Published online Jun 24, 2024. doi: 10.5306/wjco.v15.i6.755
Revised: May 9, 2024
Accepted: May 28, 2024
Published online: June 24, 2024
Processing time: 119 Days and 18 Hours
Tankyrase 2 (TNKS2) is a potential candidate molecular target for the prognosis and treatment of non-small cell lung cancer (NSCLC), but its biological functions are unclear.
To investigate the biological functions of TNKS2 in NSCLC.
Using a lentiviral vector, we generated H647 model cells with TNKS2 knockdown by RNA interference and A549 model cells with TNKS2 overexpression by tran
TNKS2 mRNA and protein expression was significantly higher in the highly malignant NCI-H647 cells, while it remained at a low level in the less malignant A549 cells. Lentivirus-mediated overexpression of TNKS2 in A549 cells resulted in a 3-fold increase in gene expression and a 1.7-fold increase in protein expression (P < 0.01). Conversely, shRNA interference targeting TNKS2 Led to an 8-fold decrease in gene expression and a 3-fold decrease in protein expression (P < 0.01) in NCI-H647 cells. Furthermore, the cell apoptosis rate was significantly reduced (50%) and cell migration rate was increased (35%) in the TNKS2 overexpression group than in the control group (P < 0.05). In contrast, shTNKS2 promoted apoptosis by more than one fold and reduced migration by 60% (P < 0.05). Immunofluorescence analysis revealed enhanced nuclear localization of β-catenin fluorescence signal associated with high TNKS2 expression levels. Western blot analysis investigating TNKS2/β-catenin-related proteins indicated consistent changes between TNKS2 and β-catenin expression in lung cancer cells, whereas Axin displayed an opposite trend (P < 0.05).
The obtained results revealed that TNKS2 may serve as an adverse prognostic factor and a potential therapeutic target in NSCLC.
Core Tip: This study provides a comprehensive overview of the role of tankyrase 2 (TNKS2) overexpression, which fac
- Citation: Wang Y, Zhang YJ. Tankyrase 2 promotes lung cancer cell malignancy. World J Clin Oncol 2024; 15(6): 755-764
- URL: https://www.wjgnet.com/2218-4333/full/v15/i6/755.htm
- DOI: https://dx.doi.org/10.5306/wjco.v15.i6.755
Lung cancer has the highest fatality rate among all cancers worldwide[1,2], and non-small cell lung cancer (NSCLC) is the most common histological subtype, accounting for approximately 85% of all lung cancer cases worldwide[3]. Despite considerable advancements in lung cancer diagnosis and therapy (such as surgery, chemotherapy, and radiation the
Dysregulated β-catenin expression plays a causal role in the pathogenesis of NSCLC[6]. In the canonical activation of the Wnt/β-catenin signal, β-catenin is phosphorylated and degraded, after which it undergoes nuclear translocation and interacts with TCF4/LEF, promoting the transcription of downstream target genes[7]. The poly (ADP-ribose) polymerase (PARP) tankyrase (TNKS) plays a key role in the carcinogenic Wnt/β-catenin signaling pathway, which can not only promote the accumulation of cytoplasmic β-catenin but also participate in the process of driving the transcription of proto-oncogenes[8]. TNKS are a family of enzymes belonging to the PARP superfamily which is located on chromosome 8 and contains 1327 amino acids. TNKS’ primary structure includes four domains: The C-terminal catalytic PARP domain that mediates the poly(ADP-ribose) addition to its substrates, a sterile alpha module responsible for the homo- and hetero-oligomer formation, the ankyrin domain divided into five clusters (ARC 1-5) which serve as the substrate binding site, and the His, Pro, and Ser rich domain with unknown function at the N-terminus[9]. TNKS performs various fun
Expression of the TNKS2 gene, which is located on 10q23.32, enhances β-catenin activation via induction of axin degradation[8]. The Wnt/β-catenin pathway is a promising target for NSCLC[11]. TNKS2 is widely distributed and is involved in the regulation of various physiological and pathological processes, including cell growth[12], signal tran
The NSCLC cell lines were purchased from the Cell Bank of the Chinese Academy of Sciences. TNKS2 expression was high in the H647 cells and low in the A549 cells, as confirmed by real-time reverse transcriptase-polymerase chain reaction (RT-qPCR) and western blot analyses. Briefly, H647 cells were cultured in an RPMI-1640 medium (Corning, Shanghai, China) supplemented with 10% fetal bovine serum (FBS; Gibco, NY, United States). A549 cells were cultured in DMEM (Corning) containing 10% FBS. All the cells were grown in an incubator with saturated humidity at 37 °C and 5% CO2.
