Published online Dec 10, 2014. doi: 10.5306/wjco.v5.i5.883
Revised: September 13, 2014
Accepted: October 1, 2014
Published online: December 10, 2014
Processing time: 344 Days and 21.5 Hours
The purpose of this study was the overview of current knowledge regarding the use of survivin and its isoforms in prognosis and treatment of breast cancer. An advanced search of Medline was performed using the following search strategy: “(survivin isoforms) OR (survivin transcript variants) AND (breast cancer) AND (neoplasm OR tumor OR cancer OR carcinoma)”. Relevant studies were retrieved and processed thoroughly in order to analyze the related data. Besides wild-type survivin full-length transcript, another six splice variants have been identified. Overexpression of survivin and its isoforms leads to shorter overall and disease-free survival; the transcript variants are correlated with apoptosis and could assist prognosis prediction. It has been proved through numerous studies that inhibiting survivin isoforms might become a promising target of drug therapy of carcinomas. Use of small molecule YM155 could offer new therapy for triple negative breast cancer patients, while, chemotherapy with 5-fluorouracil + epirubicin + cyclophosphamide and Tax-Epi could be guided by survivin splice variants measurements. Survivin transcript variants could become prognostic biomarkers and could provide information about clinical management of patients suffering from breast cancer.
Core tip: Besides wild type survivin full length transcript, another six splice variants have been identified. Overexpression of survivin and its isoforms leads to shorter overall and disease-free survival; the transcript variants are positively correlated with apoptosis and could assist prognosis prediction. It has been proved through numerous studies that inhibiting survivin isoforms might become a promising target of drug therapy of carcinomas. Use of small molecule YM155 could offer new therapy for triple negative breast cancer patients while, chemotherapy with 5-fluorouracil + epirubicin + cyclophosphamide and Tax-Epi could be guided by survivin splice variants measurements. Survivin transcript variants could become prognostic biomarkers and could provide information about clinical management of patients suffering from breast cancer.
- Citation: Pavlidou A, Kroupis C, Dimas K. Association of survivin splice variants with prognosis and treatment of breast cancer. World J Clin Oncol 2014; 5(5): 883-894
- URL: https://www.wjgnet.com/2218-4333/full/v5/i5/883.htm
- DOI: https://dx.doi.org/10.5306/wjco.v5.i5.883
Globally, breast cancer is the most common type of non-skin human malignancy and the second cause of cancer-related deaths amongst women in the Western developed World. Breast cancer is responsible for 22.9% of all new cancer cases among women worldwide and 13.7% of cancer deaths[1]. There are numerous factors associated with the occurrence of breast cancer, such as: genetic susceptibility arising from mutations in high/moderate penetrance genes (such as BRCA1/2, PALB2, CHEK2, BRIP1, RAD50, NSB1 etc.)[2-3], hormone-associated reproductive factors such as increased menstrual cycles arising from either earlier age at menarche or later age at menopause or decreased parity, older age at first birth and use of hormone therapy; consumption of alcohol and type of diet; obesity; exposure to radiation; atypical hyperplasia of the mammary gland[4].
Inhibition of apoptosis causes tumorigenesis through cell survival resulting in accumulation of genetic mutations that lead normal tissues to transformation. Apoptosis is a tightly controlled procedure of cellular death, which is crucial for tissue homeostasis. Proteins of the bcl-2 family and the inhibitors of apoptosis (IAP) family belong to key regulators of apoptosis[5].
The main characteristics of the IAP family are the baculovirus IAP repeat (BIR) domains and the blocking of apoptosis by inhibiting directly caspases and procaspases. Till today eight proteins of IAP family were identified, survivin corresponds to the baculoviral IAP repeat-containing 5 domain (BIRC5). Among them, survivin and XIAP have attracted the research interest as therapeutic targets for various malignancies[6].
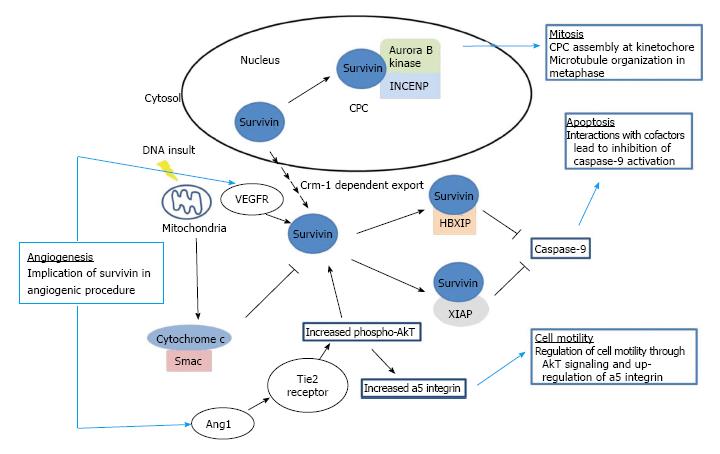
Survivin is the smallest member of IAP family and is a multifunctional protein, which participates in the control of apoptosis, angiogenesis and proliferation (Figure 1). Additionally, since survivin is a member of the family of the chromosomal passenger complex (CPC) proteins, it interacts with Borealin, the Aurora B kinase and the inner centromere protein (INCENP) in order to carry out substantial roles in cell division[7]. Recently, a critical role for survivin in the control of autophagy was also reported[8]. Survivin, normally expressed during embryonic and fetal development, is downregulated in adult tissues and overexpressed in a variety of human cancers[9-12]. Inhibition of apoptosis by survivin is a predictor of poor prognosis and shorter survival in patients suffering from various carcinomas[13].
In this review, we analysed the literature data regarding the correlation of survivin isoforms with clinicopathological characteristics of breast cancer and their prognostic significance in order to explore the inclusion of survivin isoforms -besides wild-type survivin- as prognostic biomarkers in emerging multiparameter technologies examining tissue RNA expression (analogous to Oncotype, Mammaprint, PAM50[14,15] or their clinical use in target-oriented therapies.
The survivin gene is located on chromosome 17q25 and up to seven alternatively spliced surviving transcripts have been detected so far[7]. Alternative splicing of precursor messenger RNA (mRNA) is a process by which the exons are connected, so they could generate different mRNAs and proteins. Alternative splicing is a significant procedure which maintains the diversity of the genome. Over 95% of the human genes produce different splice variants which in many occasions present opposite tasks[16]. Recently there has been evidence that differences in the splicing process can provoke myeloid cancer[17], while comprehending this process is important in designing new therapies for cancer. One more research uncovered the fact that a common regulatory network is responsible for the simultaneous expression of a number of functionally associated splice isoforms[18], which supports the idea that we should consider better as therapeutic targets the whole group of different transcript variants or their common regulatory network, rather than targeting on a single gene product[19].
