Published online Nov 24, 2024. doi: 10.5306/wjco.v15.i11.1412
Revised: August 17, 2024
Accepted: September 9, 2024
Published online: November 24, 2024
Processing time: 203 Days and 18.6 Hours
Colorectal cancer (CRC) causes many deaths worldwide. Synaptotagmin binding cytoplasmic RNA interacting protein (SYNCRIP) is an RNA-binding protein that plays an important role in multiple cancers by epigenetically targeting some genes. Our study will examine the expression, potential effect, biological function and clinical value of SYNCRIP in CRC.
To examine the expression, potential effect, biological function and clinical value of SYNCRIP in CRC.
The expression of SYNCRIP was examined by immunohistochemistry arrays and high-throughput data. The effect of SYNCRIP gene in CRC cell growth was evaluated by CRISPR-Cas9 technology. The target genes of SYNCRIP were calculated using various algorithms, and the molecular mechanism of SYNCRIP in CRC was explored by mutation analysis and pathway analysis. The clinical value of SYNCRIP in prognosis and radiotherapy was re
The protein and mRNA levels of SYNCRIP were both highly expressed in CRC samples compared to nontumorous tissue based on 330 immunohistochemistry arrays and 3640 CRC samples. Cells grew more slowly in eleven CRC cell lines after knocking out the SYNCRIP gene. SYNCRIP could epigenetically target genes to promote the occurrence and development of CRC by boosting the cell cycle and affecting the tumor microenvironment. In addition, CRC patients with high SYNCRIP expression are more sensitive to radiotherapy.
SYNCRIP is upregulated in CRC, and highly expressed SYNCRIP can accelerate CRC cell division by exerting its epigenetic regulatory effects. In addition, SYNCRIP is expected to become a potential biomarker to predict the effect of radiotherapy.
Core Tip: Synaptotagmin binding cytoplasmic RNA interacting protein (SYNCRIP) is regulated by enhancers and POLR2A and highly expressed in colorectal cancer (CRC). Highly expressed SYNCRIP can reduce CD8+ T cells and accelerate CRC cell division by exerting its epigenetic regulatory effects. These CRC cells are sensitive to radiation therapy. Finally, SYNCRIP is expected to become a biomarker in CRC patients.
- Citation: Li H, Huang HQ, Huang ZG, He RQ, Fang YY, Song R, Luo JY, Zeng DT, Qin K, Wei DM, Chen G. Potential regulatory mechanism and clinical significance of synaptotagmin binding cytoplasmic RNA interacting protein in colorectal cancer. World J Clin Oncol 2024; 15(11): 1412-1427
- URL: https://www.wjgnet.com/2218-4333/full/v15/i11/1412.htm
- DOI: https://dx.doi.org/10.5306/wjco.v15.i11.1412
Colorectal cancer (CRC) accounts for approximately ten percent of diagnosed cancers and ranks second in total cancer deaths according to Cancer Statistics 2022[1]. The proximal colon is the most common site of all CRC, accounting for approximately 41%, with the distal colon accounting for 22% and the rectum accounting for 28%[2]. The risk factors for CRC include increasing age and genetic, lifestyle, diet and environmental factors[3-5]. In China, the incidence of CRC increases rapidly in individuals of all ages, especially in people over 50, and it shows an increasing tendency year by year[6]. The prevailing therapeutic methods for CRC are surgery, neoadjuvant chemotherapy, adjuvant chemotherapy and radiotherapy. Surgery is the major treatment for patients with early CRC, but it lacks sufficient effect in advanced cases with metastasis or lymphatic metastasis[7,8]. And molecular genetic material can predict and influence the occurrence and development of malignant tumors[9-13]. Therefore, it is important to explore the pathogenesis of CRC and find a potential therapeutic target for patients with CRC.
Synaptotagmin binding cytoplasmic RNA interacting protein (SYNCRIP), also known as hnRNP-Q or NSAP1, is an RNA-binding protein (RBP) with extensive expression in multiple organisms and tissues and plays a role in several cellular pathways and diseases[14,15]. SYNCRIP participates in many biological processes, such as neuronal, myeloid leukemia stem cell and muscular development. It is usually differentially expressed in various tissues and associated with immune response disorders, cardiomyopathies and neuronal degeneration disorders[16-18]. Additionally, SYNCRIP has been reported to be associated with cancers[19]. In particular, the expression of SYNCRIP is associated with a poor pro
Our study was approved by the medical ethics committee of the First Affiliated Hospital of Guangxi Medical University. The tissue microarray was purchased from Antomics, Inc. (Richmond, CA, United States), and all the patients had co
Acquisition and integration of RNA-seq and RNA-array data of CRC: To summarize the expression level of SYNCRIP in CRC and nontumor tissues, SYNCRIP expression profiles were assembled from multiple high-throughput platforms. The search strategies were set to ((colorectal) OR (colon) OR (rectum)) AND ((carcinoma) OR (cancer) OR (malignant)). The inclusion criteria were as follows: (1) Homo species; (2) Primary CRC tissue; (3) Nontumorous samples as the control group; and (4) A sample size greater than or equal to three. The original or processed expression matrix of CRC was downloaded from websites. All the expression matrices were standardized by logarithmic transformation. After ex
The search strategy was also used to screen the epigenetic regulation data of CRC. The inclusion criteria for chromatin immunoprecipitation (ChIP)-seq or assay for transposase-accessible chromatin (ATAC)-seq were as follows: (1) Homo species; (2) primary CRC tissue or cell lines; and (3) ChIP-seq data modified by H3K27ac and H3K4me1. The Cistrome Data Browser (http://cistrome.org/db/#/) online database could manage the epigenetic data.
SYNCRIP expression in CRC: The data distribution of SYNCRIP in each dataset was determined and tested by the Kolmogorov-Smirnov test. The expression difference of SYNCRIP between CRC and noncancerous tissue in each exp
The CRC cell lines were obtained from Dep Map database (https://depmap.org/portal/). Gene effect score of SYNCRIP was calculated by CERES algorithm to evaluate the effect of SYNCRIP gene on the survival and proliferation in CRC cell lines. A negative gene effect score implies that knocking out the SYNCRIP gene suppresses the growth of CRC cell lines.
The mutation status of SYNCRIP in CRC: To detect the mutation status (nonsense mutation, base replacement, base deletion, copy number variation, and so on) of SYNCRIP in multiple CRC cohort studies based on genomics, we used the cancer genome atlas (TCGA)-based online tool cBioPortal (http://www.cbioportal.org/) to calculate the mutation rate and generate the diagram. The prognostic value of SYNCRIP-mutated patients with CRC was also calculated and shown by Kaplan-Meier plots.
The association between SYNCRIP and decisive factors of CRC: The decisive factors affecting the development and onset of CRC include the tumorous immune microenvironment, tumor mutational burden (TMB), and microsatellite instability (MSI). In this study, we explored the correlation between SYNCRIP and carcinogenic factors to determine the cause of the carcinogenesis process of CRC.
Recent studies have shown that TMB is a significant index to evaluate the degree of tumor mutation. Accordingly, the somatic mutation data of colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) were downloaded from GDC-TCGA; the TMB of COAD and READ were assessed via the maftools package in R software. The correlation of SYNCRIP and TMB in the COAD and READ cohorts was computed using Pearson correlation analysis; the results are shown on scatter diagrams via the ggplot2 package.
MSI is known as a risk factor for CRC; hence, the association between MSI and SYNCRIP was revealed based on high-throughput data. COAD and READ datasets from TCGA were used to calculate MSI, and the relationship between SYNCRIP and MSI was assessed by Pearson correlation analysis.
COAD and READ datasets from TCGA were used to calculate the immune microenvironments, such as CD8+ T cells, CD4+ T cells, and neutrophils, which were estimated by the CIBERSORT algorithm. The expression difference of immune items in CRC and nontumorous samples was calculated by t test, and the relation between SYNCRIP and immune items was calculated by Pearson correlation analysis. The immune items that were only differentially regulated in CRC and associated with SYNCRIP were considered pivotal immune microenvironments.