A549 and H647 cells were seeded in 12-well plates (Fisher Scientific, United States) at a density of approximately 40%. On day 2 of the culture, H647 cells were infected with the corresponding shRNA (shRNA sequences designed by Invitrogen Life Technologies). The shRNA sequences (Table 1) were designed to target different coding regions of the human TNKS2 mRNA sequence [GenBank Accession No. NM_025235.3], and A549 cells were infected with a lentivirus for TNKS2 overexpression. After 48 h of infection, the cells were selected with 1 μg/mL of puromycin for 48 h. Transfection effi
| Sequences of shRNA | RT-qPCR primers | ||
| shNC | 5′-GCTCAATCCCGACAGTAGAGT-3′ | TNKS2 | GAPDH |
| shTNKS2#1 | 5′- GCGGAAAGACGTAGTTGAATA-3′ | Forward 5′-GCTGAGCCAACCATCCGAAAT-3′ | Forward 5′-GGAGCGAGATCCCTCCAAAAT-3′ |
| shTNKS2#2 | 5′- GACCCCAATGCTCG AGATAAT-3′ | Reverse 5′-ACTTGCGTGGCAGTTGACA-3′ | Reverse 5′-GGCTGTTGTCATACTTCTCATGG-3′ |
RNA was extracted from the cells using an RNAiso Plus kit (TaKaRa, 9108). Eppendorf tubes and tips were soaked in 0.1% diethylpyrocarbonate, and the total RNA was digested using DNase I (TaKaRa, 2270A). After determining RNA purity and concentration, it was reverse-transcribed to cDNA using the RevertAid First-Strand cDNA Synthesis Kit (Thermo, K1622). The reverse transcription system included 5 µL total RNA, 4 µL 5 × TransScript All-in-One SuperMix for qPCR, 1 µL gDNA Remover, and 10 µL Rnase-free double-distilled water (ddH2O). qPCR was performed using the Sso Advance Universal SYBR Green Supermix (Bio-Rad, cat 172-5274). The qPCR reaction system is listed as follows: SybrGreen qPCR Master Mix (2 ×) 10 µL, forward primer (10 uM) 0.4 µL, reverse primer (10 uM) 0.4 µL, ddH20 7.2 µL, and template (cDNA) 2 µL. The primer sequences are shown in Table 1. The PCR conditions were set as follows: 3-min predenaturation at 95 °C, followed by 45 cycles, each consisting of 7-s denaturation at 95 °C, 10-s annealing at 57 °C, and 15-s extension at 72 °C. The relative expression levels were calculated by the 2–ΔΔCt method, using homo GAPDH as the internal reference.
Cells were lysed in a RIPA buffer (CWBIO, CW2334S) containing protease and phosphatase inhibitors (CWBIO, CW2383S). Protein samples were quantified using a bicinchoninic acid assay protein assay kit (Sigma-Aldrich). Briefly, the protein samples (10 µg) were subjected to 10% SDS-PAGE and transferred onto polyvinylidene fluoride membranes (Bio-Rad). The membranes were blocked with 5% non-fat milk for 1 h at 37 °C. The membranes were then incubated with primary antibodies against TNKS2 (1:1000 dilution; Santa Cruz Biotechnology, sc-365897) or the loading control GAPDH (1:2000; Abcam, ab8245) at 4 °C overnight, before incubation with a secondary goat anti-mouse IgG (1:1000; Biosharp, BL001A). Immunoreactivity was detected using an ECL kit and analyzed using ImageJ software (NIH, United States).
An annexin V-APC/7-ADD apoptosis kit (Lianke, 70-AP105-100) was used to detect apoptosis. After the cells in the logarithmic growth phase were trypsinized, they were centrifuged at 500 × g for 5 min. The cells were seeded in wells of a six-well plate (8 × 104 cells/well) and incubated overnight. On day 2, they were centrifuged at 500 × g for 5-10 min at room temperature (22-25 °C) and collected for flow cytometry. After washing twice with 1 × phosphate-buffered saline (PBS; precooled at 4 °C), the cells were centrifuged at 500 × g for 5-10 min. They were then resuspended in 1 × binding buffer (diluted with ddH2O), and the cell density was adjusted to 1 × 106-1 × 107 cells/mL. Thereafter, 100 μL of the cell suspension was transferred to a flow tube, so that each tube contained approximately 1 × 105 to 1 × 106 cells. Following this, 5 μL Annexin V-APC and 10 μL 7-ADD were added to each tube, and the mixture was incubated at room tem
The cells were resuspended in PBS containing 5% fetal calf serum (FCS) before seeding into the wells of a six-well plate (Thermo, United States) at a density of 1 × 107/well. A carboxyfluorescein diacetate succinimidyl ester (CFSE) working solution was prepared by adding 1 μL of 5 mmol/L CFSE solution to 1 mL of PBS containing 10% FCS. Thereafter, 1 mL of the CFSE working solution was added to a tube containing 1 mL of the cell suspension, and the mixture was incubated at 37 °C for 5-10 min. After washing thrice with a complete medium and centrifugation, ice-cold RPMI-1640 medium containing 10% FBS was added to the cells, which were then centrifuged at 500 × g for 5 min. Cell proliferation was determined by flow cytometry.
Logarithmic-phase cells were centrifuged at 500 × g for 5 mi and seeded into the wells of a six-well plate (5 × 105/well). After the cells were cultured to 80%-90% confluence, a 10 μL pipette tip was used to make a scratch through the cell layer. After washing with PBS to remove any non-adherent cells, the cells were cultured in a medium containing 5% serum for 24 h. At 0 and 24 h of culture, photographs of the cells (100 ×) were obtained using the Leica Application Suite software, and the scratch distance (width) was recorded. The relative cell migration rate was calculated as follows: Experimental group (scratch distance at 0 h-scratch distance at 24 h)/control group (scratch distance at 0 h-scratch distance at 24 h).
For immunofluorescence analysis, cells were centrifuged at 500 × g for 5 min and spread on glass coverslips at a density of approximately 40%. The coverslips were placed in the wells of a 24-well plate. The cells were subjected to immunofluorescence analysis the following day. Briefly, they were fixed with 4% paraformaldehyde for 10 min at room tem
All assays were independently repeated at least thrice. The representative results are presented. Data are expressed as the mean ± SD. Comparisons between different groups were performed using Student’s t-tests or one-way analysis of variance. Statistical significance was set at P < 0.05. All the statistical analyses were performed using GraphPad Prism software (version 8.0; GraphPad, Inc., San Diego, CA, United States).