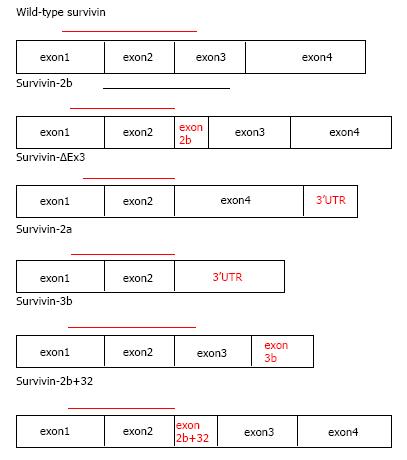
Except for wild type survivin full-length transcript, six other aberrant splice variants have been identified (Figure 2): (1) survivin-ΔΕx3 that is created from the removal of exon 3 and also contains part of the 3’ untranslated region; (2) survivin-2b that arises from the inclusion of part of intron 2 termed in this variant as “exon 2b”; (3) survivin-3b that originates from the inclusion of part of intron 3 termed in this variant as “exon 3b”; (4) survivin-2a stems from the insertions of exon 1 and 2 at the 5’ end of intron 2, 197nt of intron 2 are added, of which 195nt are noncoding[20]; (5) the recently described survivin-2b + 32 which combines intronic sequence 2b and an insertion of 32 additional nucleotides from intron 2 and therefore includes “exon 2b + 32”[5] and finally the latest addition is (6) survivin-image (SI), which consists from a part of the survivin gene (345 bp), a part of the image gene (155 bp) of eye cancer and another insertion of 7 bp. SI alternatively spliced isoform could be involved in other functional pathways related to tumorigenesis[21].
The survivin variants present differential intracellular localization that possibly regulates their antiapoptotic potential. Co-expression experiments have showed that wild-type survivin can heterodimerize with its splice variants. Heterodimer formation can lead to specific subcellular localization patterns, implying that high expression in tumor cells can lead to formation of functionally distinct survivin complexes. Co-expression of wild-type survivin with survivin-ΔEx3 results in the recruitment of these complexes to the mitochondria, where they inhibit mitochondrial dependent apoptosis[22]. The intracellular localization of survivin isoforms according to one study, depends on a Crm1-dependent nuclear export signal (NES) present in survivin, survivin-2b and survivin-3b, but absent in survivin-ΔEx3 and survivin-2a. These survivin isoforms enter the nucleus by passive diffusion since they lack an active nuclear import signal. The NES acting in consortium with an appropriate CPC formation are responsible for the cytoprotective activities of some of the survivin isoforms, as well as for their correct localization and function during cell division. Among all isoforms, only survivin-3b is cytoprotective and interacts efficiently with CPC proteins[7].
The functions of the variants are not fully understood. Survivin-ΔEx3 has been described as an anti-apoptotic protein. Survivin-2b probably has pro-apoptotic action since it possesses a truncated BIR domain and still it dimerizes readily with wild type survivin causing the reduction of the anti-apoptotic effects of wild type survivin. Survivin-3b contains a complete BIR domain and therefore has a potential anti-apoptotic activity. Survivin-2a does not have the domain involved in inhibition of apoptosis and seems to have opposed activity to wild-type survivin.
Survivin-ΔEx3, is probably associated with cell immortality, or else remains inactive or assists angiogenesis. In the same way, survivin-2a and survivin-2b are cytoprotective, proapoptotic or sometimes they seem to be inoperative by conflicting manuscripts. Current research suggested that cancer development and survival of people suffering from tumors could be associated with the differential expression of survivin transcript variants. An explanation of this hypothesis could be that survivin variants display a trans-dominant negative (TD) phenotype enacted by the creation of silent heterodimers with survivin-wild type or by the titration of CPC proteins and/or Crm1. In conclusion, survivin-3b fulfils the molecular requirements, i.e., interaction with Crm1 together with the capability to cooperate with all CPC components, demanded to perform “wt survivin-like” biological functions[7].
It should be certainly noted that is difficult to unify all data concerning survivin variants, since results are controversial, but the accumulated information could be important for designing future strategies of therapy by using specific survivin transcripts as targets.
It is possible that survivin may be of great value in the prognosis of breast cancer patients. In this part of the review, we compiled the experimental data of four studies, examining the expression of survivin transcript variants accordingly to clinicopathological characteristics of patients with breast cancer and their survival (Table 1).
| Ref. | Examined variants | Methods | Association with patient characteristics | Conclusion |
| Végran et al[22] | wt, sur2b, surΔΕx3, sur3b, sur2a | Real time qPCR | Tumor grade, estrogen receptors, lymph nodes | High expression sur3b tumors had shorter OS and DFS |
| Span et al[23] | wt, sur2b, surΔΕx3, sur3b, sur2a | Reverse transcription qPCR | Age, tumor grade, lymph nodes, steroid hormone receptors | High expression of sur2a, sur3b and wt indicate poorer prognosis |
| Ryan et al[10] | wt, sur2b, surΔΕx3 | Reverse transcription qPCR, Western blotting | Tumor size, nodal metastases, breast cancer type | Wt and surΔΕx3 were correlated posively with apoptosis |
| Pavlidou et al[4] | wt, sur2b, surΔΕx3 | Real time qPCR | Tumor grade, estrogen receptors | Sur2b/wt had significant association with estrogen receptors |
| Athanassiadou et al[24] | wt | Immunocytochemistry, immunohistochemistry | Grade, lymph nodes, tumor size | increased wt may indicate a worse prognosis |
In the study of Végran et al[22], survivin-3b variant expression provided the major findings in patient samples, presenting with inverse correlations with ten pro-apoptotic genes and five anti-apoptotic genes. These results may suggest a feedback loop at the gene levels with survivin-3b and could be a way that cancer cells use in order to counteract survivin-3b overexpression. The data are in agreement with the putative anti-apoptotic role of survivin-3b suggested initially by its amino acid sequence. The results also confirm previous observations concerning the higher expression of the antiapoptotic survivin-ΔEx3 and survivin-3b in p53-mutated breast tumors. However, some studies found out that survivin-ΔEx3 expression increases with histological grade and is more expressed in ER-negative tumors. Survivin-2b isoform is overexpressed in high-grade ER-negative and node-invasive tumors. This result is not in accordance with its putative proapoptotic role but it indicates that survivin-2b could be a marker of aggressiveness. Survivin-2a -probably due to its potential pro-apoptotic role- is more present in low-grade and non-invasive tumors. Végran et al[22] results revealed that survivin, survivin-ΔEx3, and survivin-2b have no prognostic role in breast carcinoma. A pro-apoptotic role for survivin-2a has been ascribed; however, in this study it was found to be associated with the worst prognosis of breast cancer patients.