SYNCRIP target genes and potential pathways in CRC: As an RNA binding protein, SYNCRIP combines with genes and regulates their expression. To reveal the potential pathways regulated by SYNCRIP in CRC, the SYNCRIP target genes were first extracted. Genes co expressed with SYNCRIP were calculated based on the TCGA-GTEx dataset. The genes binding by SYNCRIP were predicted via the RNAct database (https://rnact.crg.eu/), and genes with a predictive score greater than 23 were considered SYNCRIP combined genes. Ultimately, ChIP-seq data marked by SYNCRIP of CRC were retrieved in a public high-throughput database, and the SYNCRIP binding genes were calculated. The Fragments Per Kilobase of exon model per Million mapped fragments value in the ChIP-seq was standardized by Z-transform, and genes with more than twice the standard deviation value were seen as the SYNCRIP binding genes. The most reliable SYNCRIP target genes were intersected via coexpressed genes of SYNCRIP, highly expressed genes in CRC and SYNCRIP binding genes.
These target genes were utilized to identify the potential pathways and biological functions regulated by SYNCRIP. The BP, cellular component and molecular function categories were summarized by gene ontology (GO) analysis and used to represent the biological function of SYNCRIP in CRC. The potential pathways based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome databases were applied for the SYNCRIP targets.
Regulatory patterns of SYNCRIP in CRC: To further explore the mechanism of SYNCRIP in CRC, regulatory patterns at the transcriptome level were determined via high-throughput data. The Cistrome Data Browser (http://cistrome.org/db/#/) includes multiple epigenetic data and was used to excavate the regulatory patterns of SYNCRIP in CRC. The upstream transcription factors (TFs) of SYNCRIP were mined via a series of ChIP-seq data. The chromatin accessibility and transcriptional activity of SYNCRIP in CRC were explored by using ATAC-seq, H3K27ac-marked ChIP-seq and H3K4me1-marked ChIP-seq. SYNCRIP is regulated by enhancers only when the peaks are enriched in the promoter region of SYNCRIP.
Prognostic value of SYNCRIP in CRC: By mining TCGA and various high-throughput datasets, large volumes of data concerning the prognosis of SYNCRIP in patients with CRC were acquired. The patients with a higher expression of SYNCRIP were grouped into the high-risk group, and the rest were evenly defined as the low-risk group. A Cox proportional hazards model was used to calculate the hazard risk between the high-risk group and the low-risk group, and the log-rank test was used to test the prognostic value of SYNCRIP. Hazard ratio > 1 and log-rank at P < 0.05 illustrated that SYNCRIP was a risk factor in patients with CRC. All tests and Kaplan-Meier curves were carried out using the R packages survival and survminer.