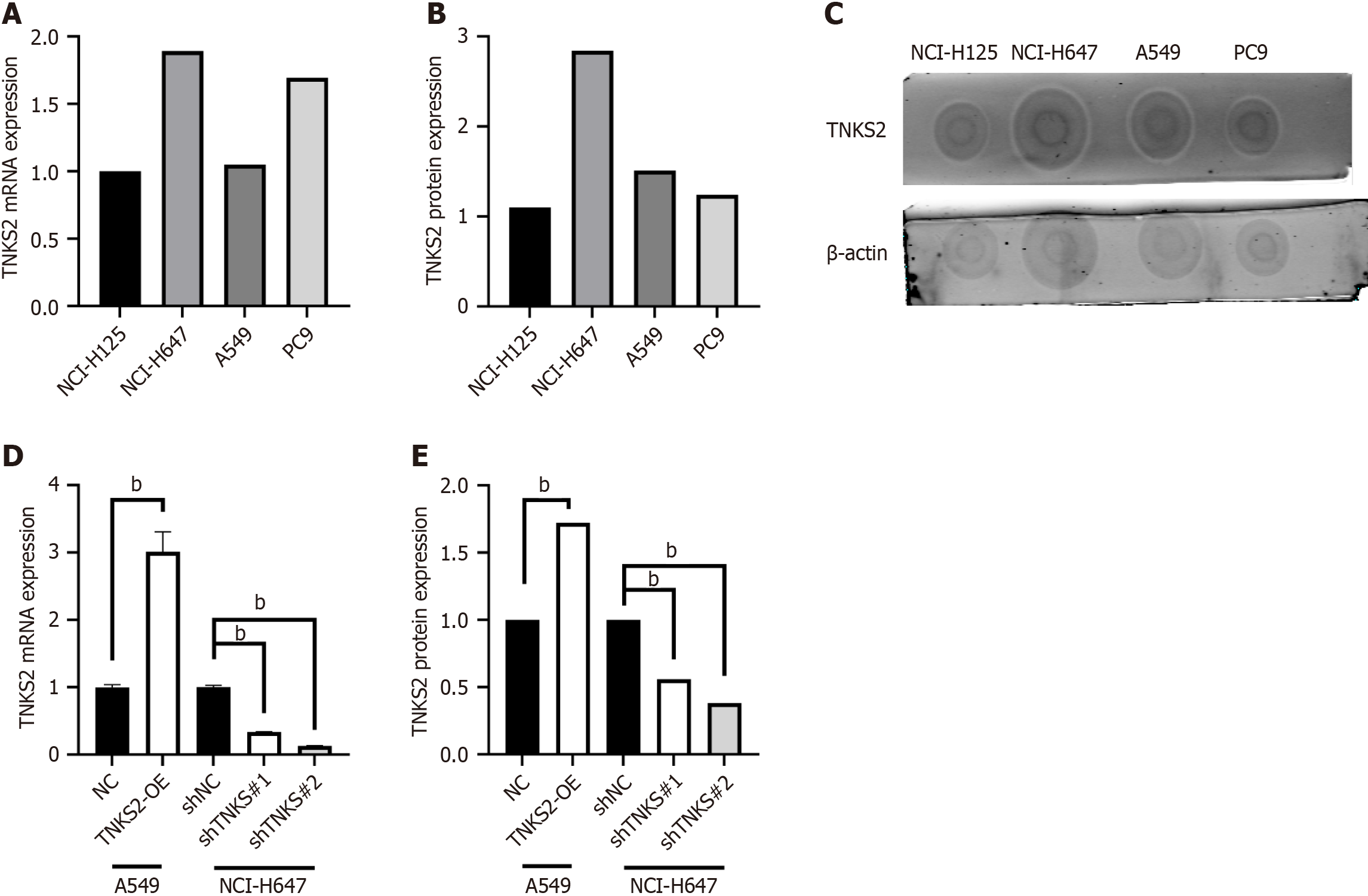
To study the role of TNKS2 in NSCLC, we first selected H647 cells, which express a high level of TNKS2, to construct a stably transfected TNKS2 interference cell line, and A549 cells, which have a low TNKS2 expression level, to be transfected with a TNKS2-overexpression lentivirus (Figure 1A-C). To avoid the off-target effects of shRNA, two shRNAs were synthesized. RT-qPCR and Western blot analyses revealed that TNKS2 mRNA and protein levels were significantly downregulated after transfection with shTNKS2. Of the two shRNAs examined, a better interference effect was observed for shTNKS2#2. Therefore, shTNKS2#2 was used for H647 cells and shNC was used as a control in the assay. A549 cells were transfected with a TNKS2-overexpression lentiviral vector or empty control vector. The transfected cells were selected using puromycin. RNA and proteins were extracted from the collected cells for subsequent use in RT-qPCR and Western blot analyses, respectively. The results of RT-qPCR and Western blot analyses revealed the successful construction of a stable TNKS2-overexpressing A549 cell line and a TNKS2 interference H647 cell line, as shown by TNKS2 overexpression at the mRNA and protein levels (Figure 1D and E).