Span et al[23] 2006 discovered that survivin-wild type, survivin-2a, and survivin-3b were associated in patient samples with poor relapse free survival. Survivin, survivin-ΔEx3 and survivin-2a variants were associated with younger age, advanced histological grade and steroid hormone-receptor-negative tumors, all factors that indicate poor prognosis for a breast cancer patient. Tumors from patients with many involved lymph nodes had lower amounts of the proapoptotic survivin-2b variant in the primary tumor than those with a limited number of involved lymph nodes. This negative correlation with lymph node status could suggest a role for survivin-2b in counteracting the antiapoptotic and/or cell cycle stimulating role of wild-type survivin. The data in this study confirm that some of the splice variants interact with each other and modify the prognosis of the patients. Specifically, high expression of the survivin-2a and survivin-3b, in addition to survivin indicate a poorer prognosis and the prognostic value of survivin is strongest in the presence of higher concentrations of these variants[23].
In the report of Ryan et al[10], both wild-type survivin and survivin-ΔEx3 mRNA correlated moderately with apoptosis in patient samples. Levels of the survivin-2b and survivin-ΔEx3 but not wild-type survivin were significantly higher in positive lymph nodes compared to the primary tumor. A weak but significant inverse relation was found between survivin-ΔEx3 and both tumor size and number of positive lymph nodes. This form of survivin was also detected more frequently in the ductal histological type compared to the lobular one. Previously, they have reported that caspase 3 levels were also significantly higher in ductal than lobular breast cancers. These findings suggest that the regulation of apoptosis is different in these two histological types[10].
In a previous study of ours (Pavlidou et al[4]) we have reported that survivin-2b, survivin-ΔEx3 and the ratio of survivin-2b/wild-type survivin showed in vivo a positive correlation with the grade of the tumor in breast cancer samples (P < 0.05). The two isoforms presented an increased expression in advanced histopathologic grade, a finding that was expected for survivin-ΔEx3 but not for survivin-2b due to its pro-apoptotic activity. The fact that the two isoforms are elevated in high grade tumors makes them possible markers of tumor aggressiveness. Perhaps, these two isoforms have important but opposite roles in cancer development. The ratio of survivin-2b/wild-type survivin is increased in the early stages I and II, a result which may be due to different and antagonistic functions of the two proteins. Also it was found a significant association between the ratio of survivin-2b/wild-type survivin and estrogen receptors, which further reinforces the fact that this particular variation of survivin mRNA could predict survival of breast cancer patients.
Nectins are cell adhesion molecules involved in epithelial cell physiology. Nectin-4 is a new tumor-associated antigen and a reliable biomarker for breast carcinoma. The in vivo association of survivin and Nectin-4 with unfavourable prognostic indicators, and with one another, suggests that these proteins may also interact in breast carcinoma in order to exert their adverse effect. In conclusion, the combined survivin and Nectin-4 expression demonstrates a strong independent association with poor prognosis[24].
The search for novel prognostic and predictive biomarkers could deter cancer patients from unnecessary, inadequate and toxic therapy. This effort has culminated in the availability of few -but useful-commercial standardized assays for breast cancer patients. Besides the routine immunohistochemical measurements of hormone receptors and the HER2 oncogene protein product, only one other effort looked at the protein level: the Mammostrat test (by Clarient Diagnostic Services, GE Healthcare) that assesses 5 molecules: SLC7A5, HTF9C, P53, NDRG1, and CEACAM5 by IHC[25]. It is not known whether the addition of survivin would strengthen the value of this approach (monoclonal antibodies to survivin now exist).
The majority of the other available assays have looked at the RNA level either with microarray expression arrays or with real-time PCR (RT-PCR) assays. The technology of microarray gene expression emerged as the best method to categorize patients based on their molecular signature and distinguish those who could use other therapy strategies. Nevertheless it is not yet inducted into the clinical practice due to some restrictions[6,26]. An FDA approved kit, Mammaprint (by Agendia Inc.) assessing the mRNA expression of 70 genes from frozen breast cancer tissues is being extensively used in order to stratify patients into two distinct groups: low risk or high risk of distant recurrence (with no intermediate results). The tumor cell percentage and an RNA integrity score are also provided. Survivin (BIRC5) was included in the genes tested in the original validation study; however it was not selected for the final 70-gene panel[27,28].
Other commercial kits employ RT-PCR assays for selected genes from RNA extracted from formalin-fixed paraffin-embedded tissues: (1) an 8-gene panel from Aviara (now Biotheranostics Inc) combining the Breast Cancer Index HOXB13:IL17BR ratio with the Molecular Grade Index consisting of the average expression of five cell cycle-associated genes (BUB1B, CENPA, NEK2, RACGAP1 and RRM2)[29]; (2) the PAM 50 assay (by ARUP Laboratories) that includes wild-type survivin among the 50 selected genes[30]; and (3) the FDA-approved Oncotype DX Breast Cancer Assay (by Genomic Health). Oncotype DX testing may aid a patient by preventing redudant chemotherapy and its connected toxicity, while reduction of chemotherapy also leads to economical changes in health systems[31,32].
It provides breast cancer patients with a Recurrence Score (RS) (a number between 0 and 100) that corresponds to a specific likelihood of breast cancer recurrence within 10 years of the initial diagnosis. This score is based on the relative expression of 16 different cancer genes measured in triplicate) normalized relative to a set of 5 reference genes within a range 0-15. A one unit increase reflects the doubling of expression. The Recurrence Score arises from the following equation[31]:
RS = + 0.47 × HER2 Group Score - 0.34 × ER Group Score + 1.04 × Proliferation Group Score + 0.10 × Invasion Group Score + 0.05 × CD68-0.08 × GSTM1 - 0.07 × BAG1
As seen in Table 2, survivin is one of the cancer genes belonging to the proliferation group and it corresponds to the highest positive factor in the RS equation (1.04). A low risk of recurrence is defined of less than 18, an intermediate risk as less than 31 and a high risk as 31 and higher.
| Proliferation group | Invasion group | Estrogen group | HER2 group | Reference genes | |
| Ki-671 | ST3 | ER | GRB7 (growth factor receptor-bound protein 7) | GSTM11 | ACTB1 |
| STK151 | CTSL2 | PR1 | HER2 [human EGF (epidermal growth factor) receptor 2] | CD68 | GAPDH1 |
| BIRC5 (Survivin)1 | Bcl2 | RPLPO1 | |||
| CCNB11 | SCUBE2 [Signal peptide CUB (complement proteins C1r/C1s, Uegf, and Bmp 1)-EGF domain-containing protein 2] | BAG1 | GUS1 | ||
| MYBL21 | TFRC1 |
In Stemmer’s et al[32] 2013 article, Oncotype DX testing seems to have significant impact on minimising chemotherapy in node positive/estrogen positive (N1+/ER+) patients with breast tumor. It has been proven that Recurrence Score is correlated with tumor grade; higher Recurrence Score results are associated with higher histologic grade. Only patients belonging to the high Recurrence Score group were prescribed for chemotherapy. The differences in the proportions of patients treated with chemotherapy between the low, intermediate, and high Recurrence Score groups were statistically significant[32]. The clinical application of Oncotype DX testing was correlated with a decrease in chemotherapy and change of treatment recommendations[32].