SYNCRIP expression profiles in CRC with radio resistance: To understand the therapeutic implications of SYNCRIP in CRC, high-throughput sequencing data of radio-treated CRC were retrieved from global laboratories. The retrieval keywords were set as follows: [(colon) OR (rectal) OR (colorectal)] AND [(carcinoma) OR (cancer) OR (malignant) OR (tumor) OR (adenoma)] AND [(radio) OR (radiotherapy) OR (radiation)]. Strict inclusion criteria were used to ensure the reliability of the data. They included: (1) Homo species; (2) Primary CRC tissue; (3) CRC cell lines; (4) Available detailed information on radiation; (5) CRC samples not treated except for radiation; (6) a sample size of the experimental and control groups greater than or equal to three; and (7) Available SYNCRIP expression.
After a series of filtrations, we enrolled available high-throughput data and stratified the samples into radioresistant and radiosensitive groups. The radiosensitivity of CRC tissues was assessed using the tumor regression score of the National Comprehensive Cancer Network guideline V2018, in which a Tumor Regression Grading score of 0 was defined as radiosensitive. In contrast, scores between 1 and 3 were radioresistant. The difference in the expression of SYNCRIP in the two groups was calculated.
The statistical analysis in our study was performed by SPSS 22.0, R software, STATA 12.0 and Graph Pad Prism 9.0. For the statistical description, the ratio was used to describe the classification variable, and the mean ± SD or median ± quartile was used to describe the continuous variable. The χ2 test and Fisher’s exact test were used to compare the differences in classification variables, and the t test and Mann-Whitney U test were used to compare the differences in continuous variables. For correlation analysis, Pearson’s method or Spearman’s method was selected according to the data distribution type. Two-sided P < 0.05 indicated a significant difference.
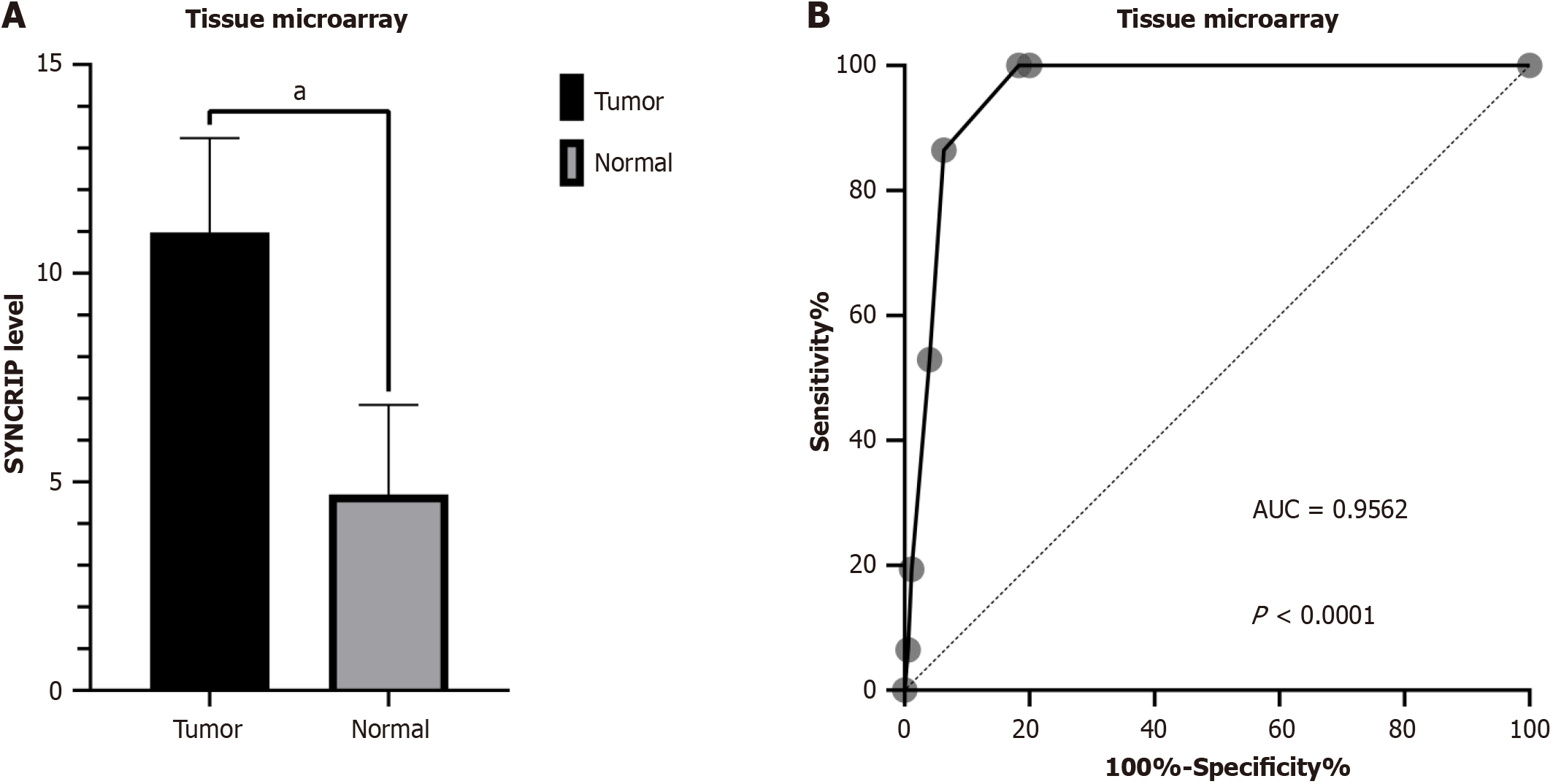
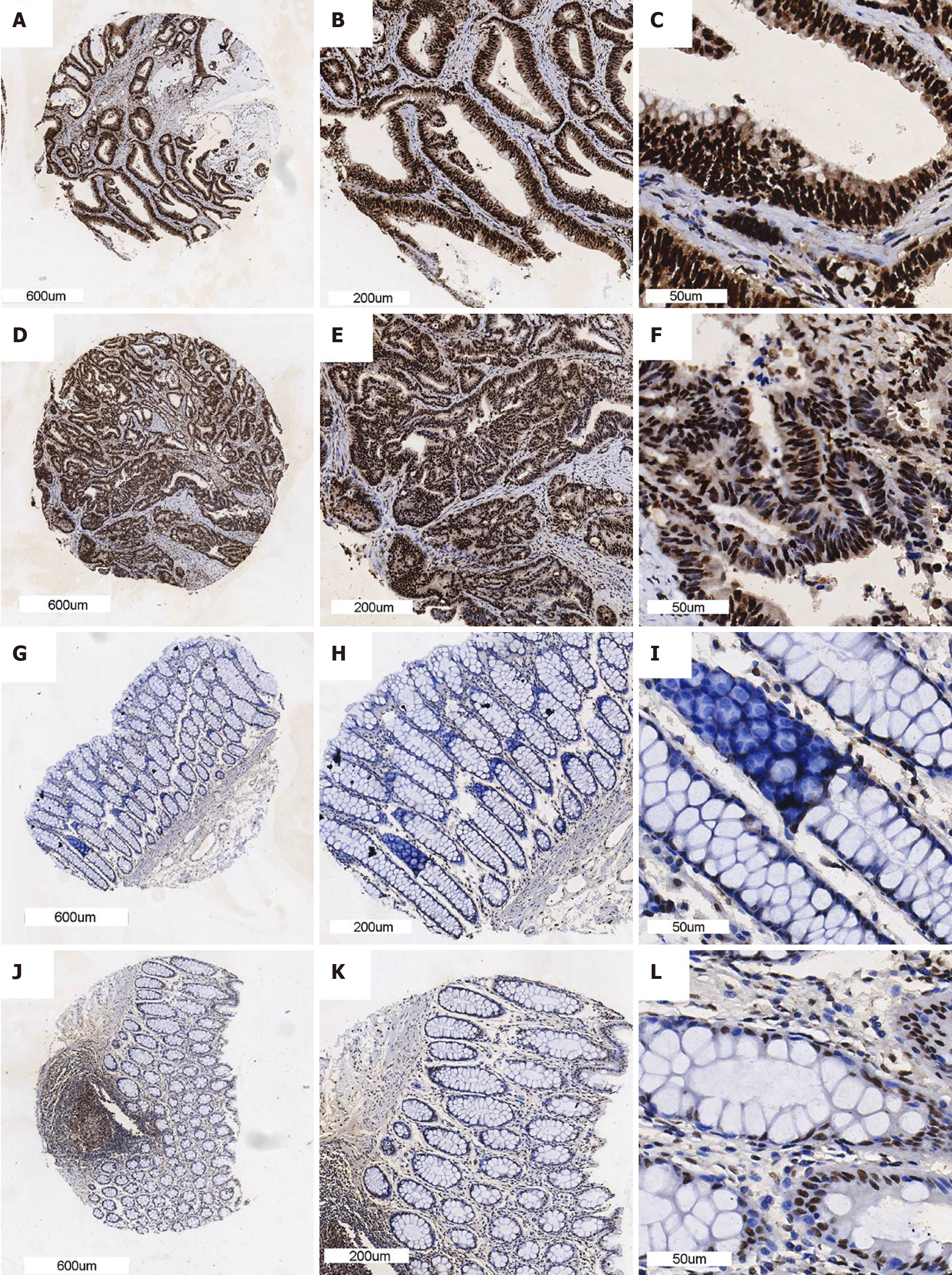