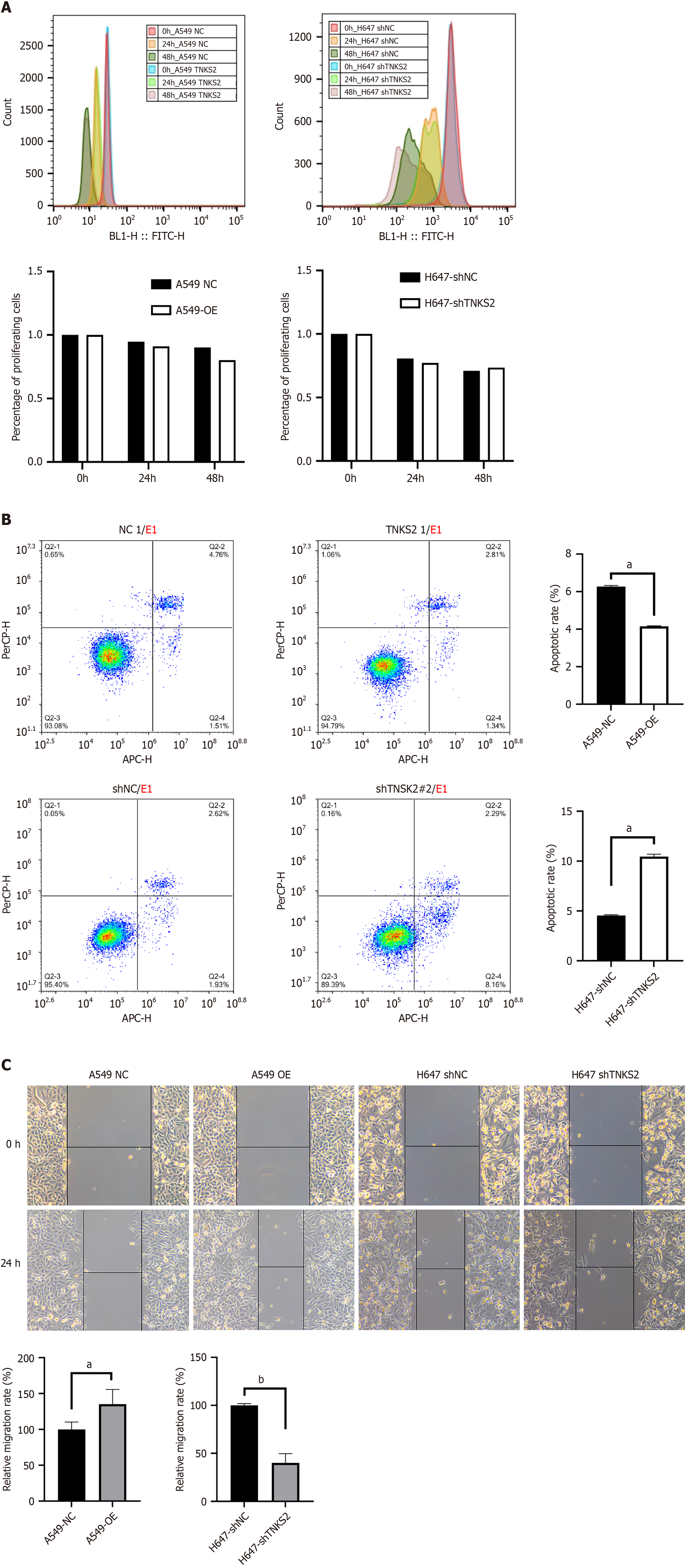
The proliferative capacity of NSCLC cells transfected with shTNKS2 and TNKS2-overexpression lentiviral vectors was assessed. Compared to the rate of proliferation seen in control cells, the proliferative ability of H647 cells was the same as that of cells with silenced TNKS2, whereas the proliferative ability of A549 cells with TNKS2 overexpression was the same as that of A549 cells (Figure 2A).
Using flow cytometry analysis, we investigated the effects of TNKS2 knockdown or overexpression on apoptosis in NSCLC cells. Our results revealed that TNKS2 knockdown led to a significant increase in the apoptosis rate of H647 cells compared to that in the control group. By contrast, TNKS2 overexpression distinctly inhibited the apoptosis of A549 cells compared to that in the control group (Figure 2B). These results indicated that TNKS2 expression is negatively correlated with apoptosis in NSCLC cells.
The scratch test was used to determine the influence of TNKS2 silencing or overexpression on the migratory ability of NSCLC cells. Our results showed that TNKS2 knockdown significantly reduced the scratch-healing ability of H647 cells, indicating that cell migration was weakened by TNKS2 knockdown. In contrast, TNKS2 overexpression significantly enhanced the scratch-healing ability of A549 cells, indicating that the migratory ability of A549 cells was enhanced by TNKS2 overexpression (Figure 2C).