In another recent study, Solin et al[33] 2013 used the so-called 12-gene Oncotype DX DCIS Score, which quantifies local recurrence risk and provides risk information independent of traditional clinical and pathologic characteristics. The treatment of ductal carcinoma in situ (DCIS; intraductal carcinoma) of the breast is variable, with concerns about both overtreatment and undertreatment. Among those 12 genes is survivin, in the group of cancer proliferation-related genes as shown in Table 2. The DCIS Score for proliferation group, where survivin belongs, arises from the following equation:
DCIS Score = + 0.31 × Proliferation Group Score - 0.08 × PR - 0.09 × GSTM1.
The DCIS Score predicts the risks of invasive local recurrence and provides information that complements traditional clinicopathologic prognostic factors for women with DCIS. The differences in the risks of developing local recurrence and invasive local recurrence between patients with a lower DCIS Score and a higher DCIS Score were statistically significant and clinically meaningful[33].
In the future, it would be very intriguing to explore the inclusion of the other survivin isoforms as prognostic biomarkers in such emerging multiparameter technologies examining tissue expression (analogous to Oncotype, Mammaprint, PAM50 etc.[14-15] since it has been proved that they possess potential importance in apoptosis and proliferation of breast cancer cells, especially when compared to wild-type full-length survivin.
A lot of studies were designed in order to find out pathways and compounds, which can interact with survivin and its isoforms, since they seem to provide promising therapeutic targets in cancer. Initially these efforts had as an obstacle the structural properties of survivin, which assigned the molecule of survivin under the label ‘‘non-druggable’’. Despite the difficulties, numerous ways concerning survivin inhibition have been utilized and depend mostly on indirect mechanisms, e.g., by interference with the expression of the survivin gene, its mRNA processing, the intracellular localization and the binding partners, or by affecting stability of survivin protein or by provoking survivin-specific immune responses[34]. Research is continuing in order to deploy drugs which cooperate with survivin and restrain its operation[34,35].
Numerous proteins and protein complexes were recognized and are necessary for the growth and survival of cancer cells. RNA interference experiments-for example- can reduce the production of these proteins and confirm the fact that they are crucial for tumor cells in order to survive[34,36].
RNA-based therapeutics comprise small interfering RNA (siRNA), antisense oligonucleotides, and microRNAs (miRNAs)[19].
Chemically-modified antisense oligonucleotides consist approximately of 20 nucleotides and their annealing to mRNA permits its cleavage by ribonuclease H. A variety of alterations in their chemical structure was achieved, like using a phosphorothioate linkage instead of natural phosphates as a backbone of nucleotides and 2′-O-methyl residues. 2′-O-methoxyethyl residues or locked nucleic acids[19,37] were also developed in order to improve pharmacological quality and structural stability. However, the chemistry-dependent toxicities due to their structures are a significant problem expected to be solved[19].
siRNA is another structure of a double-stranded RNA which is composed by 21-22 nucleotides. The antisense strand of the siRNA after interacting with multiprotein RNA-induced silencing complex, anneals to the complementary mRNA and an endonuclease cleaves the annealed mRNA[19]. There is data showing that survivin is a transcriptional target of HA-CD44 signalling, therefore the specific inhibition of CD44 expression by siRNA, resulted in a specific decrease in survivin expression. This correlated with a significant loss of breast cancer cell invasiveness. Simultaneous increased expression of both CD44 and survivin was detected in late-stage metastatic breast cancer cells, expression that was absent in normal breast tissue samples[38].
Recently, a group of small non-coding RNAs, known as microRNAs, attracted cancer researchers[39,40]. MiRNAs are small, non-coding RNAs of 19-25 nucleotides in length that are endogenously expressed in mammalian cells. miRNAs regulate gene expression post-transcriptionally, by pairing with complementary nucleotide sequences in the 3’-UTRs of specific target mRNAs. miRNAs act either as oncogenes or as tumor suppressors. More than 50% of miRNA genes are located in cancer-associated regions of the genome. Deletion or epigenetic silencing of a miRNA that e.g., normally suppresses one or more oncogenes could cause tumorigenesis, progression and invasion, as it was showed for miR-200, miR-122 and miR-203. Survivin was recognized as direct target of miR-203 in TNBC cells. Furthermore, the fact that overexpression of miR-203 can repress proliferation and migration of TNBC cells and is escorted by a reduction in BIRC5 expression, proves that miR-203 acts as a tumor suppressor in TNBC[41].
The multiple operations of survivin in individual subcellular compartments are probably fulfilled through the domain structure of the molecule and finely regulated by secondary alterations. The modifying enzymes could also become probable targets in order to investigate how the interference with survivin functions and its specific subcellular localization may be utilized in cancer therapy[34,42].
Heat shock proteins (HSPs) are molecular chaperones participating in the folding and stable protein conformation. One of their tasks is to block the creation of protein complexes and usually they are over-expressed in cancer cells. Survivin is a HSP90 client protein and the association among these proteins was used to generate preventive compounds. The formation of survivin-HSP90 complex is inhibited when shepherdin is bound to HSP90. Thus, incubation of glioblastoma cells with shepherdin triggered the irreversible destruction of mitochondria and HSP90 client proteins in the cytosol and finally tumor cell death. Targeting the action of HSP90 in different subcellular compartments might contribute a lot in cancer therapy, particularly when affecting its interaction with survivin[34].
The transcription factor GATA 1 that was found to upregulate survivin expression in breast carcinoma could be a novel target of therapy. A research group revealed that the extraction of the grape seed regulates the function of a transcription factor, which is associated with the core promoter of survivin and can finally reduce its activity[43].
Isolating antibodies specific for survivin peptides seems significant, as the structure and sequence of the epitope binding groove of the antibody could be examined and possibly additional epitopes are discovered. Also this study could define with accuracy the peptide portion of the survivin protein that is bound most efficiently and most commonly by humoral antibodies generated against survivin[19]. This will lead to more specific survivin vaccines that would elicit better immune response, generate immune memory and offer protection from tumor progression[44]. The development of antibodies is still costing a large amount of money and there are several considerations for their use[19,45].