A total of 175 cases of CRC and 155 noncancerous cases were included in the tissue microarray. The protein level of SYNCRIP in CRC was higher than that in normal colorectal tissue (P < 0.0001) (Figure 1A), and the AUC of the ROC curve was 0.9562 (Figure 1B). We randomly selected three IHC samples of both CRC and normal tissue to demonstrate the level of SYNCRIP, and the staining intensity of SYNCRIP in CRC was obviously stronger than that in normal tissue (Figure 2). In addition, SYNCRIP was expressed at low levels in CRC with lymphatic invasion compared to noninvasive CRC (P = 0.0059) but was not different in the vascular invasion group (P = 0.4361).
Collection of CRC expression data: The flow chart of high-throughput data is listed in Supplementary Figure 1. The expression matrix of the RNA-array was pretreated and normalized according to the platform, and the expression matrix of RNA-seq was standardized using transcripts per kilobase of exon model per million mapped reads. A total of 22 da
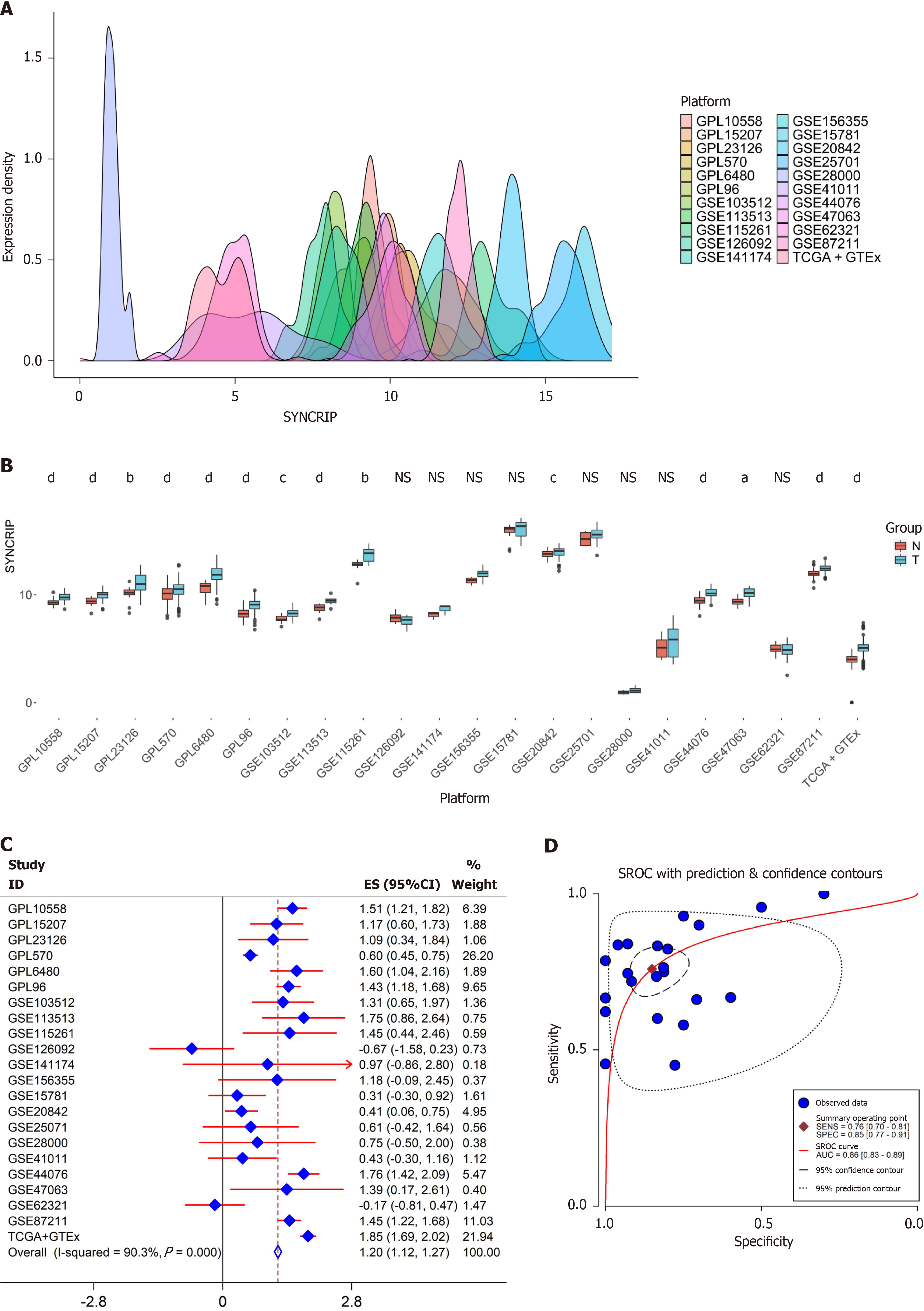
SYNCRIP expression in CRC: In the 25 expression matrix, the expression of SYNCRIP was close to a Gaussian distribution (Figure 3A). SYNCRIP was highly expressed in the CRC group with prominent statistical significance in 14 da
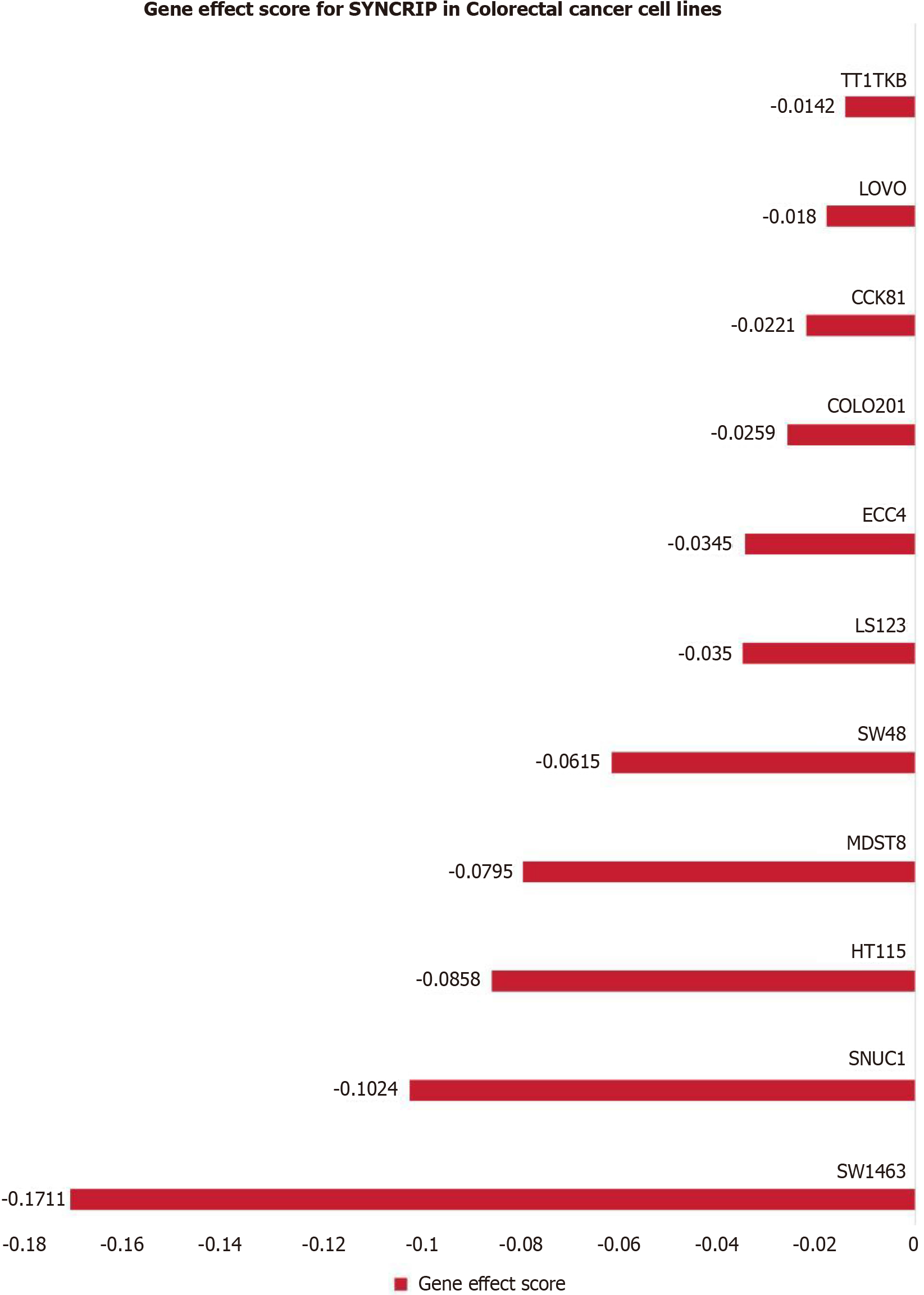
We explored CRISPR knockout screens derived from Dep Map to identify the effect of SYNCRIP on the proliferation of CRC cells. The gene effect score of SYNCRIP in eleven CRC cell lines was shown in Figure 4. The result revealed that SYNCRIP gene promoted cell growth in these eleven CRC cell lines.
The mutation status of SYNCRIP in CRC: Three clinical cohorts of CRC with gene mutation information were utilized to examine the mutation status of SYNCRIP. The mutation frequency of SYNCRIP was 4%, 1.8% and 1.9% in the Dana-Farber Cancer Institute (Cell reports 2016), TCGA (Firehose Legacy) and TCGA (Nature 2012) cohorts, respectively (Supplementary Figure 3). The results showed that SYNCRIP was not widely mutated in CRC.
The relationship between SYNCRIP and the tumor microenvironment: The expression of COAD and READ was downloaded from GDC-TCGA, and the TMB score was calculated (Supplementary Figure 4A and B). There was no correlation between SYNCRIP and the TMB of CRC (Supplementary Figure 4C and D). SYNCRIP did not disturb the mutation status of CRC. The MSI status of CRC was downloaded from the cBio Portal database, and there was no difference in the expression of SYNCRIP in the high-MSI group, low-MSI group and MSS group (Supplementary Figure 5).
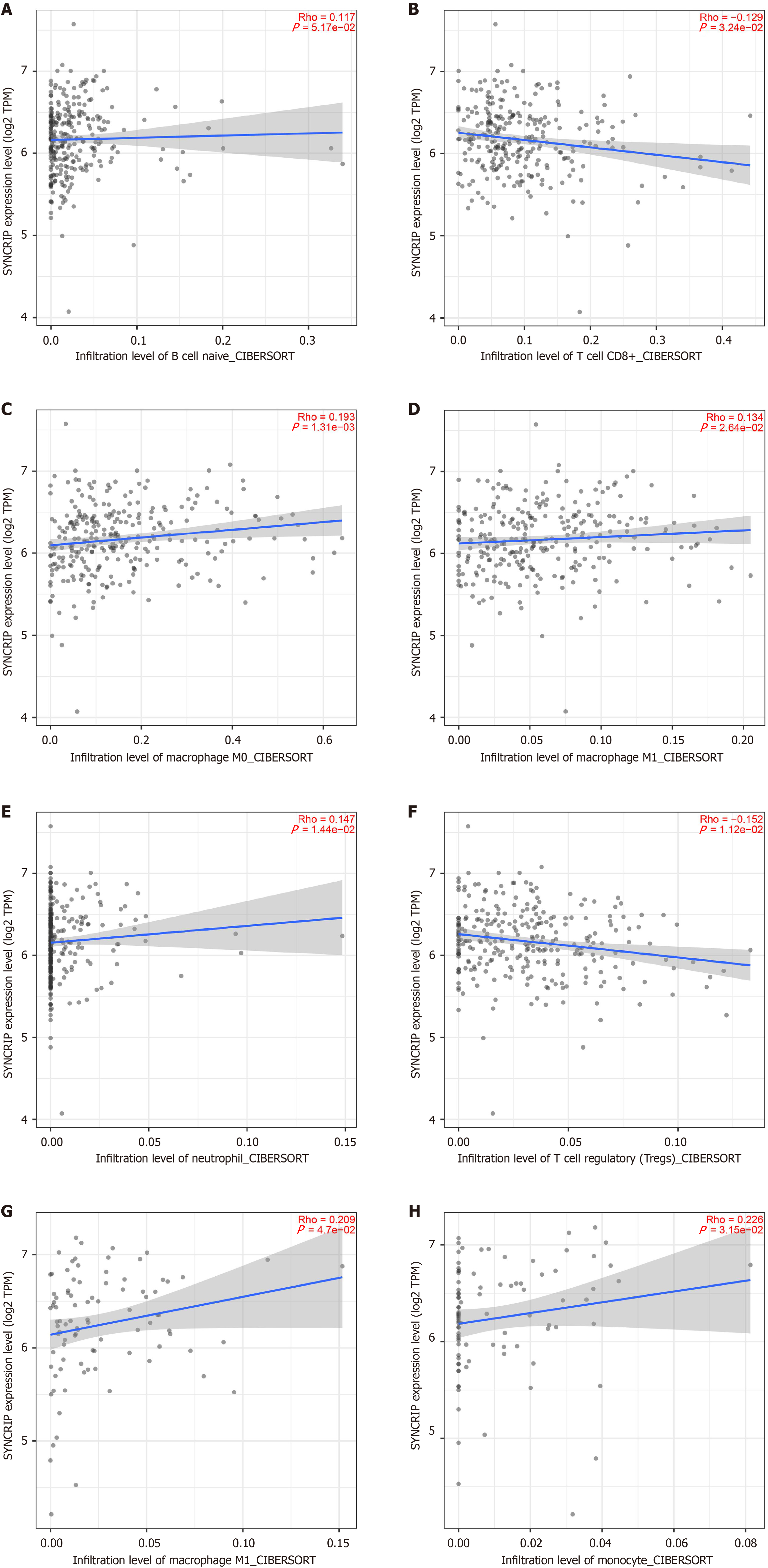
The immune microenvironment scores in COAD and READ were evaluated by using the TCGA-GTEx dataset. As Figure 4 and Figure 5 shows, in COAD and READ, SYNCRIP was positively related to naive B cells, macrophages, neutrophils and monocytes, and SYNCRIP was negatively related to CD8+ T cells and regulatory T cells (Figure 5).