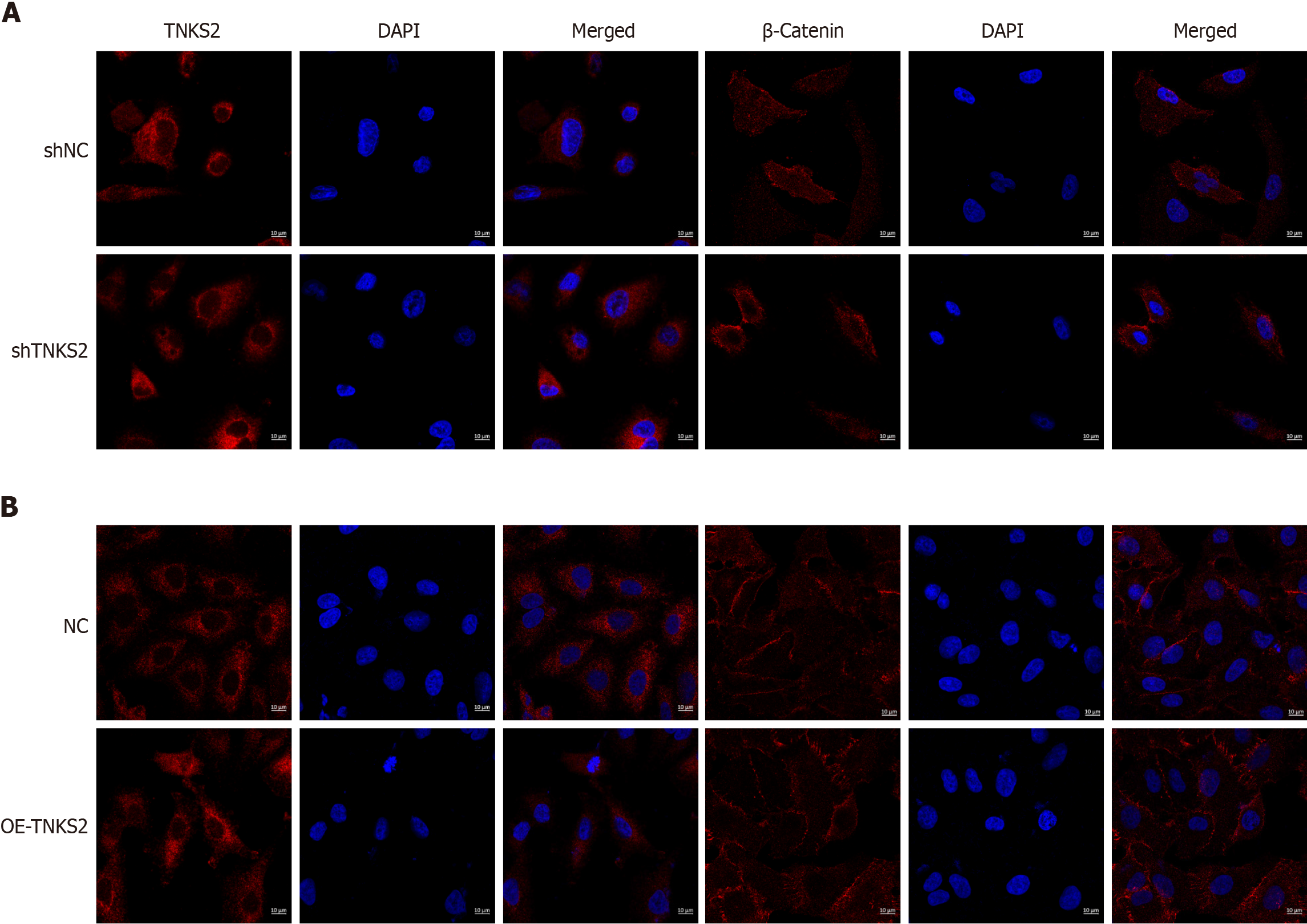
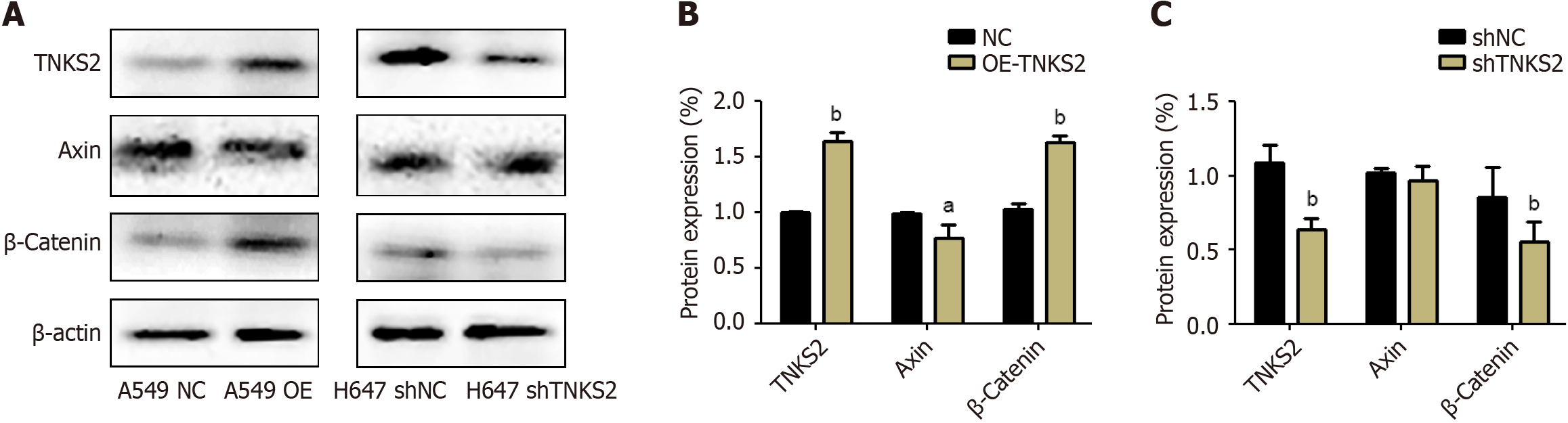
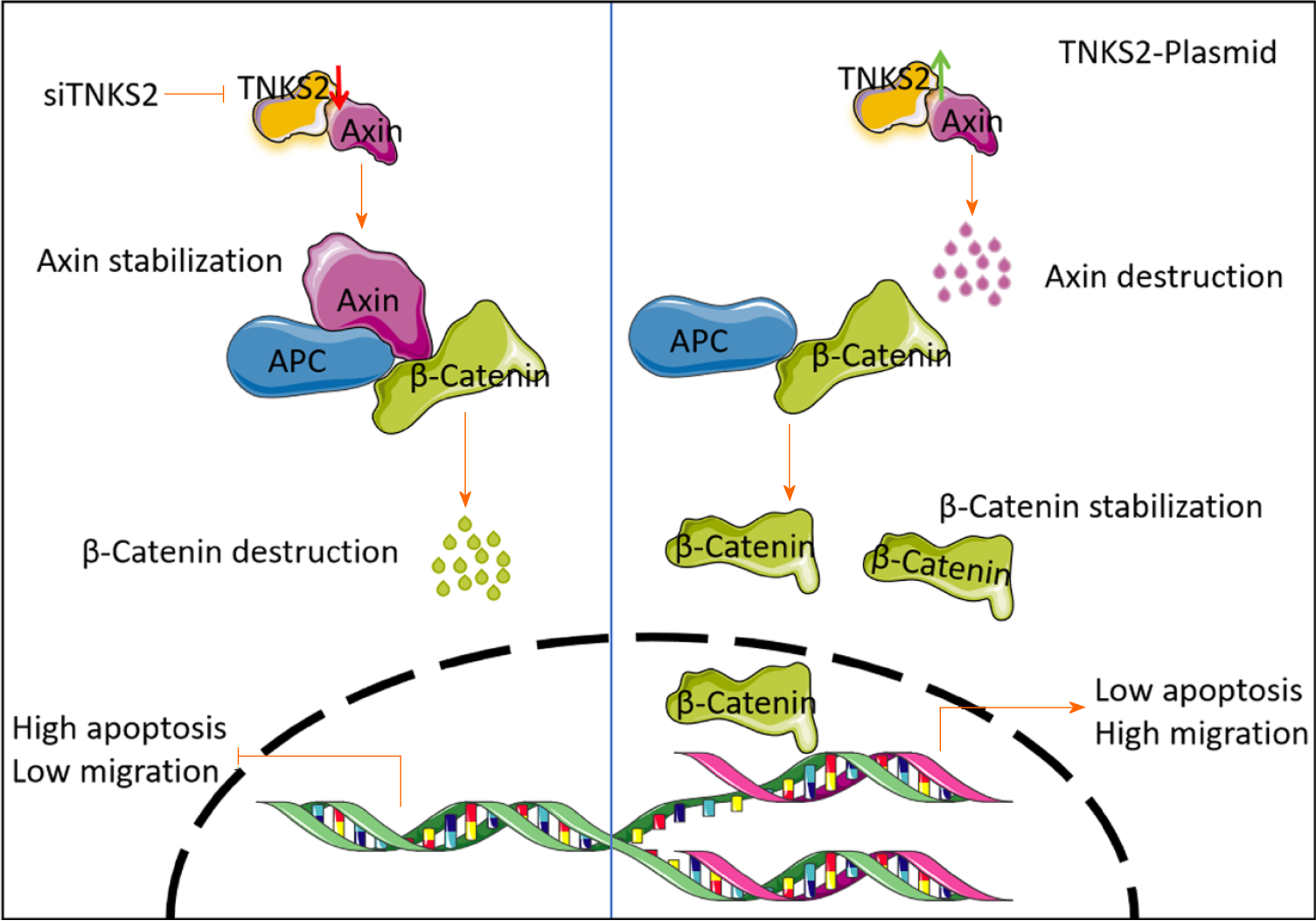
We used immunofluorescence staining to evaluate the expression of TNKS2 in NSCLC cells transfected with shTNKS2 or TNKS2-overexpression vectors. TNKS2 expression notably decreased in shTNKS2-transfected H647 cells. β-catenin protein expression was decreased in both the nucleus and cytoplasm (Figure 3A). Furthermore, immunofluorescence staining was conducted to detect the effects of TNKS2 overexpression on the expression and location of β-catenin. Conversely, its expression was upregulated in A549 cells transfected with a TNKS2-overexpression vector. Upon TNKS2 overexpression, β-catenin protein expression significantly increased in both the nucleus and cytoplasm (Figure 3B). The proteins were then extracted, and the expression of TNKS2/β-catenin-related proteins (axin and β-catenin) was analyzed by Western blotting. β-catenin expression was upregulated with the elevated expression of TNKS2, whereas axin expression was downregulated (Figure 4). These results indicated that TNKS2 overexpression promotes β-catenin activation.
Research to discover prognostic markers for NSCLC has attracted increasing attention; however, continued studies are required to identify accurate, simple, and rapid markers for NSCLC[15-17]. Optimal prognostic markers for lung cancer may be determined by conducting studies with rigorous design and large sample sizes. WNT/β-catenin signaling pathway activation has emerged as a potential therapeutic target in NSCLC[18], and there are several candidate therapeutic target proteins in the WNT/β-catenin signaling pathway[19-21]. One study conducted a focused analysis of 500 microarray probes in the Wnt receptor signaling pathway. TNKS1 and TNKS2 expression levels were 1.30- and 1.43-fold higher in the NSCLC cells than in adjacent normal lung cells in a murine model[22]. In the present study, we provided preliminary evidence to support the hypothesis that TNKS2 overexpression promotes the malignant behavior of lung cancer cells. These results suggest that TNKS2 is a feasible and clinically relevant target for the diagnosis and treatment of NSCLC.