Comprehending the molecular principles of cancer and the function of immune defense mechanisms guided to development of novel therapeutic strategies. The most significant immune cells against the development of the tumor are the differentiated cytolytic CD8+ lymphocytes (CTLs)[34]. Survivin is a tumor-associated antigen so it could be a potential target for immunotherapy[34]. During the search for immunogenic tumor antigens, spontaneous CTL responses against survivin were identified[45]. Survivin is an attractive target for vaccination since immune escape by down regulation or loss of expression of this protein would impair sustained tumor growth. The therapeutic effectiveness is improved by the availability of multiple survivin epitopes presented by different HLA restriction elements[45,46]. Survivin-derived epitopes have been exploited in vaccination so that CD4+ responses were initiated in prostate cancer and melanoma patients. Tumor response and patient survival were correlated with the activities of survivin specific T cells[34].
Radiotherapy enhances apoptosis and destroys cells once and for all. Survivin mRNA expression was inversely correlated with the sensitivity to ionizing radiation[43,47,48]. Additionally, survivin could be a radio resistance factor since its production was increased by sublethal doses of radiation[49]. Transfer of tumor-specific adenovirus-mediated PUMA gene using the survivin promoter enhances the radiosensitivity of breast cancer cells in vitro and in vivo[43]. Also, signal transducer and activator of transcription 3 (Stat 3) could be used as target therapy in sensitization by radiation for patients with breast carcinomas, because these two molecules influence survivin. Radiotherapeutic sensitization approach has not been widely used in all clinical trials, despite the fact that survivin inhibitors might be a new group of antagonists, which might enhance the effects of radiotherapy when survivin is over-expressed[43,50].
In preclinical and clinical models, a variety of experiments were performed in order to block IAPs and therefore to lead breast malignant cells to apoptosis and to decreased size even in chemoresistant tumors, reinforcing their possible future utilization as cancer therapy targets[6].
Overexpression of survivin has been shown to restrain apoptosis and to develop multi-drug resistance (MDR)[51]. MDR in cancer therapy is represented by resistance to a large variety of structurally unrelated cytotoxic compounds, resulting to insufficient death of tumor cells and chemotherapeutic failure in patients with solid tumors. Both breast cancer resistance protein (BCRP, ABCG2) and apoptosis-related molecules are associated with the evolution of MDR in tumor cells[51].
It has been shown that survivin expression is significantly higher in cell lines after treatment with anticancer drugs. High levels of HER2 or survivin are related to chemo/endocrine therapy resistance and are predicting poor clinical outcome for breast carcinomas. Moreover, it was shown that low survivin expression levels increased the sensitivity of breast cancer cells to etoposide and 5FU[43]. Prodigiosin (a bacterial metabolite), which downregulates survivin transcriptionally, could be used in the treatment of paclitaxel-resistant breast tumors[43].
It seems that p53 possesses an important role in the survivin-regulated BCRP expression: through downregulation of p53 expression, survivin reduces the inhibitory action of p53 on NF-κB (p50) and then increases the expression of BCRP. In the majority of carcinomas, overexpression of survivin and loss of wild-type p53 expression/function take place simultaneously, hence BCRP overexpression and MDR to multiple compounds such as mitoxantrone, anthracyclines, methotrexate, topoisomerase I inhibitors, gefitinib, doxorubicin (dox), and 5-fluorouracil. Consequently, suppression of survivin expression can be a novel strategy to overpower BCRP-mediated MDR in tumors[51].
1,25(OH)2D3 (active vitamin D3) regulates genes involved in calcium homeostasis and bone formation through its interaction with the vitamin D receptor (VDR). Vitamin D3 inhibits survivin and TP73 isoforms in colon and breast carcinomas. In vitro and in vivo approaches speculate that the downregulation of TP73 isoforms by 1,25(OH)2D3 could be survivin-dependent[52].
Research is still in progress in order to design new agents aiming survivin either at the genomic level or at the protein level. A prominent member of these agents is YM155 (sepantronium bromide), a tiny molecule which inhibits survivin and decreases production of survivin by binding to the C-terminal region of interleukin enhancer-binding factor 3[19]. LY2181308 is a second-generation antisense oligonucleotide with a phosphorothioate backbone and other structural alterations targeting the translation initiation site of survivin isoforms. YM155 and LY2181308 can inhibit expression of all survivin isoforms. Recently, it was found that amiloride can regulate the mechanism of different survivin splicing[19].
It has been shown that survivin can block apoptosis when agents such as tamoxifen, paclitaxel and trastuzuma are used. It was also shown that a reduction of apoptosis caused by tamoxifen occurs after increasing the expression of survivin. Recent research in mammary tumors proved that survivin acts downstream the human epidermal growth factor receptor-2 (Her2) and (Her3)/PI3K/Akt pathway, so it seems to be essential in moderating plaxitaxel impedance when Her2 is over-expressed[6]. An additional study showed that cells overexpressing survivin showed no decreased viability when treated with trastuzumab, providing evidence that survivin can overcome transtuzumab-induced cell growth inhibition[6].
Despite the fact that doxorubicin, an anthracyclic agent of chemotherapy, is extensively used in the treatment of breast cancer, the research community has little information on the role of survivin in resistance to doxorubicin[6]. Survivin-induced overexpression does not block dox-mediated lethal effects in invasive and non-invasive breast tumor cells. In the same way, silencing survivin by siRNA, with or without blocking XIAP too, cannot provoke cytotoxic stimuli and sensitize cells. In conclusion, these data imply that survivin and XIAP expression do not affect dox resistance in breast carcinomas[6].
It has been well documented that the downregulation of survivin by chemotherapeutic agents sensitizes cancer cells to TRAIL-induced apoptosis. Consistently, it was also found that nemadipine-A potentiates TRAIL-induced apoptosis by reducing survivin expression in lung cancer cells. Although, the precise mechanisms are not clear, it was recently reported that survivin expression could be up-regulated by cellular calcium level[53].
Moreover, derivatives of the natural alkaloid camptothecin have the ability to provoke proteasomal degradation of survivin in cells with defective p53 function and elevated XAFI (another tumor suppressor) expression[54].
Also, it was reported that overexpression of survivin-3b in breast tumor cell lines strongly inhibits 5-fluorouracil + epirubicin + cyclophosphamide (FEC) toxicity, a combination used widely in breast carcinoma treatment. Recently, the cytoprotective effect of survivin-3b after cisplatin treatment was reported. These results also showed for the first time the cytoprotective effect of survivin-3b after FEC treatment in less and in more aggressive cell lines by a p53-independent manner. In addition, increased expression of survivin-3b after one course of docetaxel/epirubicin treatment was associated with reduced disease free survival (DFS) of breast cancer patients. Indeed, high survivin-3b expression tumors had a shorter overall and DFS[23]. All these findings suggest that the role of survivin is probably therapy specific in resistance. Recently, the complete pathologic response to GAT therapy (dox on first day, paclitaxel and gemcitabine on second day, every 14 d for 6 cycles) underlined its relation with tumor markers and unveiled a reduction in survivin expression in tumors after treatment[6]. These results suggested that the decrease of survivin expression could be associated with the response to GAT[6].