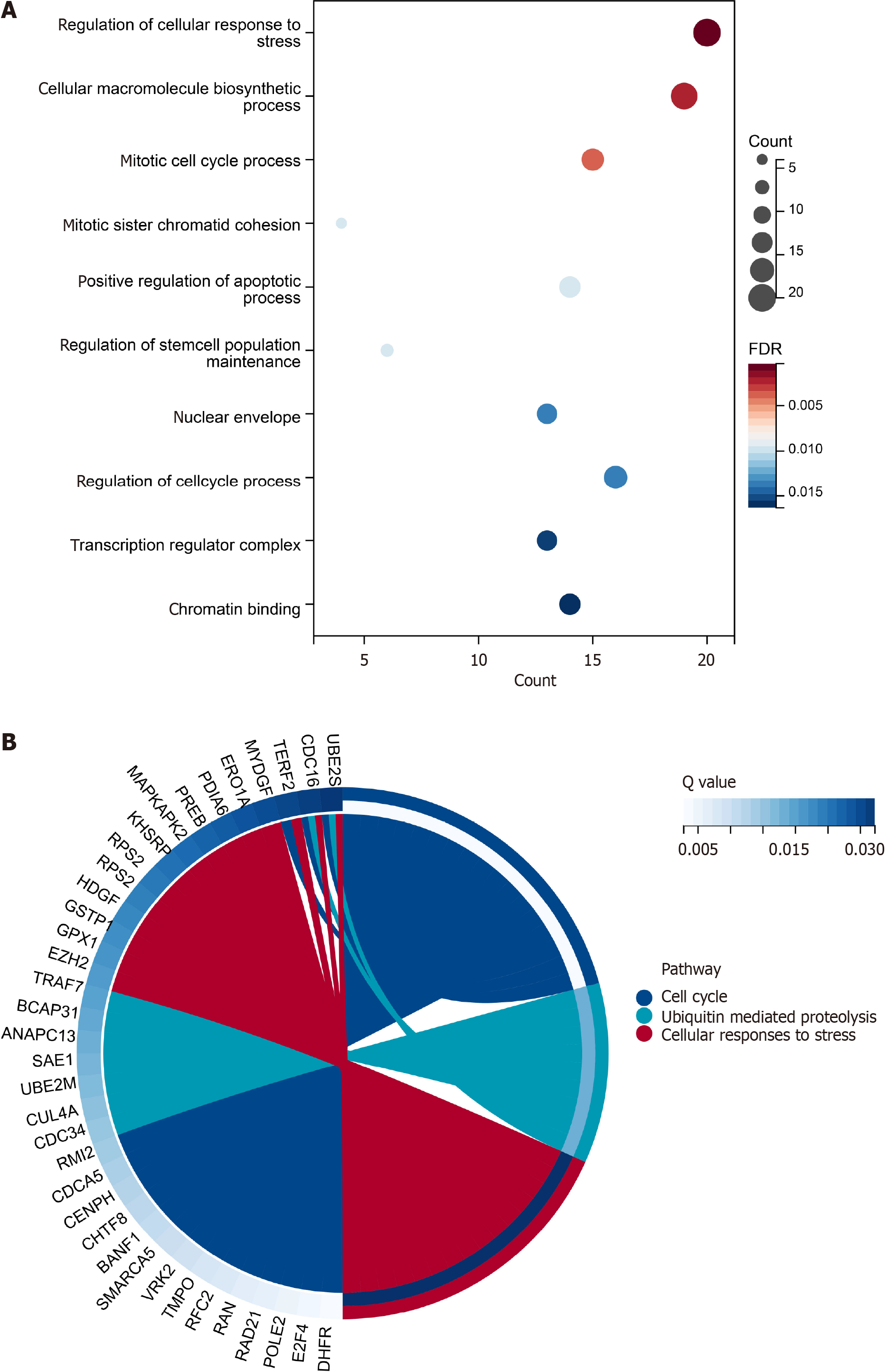
SYNCRIP target genes and potential pathways in CRC: To obtain the target genes of SYNCRIP, the co-expressed genes, SYNCRIP binding genes and highly expressed genes of CRC were calculated and intersected. A total of 15702 genes were conspicuously related to SYNCRIP with P < 0.05. A total of 2085 potential target genes with the structural domain of SYNCRIP were predicted by the RNAct database, and 6818 genes were regulated by SYNCRIP according to ChIP-seq data. After integration with 8427 highly expressed genes of CRC, a total of 180 genes were considered the most credible genes targeted by SYNCRIP.
For the GO analysis, a total of 15 items were significant, and the top three were regulation of cellular response to stress, cellular macromolecule biosynthetic process and mitotic cell cycle process (Figure 6A). For the pathway analysis, only three pathways were considered crucial pathways regulated by SYNCRIP: cell cycle (R-HSA-1640170), ubiquitin-mediated proteolysis (hsa04120) and cellular responses to stress (R-HSA-2262752) (Figure 6B). Cell cycle-related functions and stress response functions appeared in GO and pathway analyses simultaneously.