By constructing shRNA and overexpression vectors that alter the expression of TNKS2, we established NSCLC cell lines with stable knockdown or overexpression of TNKS2 and studied the correlation between TNKS2 expression and the biological behavior of NSCLC cells. Low TNKS2 expression led to increased apoptosis in NSCLC cells, while high expression had the opposite effect. Apoptosis is the orderly and autonomous death of cells that is controlled by gene expression to maintain a stable internal environment[23-25]. Unlike cell necrosis, apoptosis is not a passive process but an active one that involves the activation, expression, and regulation of a series of genes. It is not a phenomenon of auto
Malignant tumors easily metastasize through various channels, such as lymphatic vessels and blood circulation[27-29]. Migration and invasion are key processes in tumor metastasis[30-32]. In the present study, we performed a wound-healing assay to determine the effect of TNKS2 on tumor cell migration. Our results revealed that high TNKS2 expression enhanced the migration of NSCLC cells, whereas low TNKS2 expression had the opposite effect. Previous studies have consistently shown that TNKS2 accelerates the growth and invasiveness of triple-negative breast[30] and cervical cancer cells[33]. TNKS2 can dissociate the destruction complex containing axins, thereby enhancing the stability of β-catenin[34]. Activation of β-catenin contributes to the malignant phenotype of NSCLC. Herein, the knockdown and overexpression of TNKS2 inhibited and promoted, respectively, the expression of nuclear β-catenin protein in NSCLC cells, consequently confirming the antitumor potential of inhibiting these molecules in the WNT/β-catenin pathway.