Patients with local breast cancer could be cured by surgical resection combined with adjuvant therapy, including radiation, anti-estrogen therapy and Her2-targeting agents. Unfortunately, the management of metastatic breast cancer is far less successful than treatment of local disease. Metastatic cancer -despite the progress in molecular-targeted therapies- is the first cause of breast cancer death and presents a true challenge for selecting optimal treatment. Triple negative breast cancer (TNBC) is a high-risk category with negative expression of both estrogen and progesterone hormone receptors and no Her2 protein overexpression (or c-erbb2 gene amplification) that has limited therapeutic options[2]. TNBC patients represent approximately 15% of total breast cancers. Τhis cancer type may be very aggressive, with rapid tumor growth, a high incidence of metastasis, an increased possibility of distant recurrence and a higher mortality rate than other breast cancers. The need to develop novel therapeutic options that are suitable for this subgroup of patients is obvious[55].
Microtubule-targeting agents, like taxanes and vinca alkaloids are one of the most common classes of chemotherapeutic drugs for the treatment of TNBC. Survivin siRNA and a dominant-negative mutant of survivin enhanced the antitumor activity of taxanes in several types of cancer. Survivin suppression by YM155 increases susceptibility to apoptosis and enhances the disruption of mitosis, resulting in an enhanced response to microtubule-targeting agents. In previous studies, it was demonstrated that tumor regression induced by YM155 in combination with docetaxel was accompanied by a decrease in intratumoral survivin and an increase in apoptosis rate. These results support the conclusion that survivin inhibition might be an effective way to enhance the efficacies of microtubule-targeting agents against TNBC[56].
In this section of the review, we present the experimental data of three studies correlating survivin isoforms with breast cancer treatment. The group of studies examined includes the articles of Yamanaka et al[55], Boidot et al[57] and Zheng et al[58] (Table 3).
| Ref. | Examined variants | Methods | Approaches for inhibition of survivin variants | Conclusion |
| Yamanaka et al[55] | wt, sur2b, surΔΕx3, sur3b | Real time qPCR | YM155 | Survivin suppressing activity of YM155 may offer novel therapeutic option in TNBC |
| Boidot et al[57] | wt, sur2b, surΔΕx3, sur3b, sur2a | Real time qPCR | FEC/Tax-Epi | Alternative survivin transcript expression levels may be predictive markers in FEC and Tax-Epi treatment |
| Zheng et al[58] | wt, sur2b, surΔΕx3 | MMT, flow cytmetry | Recombinant plasmids pGEM-T | Feasibility of targeting wt and surΔΕx3 in treating breast cancer |
Yamanaka et al[55], by using cell lines and mouse models, underscored the need for extensive investigation of the role of YM155 in breast cancer treatment and to develop this compound as a novel therapeutic option for metastatic breast cancer patients. YM155 downregulated both levels of survivin mRNA and protein. RT-PCR revealed that YM155 suppressed the expression of survivin-2b, survivin-ΔEx3, and survivin-3b isoforms in human TNBC cells. The decrease of survivin, which was induced by YM155, was accompanied by spontaneous apoptosis. YM155 caused tumor regression with negligible systemic toxicity as evidenced by an absence of body weight loss. Furthermore, in this study there is the first evidence that YM155 may have therapeutic value in reducing the spontaneous metastasis of human TNBC cells in vivo. Considering the fact that survivin is overexpressed in high grade invasive and metastatic human tumors, it is suggested that dysregulation of survivin expression may confer an ability to evade immune responses and physical barriers to invasion of normal tissues. On the contrary, breast cancer cells may remain sensitive to apoptosis induction when survivin is downregulated by YM155, even after distant metastasis has been established. Taken together, therapeutic targeting of the survivin pathway may be beneficial for the treatment of TNBC patients[55].
In their study, Boidot et al[57] examined with a real-time quantitative PCR technique, five survivin isoforms in breast cancer patients that were using as treatment either the combination of docetaxel + epirubicin (Tax-Epi) or the combination of FEC. Before therapy, survivin-2a was considerably higher in resistant than in sensitive tumors in the FEC treatment arm, suggesting for the first time that survivin-2a may be involved in resistance to FEC treatment. This result may be consistent with the finding that survivin can heterodimerize with its splice variants causing specific subcellular localisation patterns leading to formation of the functionally distinct survivin complexes. The ratio of survivin-ΔEx3 to wild type was also higher in sensitive than in resistant tumors in the Tax-Epi treatment arm. Increased measurements of survivin-3b after only one course of chemotherapy were significantly correlated with resistance in the FEC regimen cluster, and the ratios of survivin-ΔEx3 and survivin-2b to wild type were significantly higher in sensitive than in resistant tumors in the Tax-Epi treatment arm. Especially, elevated expression and ratio of survivin-3b, after one course of Tax-Epi, showed correlation with decreased DFS and with reduced overall survival of the patients. These results indicate that an imbalance in the alternative transcript ratios may render the cells resistant or sensitive to apoptosis. They also show for the first time that measurements of alternative survivin transcript levels may become useful predictive biomarkers in FEC and Tax-Epi treatment in breast carcinomas[57].
During their research Zheng et al[58] 2011 created four vectors by merging (1) the antisense gene of survivin; (2) the survivin gene (T34A); (3) the antisense gene of survivin-ΔEx3; and (4) the survivin-2b gene with enhanced green fluorescent protein gene (eGFP) in cell lines. Their data suggested that using survivin as a target had great effects on blocking cell development and promoting apoptosis. Utilizing survivin-ΔΕx3 as a target resulted in reducing the anti-tumor action. This study uncovered the fact that the use of survivin as a target, by antisense RNA or survivin gene (T34A), was almost in the same manner efficient in prohibiting cell development and urging cell apoptosis in breast cancer cells (B-Cap-37). The advantage of survivin (T34A) could be that the dominant negative mutant competed with survivin, thus leading to phosphorylation-defective survivin. Antisense survivin-ΔEx3 notably prohibited the proliferation and promoted the apoptosis of breast cancer cells in vitro. These data suggest that restraining or preventing survivin may be a major step in designing drugs for breast cancer therapy and survivin-ΔΕx3 may as well become a useful target for drugs against breast carcinomas[59].