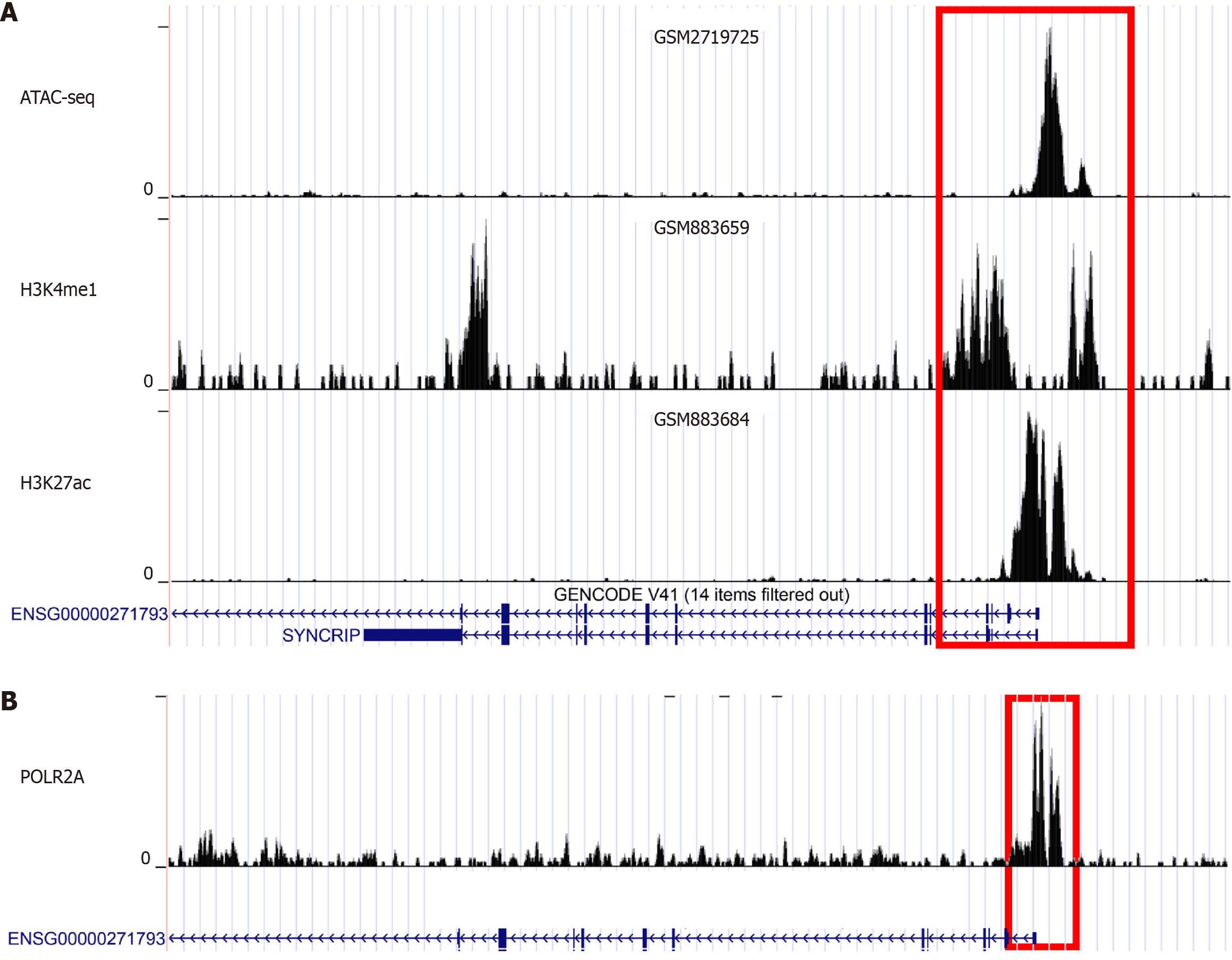
The upstream regulatory mechanism of SYNCRIP in CRC: The two major factors affecting SYNCRIP expression were enhancers and TFs. To explore the regulatory status of enhancers, an ATAC-seq sample (GEO number: GSM2719725) was used to explore the chromatin accessibility of SYNCRIP. The results showed that SYNCRIP had notable transcriptional activity in CRC (Figure 7A). SYNCRIP was regulated by enhancers in CRC because the peaks of H3k27ac (GEO number: GSM883684) and H3k4me1 (GEO number: GSM883659) were abundantly enriched in the promoter region of SYNCRIP (Figure 7A). POLR2A was the strongest regulatory TF for SYN
A total of five cohorts with outcome information on CRC were enrolled to explore the prognostic value of CRC. SYNCRIP had no prognostic value in patients with CRC (Supplementary Figure 6).
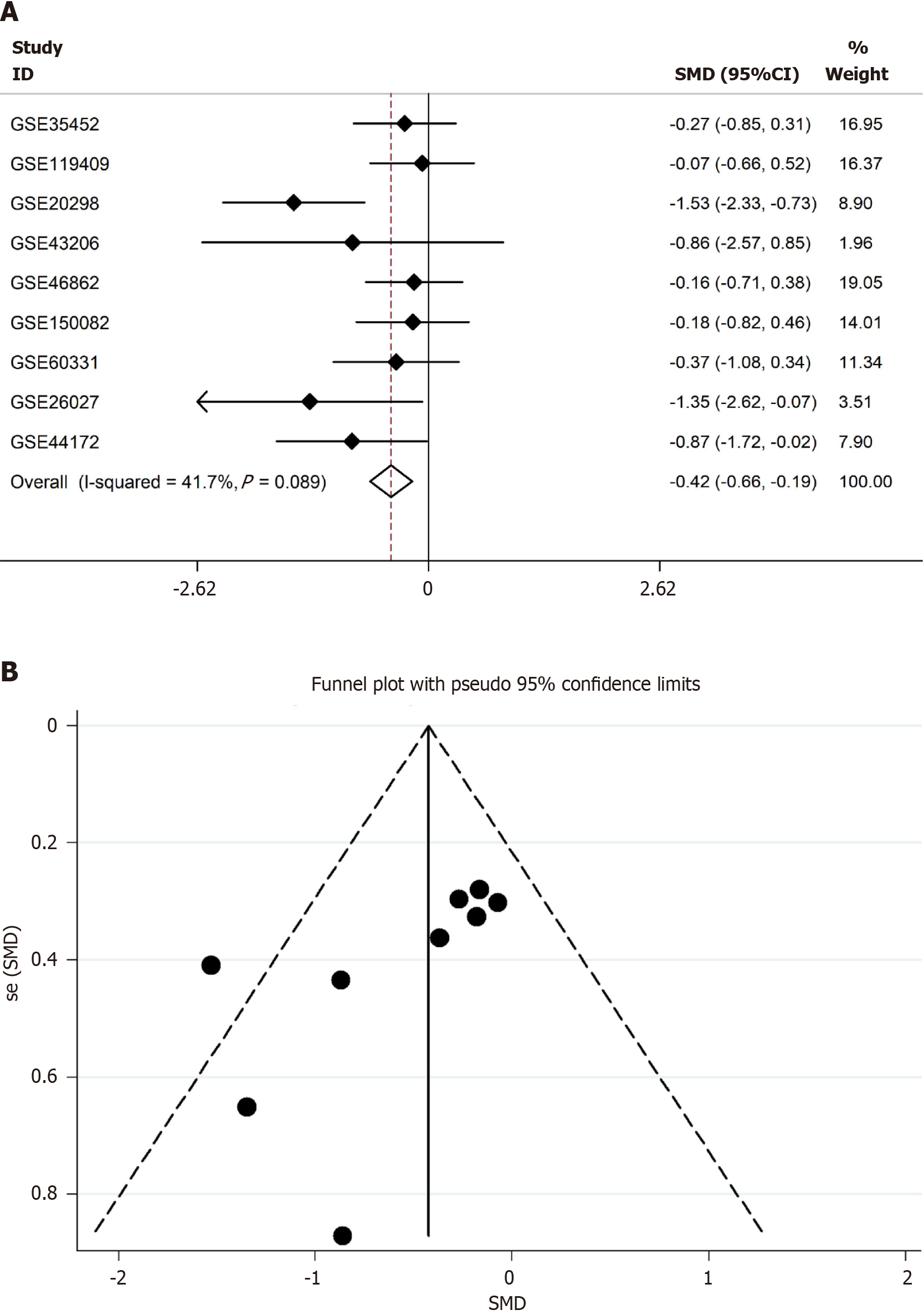
After filtering, the radiotherapy datasets were downloaded and processed. A total of nine datasets were used, including six clinical cohorts and three cytology experiments, with 218 radioresistant samples and 121 radiosensitive samples. SYNCRIP was a radiosensitivity gene in CRC with an SMD = -0.42 (95%CI: -0.66 to -0.19) (Figure 8A). No heterogeneity existed in the SMD model, and the fixed effects model was used (Figure 8B).
In our study, SYNCRIP was considered an oncogene in CRC based on its high protein and mRNA expression levels. As an RBP, SYNCRIP can target various genes and promote tumor onset and development by affecting the cell cycle and stress response-related pathways and affecting the immune microenvironment. In epigenetics, highly expressed SYNCRIP is regulated by enhancers and POLR2A. In addition, SYNCRIP may accelerate the sensitivity of radiotherapy for patients with CRC.
