of microenvironment essentially affects the therapeutic outcome[102]. The 2D monolayered and 3D coculture cellular models lack illustrated characteristics of natural 3D tissues in vivo[103]. 2D monolayered culture has the increased drug diffusion properties which do not match with the natural tumour character. A lot of drugs have their site of action inside the cells and hence their penetration is very important for effectiveness. This character of cell culture models explains the importance of three-dimensional arrangements for the proper success of the therapy.
To overcome the drawback of the cell culture models various alternative animal models were developed, e.g., genetically altered and immunocompromised mice models. Animal models have contributed enormously to the present understanding of cancer, however, they could not reflect the actual pathophysiology involve in disease progression because of the species differences[104].
To overcome the hurdles of simulating the exact complex tumour microenvironment in cell culture, 3D printing technology was adapted to produce the 3D bio-printed tissues and organs. Similarly, 3D printing technology could be easily utilised to produce the 3D tumour models which subsequently could be utilised to study the cancer biology and anticancer drugs[105,106]. Various techniques, such as cell-seeding 3D scaffolds, hydrogel embedding, multicellular spheroids, cell patterning and microfluidic chips have been explored for the construction of 3D tumour models in vitro[76].
Several advances in 3D printing technology and stem cell research offers unique opportunity for the construction of complex organs and tumours. The 3D printed organs and tumour models essentially simulate the exact physiological and pathophysiological microenvironments. The exact recreation of the tumour microenvironment facilitates the better understanding of the disease[107,108].
Till date, very few reports have been published describing the 3D printed tumour models. Zhao et al[93] demonstrated the use of HeLa cells in gelatin/alginate/fibrinogen hydrogels to bio-print the 3D in vitro models of cervical tumours. When compared with 2D cell culture model, 3D printed tumour model have shown 90% proliferation rate. Zhao et al[93] also observed the increased expression of matrix metalloprotease protein and chemoresistance in 3D printed tumour models when compared with 2D cell culture model. Work of Zhao et al[93] is just one example of the advancement of 3D bio-printed tumour model, with further advancement in 3D printing technology, a revolution in the field of cancer research is on the corner.
CONCLUSION
The 3D bio-printing of tissue and organ models is a developing field in which several ground-breaking results have been obtained over the past few years. The 3D-bioprinted tissue constructs are being prepared not only for the solid organ transplantation but also for use in drug discovery process. Fabrication of the realistic tissues, organs and tumour models with the help of the various 3D bio-printing techniques is now possible. Extrapolation of the results obtained from the cell culture and animal models are not trustworthy because of the species differences. This challenge of species difference could be overcome by printing the 3D tissues and organs from the human cells. The 3D printed tumour model fabricated from the human tumour cell lines will definitely revolutionise the oncology research. The 3D printing is the very precise which could be demonstrated by its (inkjet printer) use in transfecting genes into cells[109,110]. In coming days, 3D bio-printed tissues and organs will find its way in the pharmacological and toxicological testing of the molecules under drug development process. Bio-printing has the potential to change the way the drug enters the clinical trials after preclinical studies. The 3D printing not only has the capability to improve the attrition rate of the clinical trials but will also reduce the cost and time required in the drug discovery process. This is possible because of the speedy identification of the efficient candidate molecule. Use of 3D bio-printed models will eliminate the need of animal models and hence the data obtained in the preclinical studies will be more trustworthy.
Most published results are the early prediction and only a few studies methodologically explored the developmental method parameters. Standardisation and optimisation of the printing process parameters are essential for the successful adaption of the 3D printed tissues and organs to use them in drug development process. This is possible to achieve to by establishing the relationship between structural and functional parameters. Moreover, modern fabrication schemes rely on mathematical modelling and computer simulations for optimising the process design and making predictions[107,109]. Therefore the performance of the tissue constructs could be predicted virtually using computer simulations before actually printing the construct.
Stem cells already have revolutionised the field of regenerative medicine and have very important role to play in the construction of 3D tissue, organs and tumour models. Stem cells (e.g., induced pluripotent stem cells) offer greater possibility for fabricating complex constructs because of their ability to differentiate in various another kind of cells, as highlighted by various research groups[107,109,111,112]. However, some issues need to be fixed before stem cells can be used for 3D bio-printing. This issue includes optimisation of the cellular microenvironment to combine the advantages of cell attachment, cell stimulation and mechanical stability to mimic the in vivo environment to the highest degree.
Printed 3D models match closely with the natural organs and when compared with the cell culture models. Novel 3D cell printing technology may help to develop the tumour models in vitro which will be more useful in studying cancer cell biology. Although, 3D bio-printing techniques are still in their infancy, they offer potential to overcome many challenges associated with the production of complex tissues and organs. This technique is a promising tool for replacing current and often misleading results obtained from cell culture and animal based screening of pharmaceuticals. Interdisciplinary research and collaboration of the researcher from the various field are required to overcome the hurdles before 3D bio-printed concept accepted by the institutional and pharmaceutical researchers. To be successful, we will have to sort-out the progressive challenges of 3D bio-printing, including cell sources and biocompatible material requirements, proper vascularization and autonomous maturation and continuous functionality of the construct.