The present study has certain limitations. It only included in vitro experiments. As the lungs are the major respiratory organs of the human body, the growth environment and oxygen concentration of lung cancer cells cultured in vitro are different from those of lung tissues in vivo. Therefore, our results do not reflect the specific effects of TNKS2 on cells in vivo. Additionally, future studies need to include a more detailed and in-depth analysis to explore the mechanism of TNKS2/β-catenin or other pathways.
The results of the present study provide insights into the biological functions of TNKS2 in NSCLC cells. Furthermore, the obtained findings indicate that TNKS2 may affect the proliferation, apoptosis, and migration of lung cancer cells through the TNKS2/β-catenin pathway. These findings may be useful in the development of therapies targeting TNKS2 for the treatment of NSCLC.
Taken together, our results demonstrate that TNKS2 is a promising prognostic factor and therapeutic target for NSCLC. Furthermore, TNKS2 overexpression is associated with the malignant behavior of NSCLC cells (as shown in Figure 5); however, the underlying molecular mechanisms require further investigation.
| 1. | Miller KD, Nogueira L, Devasia T, Mariotto AB, Yabroff KR, Jemal A, Kramer J, Siegel RL. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin. 2022;72:409-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 621] [Cited by in RCA: 1748] [Article Influence: 437.0] [Reference Citation Analysis (0)] |
| 2. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4235] [Cited by in RCA: 11988] [Article Influence: 2997.0] [Reference Citation Analysis (9)] |
| 3. | Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1607] [Cited by in RCA: 3429] [Article Influence: 428.6] [Reference Citation Analysis (0)] |
| 4. | Zhu X, Chen L, Liu L, Niu X. EMT-Mediated Acquired EGFR-TKI Resistance in NSCLC: Mechanisms and Strategies. Front Oncol. 2019;9:1044. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 161] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 5. | Jin S, He J, Zhou Y, Wu D, Li J, Gao W. LncRNA FTX activates FOXA2 expression to inhibit non-small-cell lung cancer proliferation and metastasis. J Cell Mol Med. 2020;24:4839-4849. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 6. | Wang JY, Wang X, Wang XJ, Zheng BZ, Wang Y, Liang B. Curcumin inhibits the growth via Wnt/β-catenin pathway in non-small-cell lung cancer cells. Eur Rev Med Pharmacol Sci. 2018;22:7492-7499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 28] [Reference Citation Analysis (0)] |
| 7. | Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst. 2014;106:djt356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 375] [Cited by in RCA: 430] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 8. | Mariotti L, Templeton CM, Ranes M, Paracuellos P, Cronin N, Beuron F, Morris E, Guettler S. Tankyrase Requires SAM Domain-Dependent Polymerization to Support Wnt-β-Catenin Signaling. Mol Cell. 2016;63:498-513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 85] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 9. | Zamudio-Martinez E, Herrera-Campos AB, Muñoz A, Rodríguez-Vargas JM, Oliver FJ. Tankyrases as modulators of pro-tumoral functions: molecular insights and therapeutic opportunities. J Exp Clin Cancer Res. 2021;40:144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 10. | Damale MG, Pathan SK, Shinde DB, Patil RH, Arote RB, Sangshetti JN. Insights of tankyrases: A novel target for drug discovery. Eur J Med Chem. 2020;207:112712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 11. | Huang JQ, Wei FK, Xu XL, Ye SX, Song JW, Ding PK, Zhu J, Li HF, Luo XP, Gong H, Su L, Yang L, Gong LY. SOX9 drives the epithelial-mesenchymal transition in non-small-cell lung cancer through the Wnt/β-catenin pathway. J Transl Med. 2019;17:143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 106] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 12. | Jia Z, Liu Y, Gao Q, Han Y, Zhang G, Xu S, Cheng K, Zou W. miR-490-3p inhibits the growth and invasiveness in triple-negative breast cancer by repressing the expression of TNKS2. Gene. 2016;593:41-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 13. | Wang H, Lu B, Castillo J, Zhang Y, Yang Z, McAllister G, Lindeman A, Reece-Hoyes J, Tallarico J, Russ C, Hoffman G, Xu W, Schirle M, Cong F. Tankyrase Inhibitor Sensitizes Lung Cancer Cells to Endothelial Growth Factor Receptor (EGFR) Inhibition via Stabilizing Angiomotins and Inhibiting YAP Signaling. J Biol Chem. 2016;291:15256-15266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 14. | Lu ML, Zhang Y, Li J, Fu Y, Li WH, Zhao GF, Li XH, Wei L, Liu GB, Huang H. MicroRNA-124 inhibits colorectal cancer cell proliferation and suppresses tumor growth by interacting with PLCB1 and regulating Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:121-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 15. | Bonaventura A, Grossi F, Montecucco F. PCSK9 is a promising prognostic marker in patients with advanced NSCLC. Cancer Immunol Immunother. 2020;69:491-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 16. | Khandelwal A, Seam RK, Gupta M, Rana MK, Prakash H, Vasquez KM, Jain A. Circulating microRNA-590-5p functions as a liquid biopsy marker in non-small cell lung cancer. Cancer Sci. 2020;111:826-839. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 17. | Wu DM, Deng SH, Zhou J, Han R, Liu T, Zhang T, Li J, Chen JP, Xu Y. PLEK2 mediates metastasis and vascular invasion via the ubiquitin-dependent degradation of SHIP2 in non-small cell lung cancer. Int J Cancer. 2020;146:2563-2575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 18. | Dai FQ, Li CR, Fan XQ, Tan L, Wang RT, Jin H. miR-150-5p Inhibits Non-Small-Cell Lung Cancer Metastasis and Recurrence by Targeting HMGA2 and β-Catenin Signaling. Mol Ther Nucleic Acids. 2019;16:675-685. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 85] [Article Influence: 12.1] [Reference Citation Analysis (1)] |