All the biological functions of SYNCRIP, such as regulating alternative splicing and epigenetic events, are embodied in protein[22,23]. Therefore, the protein level of SYNCRIP in CRC was examined and verified by IHC. From the pathomorphology, we can clearly observe that the SYNCRIP protein level in CRC is higher than that in nontumorous tissue. Furthermore, we used a nonparametric test to examine the difference in IHC as a semiquantitative method. The protein level was transformed to a score based on staining intensity and staining percentage. The high level of SYNCRIP in CRC compared to nontumorous tissue was confirmed by statistical analysis.
In addition to our in-house IHC data, public data should also be used based on evidence-based medicine. High-throughput array and sequence methods have been widely used for gene expression profiling in recent years. Therefore, the expression of SYNCRIP mRNA was also explored by high-throughput data. We excavated multiple RNA arrays and RNA-seq data of CRC from various technology platforms. In the field of evidence-based medicine, heterogeneity often leads to biased results. Hence, the batch effects of datasets from the same platform were erased, and each dataset represented one high-throughput platform. The SMD model is always used in evidence-based medicine, and it was also used in our study. The SMD = 1.2 > 1 means that SYNCRIP is highly expressed in CRC. CRISPR-Cas9 viability screens has been used to discover new therapeutic targets for cancer in recent years. Therefore, the dependency score of SYNCRIP was calculated by CERES algorithm to identify the effect of SYNCRIP in CRC cell lines. In our study, we found that knockdown of the SYNCRIP gene inhibited the proliferation of eleven CRC cell lines. To date, the high expression status of SYNCRIP in CRC has been verified by protein, mRNA and gene-dependency analyses. Hence, we think that SYNCRIP can promote the tumorous process in CRC.
To understand the molecular mechanism of SYNCRIP in CRC, we first calculated the mutation status of SYNCRIP in CRC. SYNCRIP had no significant mutation in CRC and had no significant relation to the TMB of CRC. Therefore, we think that SYNCRIP can steadily play a role in CRC patients. Then, the immune microenvironments and the potential pathways of SYNCRIP were calculated. Interestingly, SYNCRIP is negatively related to CD8+ T cells in CRC. CD8+ T cells play an antitumor role in many malignant tumors[24,25], and SYNCRIP may reduce CD8+ T cells to promote tumor proliferation in CRC. Because SYNCRIP is an RNA binding protein, it epigenetically targets some genes and promotes cancer proliferation in CRC. Thus, the target genes of SYNCRIP were identified by computational biology methods and utilized to carry out GO and KEGG analyses. We clearly observed that in both GO enrichment analysis and KEGG pathway analysis, SYNCRIP and its target genes participate in cell cycle-related functions. An active cell cycle is the necessary condition for proliferation and migration in all cancers[26,27]. Therefore, SYNCRIP can reduce CD8+ T cells and target some genes to boost tumor cell mitosis and promote proliferation and invasion in CRC.
After probing the downstream regulatory pattern and molecular mechanism of SYNCRIP, the upstream regulatory mechanism of SYNCRIP was extended. The mutation status of SYNCRIP in CRC is stable, so it is possibly regulated by epigenetic regulation, such as enhancers and TFs. To confirm the regulation of SYNCRIP and enhancers and TFs, both potential enhancers and TFs were investigated by using high-throughput ChIP-seq data. H3k27ac and H3K4me1 are specific biomarkers of active enhancers[28,29], and the promoter region of SYNCRIP is enriched by H3k27ac- and H3K4me1-marked ChIP-seq samples. Therefore, the expression of SYNCRIP is heightened by enhancers in CRC. We previously reported that enhancers can promote CLDN1 levels in CRC and increase sensitivity to radiation by affecting the cell cycle pathway, so enhancers are considered radio-related biomarkers in CRC patients[28]. In addition, from the ChIP-seq data, we also observed that SYNCRIP is regulated by POLR2A. As a TF, POLR2A encodes the largest subunit of RNA polymerase II and plays an important role in many tumors[30-32]. POLR2A is an oncogene and has been proven to be a therapeutic target in a variety of malignant tumors, especially in CRC[33-35]. In our study, the expression of SYNCRIP was boosted by enhancers and POLR2A, and it may become a potential target for predicting and treating CRC.
Radiotherapy is the frequently used method to kill tumor cells in CRC. Some studies have reported that low levels of CD8+ T cells and an active mitotic state in CRC can increase sensitivity to radiation therapy. Dayson Moreira et al reported that in head and neck squamous cell carcinoma, the CD8+ T-cell level is reduced when patients undergo radiotherapy[36]. The CD8+ T-cell level is also significantly lower in CRC patients with radiation treatment[37]. In addition, tumor cells in the mitotic stage are conspicuously sensitive to radiation[38,39]. Then, the expression of SYNCRIP in CRC patients treated with radiotherapy was calculated. Interestingly, the expression of SYNCRIP in the radio-sensitivity group was higher than that in the radio-resistance group. Therefore, we consider that SYNCRIP can reduce CD8+ T cells and activate CRC cells into a high-rate division status to increase sensitivity to radiation. In future clinical work, we can use SYNCRIP staining IHC to determine the sensitivity of CRC patients to radiotherapy.
In conclusion, SYNCRIP is regulated by enhancers and POLR2A and highly expressed in CRC. Highly expressed SYNCRIP can reduce CD8+ T cells and accelerate CRC cell division by exerting its epigenetic regulatory effects. These CRC cells are sensitive to radiation therapy. Finally, SYNCRIP is expected to become a biomarker in CRC patients.
The authors thank the Guangxi Zhuang Autonomous Region Clinical Medicine Research Center for Molecular Pathology and Intelligent Pathology Precision Diagnosis for the technical support of computational pathology and clinical pathology.