| 1. | Ferlay J, Shin HR, Bray F. GLOBOCAN 2008 v2.0, cancer incidence and mortality worldwide: IARC CancerBase no. 10 [internet]. Lyon, France: International Agency for Research on Cancer 2010; Available from: http: //globocan.iarc.fr. Accessed: June 5, 2013.. |
| 2. | Poumpouridou N, Kroupis C. Hereditary breast cancer: beyond BRCA genetic analysis; PALB2 emerges. Clin Chem Lab Med. 2012;50:423-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 3. | Shuen AY, Foulkes WD. Inherited mutations in breast cancer genes--risk and response. J Mammary Gland Biol Neoplasia. 2011;16:3-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 39] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 4. | Pavlidou A, Kroupis C, Goutas N, Dalamaga M, Dimas K. Validation of a real-time quantitative polymerase chain reaction method for the quantification of 3 survivin transcripts and evaluation in breast cancer tissues. Clin Breast Cancer. 2014;14:122-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 5. | Pavlidou A, Dalamaga M, Kroupis C, Konstantoudakis G, Belimezi M, Athanasas G, Dimas K. Survivin isoforms and clinicopathological characteristics in colorectal adenocarcinomas using real-time qPCR. World J Gastroenterol. 2011;17:1614-1621. [PubMed] [DOI] [Full Text] |
| 6. | Nestal de Moraes G, Vasconcelos FC, Delbue D, Mognol GP, Sternberg C, Viola JP, Maia RC. Doxorubicin induces cell death in breast cancer cells regardless of Survivin and XIAP expression levels. Eur J Cell Biol. 2013;92:247-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 7. | Knauer SK, Bier C, Schlag P, Fritzmann J, Dietmaier W, Rödel F, Klein-Hitpass L, Kovács AF, Döring C, Hansmann ML. The survivin isoform survivin-3B is cytoprotective and can function as a chromosomal passenger complex protein. Cell Cycle. 2007;6:1502-1509. [PubMed] |
| 8. | Roca H, Varsos Z, Pienta KJ. CCL2 protects prostate cancer PC3 cells from autophagic death via phosphatidylinositol 3-kinase/AKT-dependent survivin up-regulation. J Biol Chem. 2008;283:25057-25073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 9. | Mita AC, Mita MM, Nawrocki ST, Giles FJ. Survivin: key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res. 2008;14:5000-5005. [PubMed] |
| 10. | Ryan B, O’Donovan N, Browne B, O’Shea C, Crown J, Hill AD, McDermott E, O’Higgins N, Duffy MJ. Expression of survivin and its splice variants survivin-2B and survivin-DeltaEx3 in breast cancer. Br J Cancer. 2005;92:120-124. [PubMed] |
| 11. | Xie D, Zeng YX, Wang HJ, Wen JM, Tao Y, Sham JS, Guan XY. Expression of cytoplasmic and nuclear Survivin in primary and secondary human glioblastoma. Br J Cancer. 2006;94:108-114. [PubMed] |
| 12. | Virrey JJ, Guan S, Li W, Schönthal AH, Chen TC, Hofman FM. Increased survivin expression confers chemoresistance to tumor-associated endothelial cells. Am J Pathol. 2008;173:575-585. [PubMed] |
| 13. | Kawasaki H, Toyoda M, Shinohara H, Okuda J, Watanabe I, Yamamoto T, Tanaka K, Tenjo T, Tanigawa N. Expression of survivin correlates with apoptosis, proliferation, and angiogenesis during human colorectal tumorigenesis. Cancer. 2001;91:2026-2032. [PubMed] |
| 14. | Albain KS, Barlow WE, Shak S, Hortobagyi GN, Livingston RB, Yeh IT, Ravdin P, Bugarini R, Baehner FL, Davidson NE. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 2010;11:55-65. [PubMed] |
| 15. | Kittaneh M, Montero AJ, Glück S. Molecular profiling for breast cancer: a comprehensive review. Biomark Cancer. 2013;5:61-70. [PubMed] |
| 16. | Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470-476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4232] [Cited by in RCA: 3981] [Article Influence: 221.2] [Reference Citation Analysis (0)] |
| 17. | Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1445] [Cited by in RCA: 1677] [Article Influence: 111.8] [Reference Citation Analysis (0)] |
| 18. | Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142:625-636. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 268] [Cited by in RCA: 270] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 19. | Miura K, Fujibuchi W, Unno M. Splice isoforms as therapeutic targets for colorectal cancer. Carcinogenesis. 2012;33:2311-2319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 20. | Caldas H, Honsey LE, Altura RA. Survivin 2alpha: a novel Survivin splice variant expressed in human malignancies. Mol Cancer. 2005;4:11. [PubMed] |
| 21. | Zheng W, Ma X, Wei D, Wang T, Ma Y, Yang S. Molecular cloning and bioinformatics analysis of a novel spliced variant of survivin from human breast cancer cells. DNA Seq. 2005;16:321-328. [PubMed] |
| 22. | Végran F, Boidot R, Bonnetain F, Cadouot M, Chevrier S, Lizard-Nacol S. Apoptosis gene signature of Survivin and its splice variant expression in breast carcinoma. Endocr Relat Cancer. 2011;18:783-792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 23. | Span PN, Tjan-Heijnen VC, Heuvel JJ, de Kok JB, Foekens JA, Sweep FC. Do the survivin (BIRC5) splice variants modulate or add to the prognostic value of total survivin in breast cancer? Clin Chem. 2006;52:1693-1700. [PubMed] |
| 24. | Athanassiadou AM, Patsouris E, Tsipis A, Gonidi M, Athanassiadou P. The significance of Survivin and Nectin-4 expression in the prognosis of breast carcinoma. Folia Histochem Cytobiol. 2011;49:26-33. [PubMed] |
| 25. | Bartlett JM, Thomas J, Ross DT, Seitz RS, Ring BZ, Beck RA, Pedersen HC, Munro A, Kunkler IH, Campbell FM. Mammostrat as a tool to stratify breast cancer patients at risk of recurrence during endocrine therapy. Breast Cancer Res. 2010;12:R47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 86] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 26. | Lavasani MA, Moinfar F. Molecular classification of breast carcinomas with particular emphasis on “basal-like” carcinoma: a critical review. J Biophotonics. 2012;5:345-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 27. | van’t Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452:564-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 365] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 28. | van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999-2009. [PubMed] |
| 29. | Habel LA, Sakoda LC, Achacoso N, Ma XJ, Erlander MG, Sgroi DC, Fehrenbacher L, Greenberg D, Quesenberry CP. HOXB13: IL17BR and molecular grade index and risk of breast cancer death among patients with lymph node-negative invasive disease. Breast Cancer Res. 2013;15:R24. [PubMed] |
| 30. | Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, Davies SR, Snider J, Stijleman IJ, Reed J. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res. 2010;16:5222-5232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 544] [Cited by in RCA: 593] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 31. | Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817-2826. [PubMed] |