| 19. | He W, He S, Wang Z, Shen H, Fang W, Zhang Y, Qian W, Lin M, Yuan J, Wang J, Huang W, Wang L, Ke Z. Astrocyte elevated gene-1(AEG-1) induces epithelial-mesenchymal transition in lung cancer through activating Wnt/β-catenin signaling. BMC Cancer. 2015;15:107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 66] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 20. | Lin S, Zhen Y, Guan Y, Yi H. Roles of Wnt/β-Catenin Signaling Pathway Regulatory Long Non-Coding RNAs in the Pathogenesis of Non-Small Cell Lung Cancer. Cancer Manag Res. 2020;12:4181-4191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 21. | Xue S, Wu W, Wang Z, Lu G, Sun J, Jin X, Xie L, Wang X, Tan C, Wang W, Ding X. Corrigendum: USP5 Promotes Metastasis in Non-Small Cell Lung Cancer by Inducing Epithelial-Mesenchymal Transition via Wnt/β-Catenin Pathway. Front Pharmacol. 2020;11:948. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 22. | Lim W, Ridge CA, Nicholson AG, Mirsadraee S. The 8(th) lung cancer TNM classification and clinical staging system: review of the changes and clinical implications. Quant Imaging Med Surg. 2018;8:709-718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 189] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 23. | Guo Y, Tan J, Miao Y, Sun Z, Zhang Q. Effects of Microvesicles on Cell Apoptosis under Hypoxia. Oxid Med Cell Longev. 2019;2019:5972152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 24. | Pistritto G, Trisciuoglio D, Ceci C, Garufi A, D'Orazi G. Apoptosis as anticancer mechanism: function and dysfunction of its modulators and targeted therapeutic strategies. Aging (Albany NY). 2016;8:603-619. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 607] [Cited by in RCA: 1198] [Article Influence: 133.1] [Reference Citation Analysis (0)] |
| 25. | Xu X, Lai Y, Hua ZC. Apoptosis and apoptotic body: disease message and therapeutic target potentials. Biosci Rep. 2019;39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 223] [Cited by in RCA: 669] [Article Influence: 95.6] [Reference Citation Analysis (0)] |
| 26. | Liu G, Pei F, Yang F, Li L, Amin AD, Liu S, Buchan JR, Cho WC. Role of Autophagy and Apoptosis in Non-Small-Cell Lung Cancer. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 247] [Cited by in RCA: 293] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 27. | Liu Z, Liang H, Lin J, Cai X, Pan Z, Liu J, Xie X, Li C, Cheng B, Zhao Y, He J, Liang W. The incidence of lymph node metastasis in patients with different oncogenic driver mutations among T1 non-small-cell lung cancer. Lung Cancer. 2019;134:218-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 28. | Yang J, Peng A, Wang B, Gusdon AM, Sun X, Jiang G, Zhang P. The prognostic impact of lymph node metastasis in patients with non-small cell lung cancer and distant organ metastasis. Clin Exp Metastasis. 2019;36:457-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 29. | Yang XY, Liao JJ, Xue WR. FMNL1 down-regulation suppresses bone metastasis through reducing TGF-β1 expression in non-small cell lung cancer (NSCLC). Biomed Pharmacother. 2019;117:109126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 30. | Gong R, Lin W, Gao A, Liu Y, Li J, Sun M, Chen X, Han S, Men C, Sun Y, Liu J. Forkhead box C1 promotes metastasis and invasion of non-small cell lung cancer by binding directly to the lysyl oxidase promoter. Cancer Sci. 2019;110:3663-3676. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 31. | He W, Zhang H, Wang Y, Zhou Y, Luo Y, Cui Y, Jiang N, Jiang W, Wang H, Xu D, Li S, Wang Z, Chen Y, Sun Y, Zhang Y, Tseng HR, Zou X, Wang L, Ke Z. CTHRC1 induces non-small cell lung cancer (NSCLC) invasion through upregulating MMP-7/MMP-9. BMC Cancer. 2018;18:400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 32. | Yu M, Chen Y, Li X, Yang R, Zhang L, Huangfu L, Zheng N, Zhao X, Lv L, Hong Y, Liang H, Shan H. YAP1 contributes to NSCLC invasion and migration by promoting Slug transcription via the transcription co-factor TEAD. Cell Death Dis. 2018;9:464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 84] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 33. | Kang HW, Wang F, Wei Q, Zhao YF, Liu M, Li X, Tang H. miR-20a promotes migration and invasion by regulating TNKS2 in human cervical cancer cells. FEBS Lett. 2012;586:897-904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 94] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 34. | Pollock K, Liu M, Zaleska M, Meniconi M, Pfuhl M, Collins I, Guettler S. Fragment-based screening identifies molecules targeting the substrate-binding ankyrin repeat domains of tankyrase. Sci Rep. 2019;9:19130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/