| 32. | Stemmer SM, Klang SH, Ben-Baruch N, Geffen DB, Steiner M, Soussan-Gutman L, Merling S, Svedman C, Rizel S, Lieberman N. The impact of the 21-gene Recurrence Score assay on clinical decision-making in node-positive (up to 3 positive nodes) estrogen receptor-positive breast cancer patients. Breast Cancer Res Treat. 2013;140:83-92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 33. | Solin LJ, Gray R, Baehner FL, Butler SM, Hughes LL, Yoshizawa C, Cherbavaz DB, Shak S, Page DL, Sledge GW. A multigene expression assay to predict local recurrence risk for ductal carcinoma in situ of the breast. J Natl Cancer Inst. 2013;105:701-710. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 368] [Cited by in RCA: 386] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 34. | Groner B, Weiss A. Targeting survivin in cancer: novel drug development approaches. BioDrugs. 2014;28:27-39. [PubMed] |
| 35. | Weiss A, Brill B, Borghouts C, Delis N, Mack L, Groner B. Survivin inhibition by an interacting recombinant peptide, derived from the human ferritin heavy chain, impedes tumor cell growth. J Cancer Res Clin Oncol. 2012;138:1205-1220. [PubMed] |
| 36. | Manning BD. Challenges and opportunities in defining the essential cancer kinome. Sci Signal. 2009;2:pe15. [PubMed] |
| 37. | Deleavey GF, Damha MJ. Designing chemically modified oligonucleotides for targeted gene silencing. Chem Biol. 2012;19:937-954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 461] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 38. | Abdraboh ME, Gaur RL, Hollenbach AD, Sandquist D, Raj MH, Ouhtit A. Survivin is a novel target of CD44-promoted breast tumor invasion. Am J Pathol. 2011;179:555-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 39. | Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer. 2010;46:298-311. [PubMed] |
| 40. | Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775-789. [PubMed] |
| 41. | Wang C, Zheng X, Shen C, Shi Y. MicroRNA-203 suppresses cell proliferation and migration by targeting BIRC5 and LASP1 in human triple-negative breast cancer cells. J Exp Clin Cancer Res. 2012;31:58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 164] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 42. | Holloway MP, Altura RA. Targeting survivin’s co-conspirators: do alternative methods of trapping survivin in the nucleus have potential in triple-negative breast cancer therapy? Future Oncol. 2012;8:907-909. [PubMed] |
| 43. | Jha K, Shukla M, Pandey M. Survivin expression and targeting in breast cancer. Surg Oncol. 2012;21:125-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 129] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 44. | Friedrichs B, Siegel S, Andersen MH, Schmitz N, Zeis M. Survivin-derived peptide epitopes and their role for induction of antitumor immunity in hematological malignancies. Leuk Lymphoma. 2006;47:978-985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 45. | Reker S, Meier A, Holten-Andersen L, Svane IM, Becker JC, thor Straten P, Andersen MH. Identification of novel survivin-derived CTL epitopes. Cancer Biol Ther. 2004;3:173-179. [PubMed] |
| 46. | Tabrizi MA, Bornstein GG, Klakamp SL, Drake A, Knight R, Roskos L. Translational strategies for development of monoclonal antibodies from discovery to the clinic. Drug Discov Today. 2009;14:298-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 20] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 47. | Asanuma K, Moriai R, Yajima T, Yagihashi A, Yamada M, Kobayashi D, Watanabe N. Survivin as a radioresistance factor in pancreatic cancer. Jpn J Cancer Res. 2000;91:1204-1209. [PubMed] |
| 48. | Papanikolaou V, Iliopoulos D, Dimou I, Dubos S, Kappas C, Kitsiou-Tzeli S, Tsezou A. Survivin regulation by HER2 through NF-κB and c-myc in irradiated breast cancer cells. J Cell Mol Med. 2011;15:1542-1550. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 49. | Bache M, Holzapfel D, Kappler M, Holzhausen HJ, Taubert H, Dunst J, Hänsgen G. Survivin protein expression and hypoxia in advanced cervical carcinoma of patients treated by radiotherapy. Gynecol Oncol. 2007;104:139-144. [PubMed] |
| 50. | Capalbo G, Rödel C, Stauber RH, Knauer SK, Bache M, Kappler M, Rödel F. The role of survivin for radiation therapy. Prognostic and predictive factor and therapeutic target. Strahlenther Onkol. 2007;183:593-599. [PubMed] |
| 51. | Wang QP, Wang Y, Wang XD, Mo XM, Gu J, Lu ZY, Pan ZL, Zhu YX. Survivin up-regulates the expression of breast cancer resistance protein (BCRP) through attenuating the suppression of p53 on NF-κB expression in MCF-7/5-FU cells. Int J Biochem Cell Biol. 2013;45:2036-2044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 52. | Díaz R, González-Sancho JM, Soldevilla B, Silva J, García JM, García V, Peña C, Herrera M, Gómez I, Bonilla F. Differential regulation of TP73 isoforms by 1α,25-dihydroxyvitamin D3 and survivin in human colon and breast carcinomas. Genes Chromosomes Cancer. 2010;49:1135-1142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 53. | Park SH, Park SJ, Kim JO, Shin JH, Kim ES, Jo YK, Kim JS, Park SJ, Jin DH, Hwang JJ. Down-Regulation of Survivin by Nemadipine-A Sensitizes Cancer Cells to TRAIL-Induced Apoptosis. Biomol Ther (Seoul). 2013;21:29-34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 54. | Tomicic MT, Kaina B. Topoisomerase degradation, DSB repair, p53 and IAPs in cancer cell resistance to camptothecin-like topoisomerase I inhibitors. Biochim Biophys Acta. 2013;1835:11-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 55. | Yamanaka K, Nakata M, Kaneko N, Fushiki H, Kita A, Nakahara T, Koutoku H, Sasamata M. YM155, a selective survivin suppressant, inhibits tumor spread and prolongs survival in a spontaneous metastatic model of human triple negative breast cancer. Int J Oncol. 2011;39:569-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 56. | Kaneko N, Yamanaka K, Kita A, Tabata K, Akabane T, Mori M. Synergistic antitumor activities of sepantronium bromide (YM155), a survivin suppressant, in combination with microtubule-targeting agents in triple-negative breast cancer cells. Biol Pharm Bull. 2013;36:1921-1927. [PubMed] |
| 57. | Boidot R, Vegran F, Lizard-Nacol S. Predictive value of survivin alternative transcript expression in locally advanced breast cancer patients treated with neoadjuvant chemotherapy. Int J Mol Med. 2009;23:285-291. [PubMed] |
| 58. | Zheng WY, Kang YY, Li LF, Xu YX, Ma XY. Levels of effectiveness of gene therapies targeting survivin and its splice variants in human breast cancer cells. Drug Discov Ther. 2011;5:293-298. [PubMed] |
| 59. | McKenzie JA, Grossman D. Role of the apoptotic and mitotic regulator survivin in melanoma. Anticancer Res. 2012;32:397-404. [PubMed] |
P- Reviewer: Kim AL, Maia CJ S- Editor: Ji FF L- Editor: A E- Editor: Lu YJ