| 1. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4235] [Cited by in RCA: 11988] [Article Influence: 2997.0] [Reference Citation Analysis (9)] |
| 2. | Thanikachalam K, Khan G. Colorectal Cancer and Nutrition. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 231] [Cited by in RCA: 520] [Article Influence: 74.3] [Reference Citation Analysis (0)] |
| 3. | Wang H, Tian T, Zhang J. Tumor-Associated Macrophages (TAMs) in Colorectal Cancer (CRC): From Mechanism to Therapy and Prognosis. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 280] [Article Influence: 56.0] [Reference Citation Analysis (2)] |
| 4. | Cho YA, Lee J, Oh JH, Chang HJ, Sohn DK, Shin A, Kim J. Genetic Risk Score, Combined Lifestyle Factors and Risk of Colorectal Cancer. Cancer Res Treat. 2019;51:1033-1040. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 5. | Azeem S, Gillani SW, Siddiqui A, Jandrajupalli SB, Poh V, Syed Sulaiman SA. Diet and Colorectal Cancer Risk in Asia--a Systematic Review. Asian Pac J Cancer Prev. 2015;16:5389-5396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 6. | Yang Y, Han Z, Li X, Huang A, Shi J, Gu J. Epidemiology and risk factors of colorectal cancer in China. Chin J Cancer Res. 2020;32:729-741. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 94] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 7. | Johdi NA, Sukor NF. Colorectal Cancer Immunotherapy: Options and Strategies. Front Immunol. 2020;11:1624. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 235] [Cited by in RCA: 324] [Article Influence: 54.0] [Reference Citation Analysis (1)] |
| 8. | Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14:89-103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 444] [Cited by in RCA: 1169] [Article Influence: 167.0] [Reference Citation Analysis (12)] |
| 9. | Sheikhnia F, Rashidi V, Maghsoudi H, Majidinia M. Potential anticancer properties and mechanisms of thymoquinone in colorectal cancer. Cancer Cell Int. 2023;23:320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 10. | Hashemi M, Esbati N, Rashidi M, Gholami S, Raesi R, Bidoki SS, Goharrizi MASB, Motlagh YSM, Khorrami R, Tavakolpournegari A, Nabavi N, Zou R, Mohammadnahal L, Entezari M, Taheriazam A, Hushmandi K. Biological landscape and nanostructural view in development and reversal of oxaliplatin resistance in colorectal cancer. Transl Oncol. 2024;40:101846. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 11. | Yang L, Atakhanova N, Arellano MTC, Mohamed MY, Hani T, Fahdil AA, Castillo-Acobo RY, Juyal A, Hussein AK, Amin AH, Pecho RDC, Akhavan-Sigari R. Translational research of new developments in targeted therapy of colorectal cancer. Pathol Res Pract. 2023;252:154888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 12. | Ni Y, Liang Y, Li M, Lin Y, Zou X, Han F, Cao J, Li L. The updates on metastatic mechanism and treatment of colorectal cancer. Pathol Res Pract. 2023;251:154837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 13. | Geng L, Wang J. Molecular effectors of radiation resistance in colorectal cancer. Precis Radiat Oncol. 2017;1:27-33. [RCA] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 14. | Khudayberdiev S, Soutschek M, Ammann I, Heinze A, Rust MB, Baumeister S, Schratt G. The cytoplasmic SYNCRIP mRNA interactome of mammalian neurons. RNA Biol. 2021;18:1252-1264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 15. | Chen Y, Chan J, Chen W, Li J, Sun M, Kannan GS, Mok YK, Yuan YA, Jobichen C. SYNCRIP, a new player in pri-let-7a processing. RNA. 2020;26:290-305. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 16. | Chen S, Miao B, Chen N, Chen C, Shao T, Zhang X, Chang L, Zhang X, Du Q, Huang Y, Tong D. SYNCRIP facilitates porcine parvovirus viral DNA replication through the alternative splicing of NS1 mRNA to promote NS2 mRNA formation. Vet Res. 2021;52:73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 17. | Hobor F, Dallmann A, Ball NJ, Cicchini C, Battistelli C, Ogrodowicz RW, Christodoulou E, Martin SR, Castello A, Tripodi M, Taylor IA, Ramos A. A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets. Nat Commun. 2018;9:831. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 99] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 18. | Semino F, Schröter J, Willemsen MH, Bast T, Biskup S, Beck-Woedl S, Brennenstuhl H, Schaaf CP, Kölker S, Hoffmann GF, Haack TB, Syrbe S. Further evidence for de novo variants in SYNCRIP as the cause of a neurodevelopmental disorder. Hum Mutat. 2021;42:1094-1100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (1)] |
| 19. | Riccioni V, Trionfetti F, Montaldo C, Garbo S, Marocco F, Battistelli C, Marchetti A, Strippoli R, Amicone L, Cicchini C, Tripodi M. SYNCRIP Modulates the Epithelial-Mesenchymal Transition in Hepatocytes and HCC Cells. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 20. | Zhang P, Cao M, Zhang Y, Xu L, Meng F, Wu X, Xia T, Chen Q, Shi G, Wu P, Chen L, Lu Z, Yin J, Cai B, Cao S, Miao Y, Jiang K. A novel antisense lncRNA NT5E promotes progression by modulating the expression of SYNCRIP and predicts a poor prognosis in pancreatic cancer. J Cell Mol Med. 2020;24:10898-10912. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 21. | Wang YC, Chang KC, Lin BW, Lee JC, Lai CH, Lin LJ, Yen Y, Lin CS, Yang SJ, Lin PC, Lee CT, Hung LY. The EGF/hnRNP Q1 axis is involved in tumorigenesis via the regulation of cell cycle-related genes. Exp Mol Med. 2018;50:1-14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 22. | Singh A. RNA-binding protein kinetics. Nat Methods. 2021;18:335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 23. | Sommer G, Heise T. Role of the RNA-binding protein La in cancer pathobiology. RNA Biol. 2021;18:218-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 24. | Raskov H, Orhan A, Christensen JP, Gögenur I. Cytotoxic CD8(+) T cells in cancer and cancer immunotherapy. Br J Cancer. 2021;124:359-367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 1171] [Article Influence: 195.2] [Reference Citation Analysis (0)] |
| 25. | Reina-Campos M, Scharping NE, Goldrath AW. CD8(+) T cell metabolism in infection and cancer. Nat Rev Immunol. 2021;21:718-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 459] [Article Influence: 91.8] [Reference Citation Analysis (0)] |
| 26. | Icard P, Fournel L, Wu Z, Alifano M, Lincet H. Interconnection between Metabolism and Cell Cycle in Cancer. Trends Biochem Sci. 2019;44:490-501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 228] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 27. | Matthews HK, Bertoli C, de Bruin RAM. Cell cycle control in cancer. Nat Rev Mol Cell Biol. 2022;23:74-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 490] [Cited by in RCA: 960] [Article Influence: 240.0] [Reference Citation Analysis (1)] |
| 28. | Chen ZX, Huang HQ, Wen JY, Qin LS, Song YD, Fang YY, Zeng DT, Huang WJ, Qin XG, Gan TQ, Luo J, Li JJ. Active Enhancer Assessment by H3K27ac ChIP-seq Reveals Claudin-1 as a Biomarker for Radiation Resistance in Colorectal Cancer. Dose Response. 2021;19:15593258211058981. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 29. | Zhang W, Ruan X, Li Y, Zhi J, Hu L, Hou X, Shi X, Wang X, Wang J, Ma W, Gu P, Zheng X, Gao M. KDM1A promotes thyroid cancer progression and maintains stemness through the Wnt/β-catenin signaling pathway. Theranostics. 2022;12:1500-1517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 90] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 30. | Xu J, Liu Y, Li Y, Wang H, Stewart S, Van der Jeught K, Agarwal P, Zhang Y, Liu S, Zhao G, Wan J, Lu X, He X. Precise targeting of POLR2A as a therapeutic strategy for human triple negative breast cancer. Nat Nanotechnol. 2019;14:388-397. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 124] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 31. | Jiang Q, Zhang J, Li F, Ma X, Wu F, Miao J, Li Q, Wang X, Sun R, Yang Y, Zhao L, Huang C. POLR2A Promotes the Proliferation of Gastric Cancer Cells by Advancing the Overall Cell Cycle Progression. Front Genet. 2021;12:688575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 32. | Mao CG, Jiang SS, Shen C, Long T, Jin H, Tan QY, Deng B. BCAR1 promotes proliferation and cell growth in lung adenocarcinoma via upregulation of POLR2A. Thorac Cancer. 2020;11:3326-3336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 33. | Li Y, Sun Y, Kulke M, Hechler T, Van der Jeught K, Dong T, He B, Miller KD, Radovich M, Schneider BP, Pahl A, Zhang X, Lu X. Targeted immunotherapy for HER2-low breast cancer with 17p loss. Sci Transl Med. 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 34. | Liu Y, Zhang X, Han C, Wan G, Huang X, Ivan C, Jiang D, Rodriguez-Aguayo C, Lopez-Berestein G, Rao PH, Maru DM, Pahl A, He X, Sood AK, Ellis LM, Anderl J, Lu X. TP53 loss creates therapeutic vulnerability in colorectal cancer. Nature. 2015;520:697-701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 192] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 35. | Errico A. Colorectal cancer: POLR2A deletion with TP53 opens a window of opportunity for therapy. Nat Rev Clin Oncol. 2015;12:374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 36. | Moreira D, Sampath S, Won H, White SV, Su YL, Alcantara M, Wang C, Lee P, Maghami E, Massarelli E, Kortylewski M. Myeloid cell-targeted STAT3 inhibition sensitizes head and neck cancers to radiotherapy and T cell-mediated immunity. J Clin Invest. 2021;131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 76] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 37. | Hsieh RC, Krishnan S, Wu RC, Boda AR, Liu A, Winkler M, Hsu WH, Lin SH, Hung MC, Chan LC, Bhanu KR, Srinivasamani A, De Azevedo RA, Chou YC, DePinho RA, Gubin M, Vilar E, Chen CH, Slay R, Jayaprakash P, Hegde SM, Hartley G, Lea ST, Prasad R, Morrow B, Couillault CA, Steiner M, Wang CC, Venkatesulu BP, Taniguchi C, Kim YSB, Chen J, Rudqvist NP, Curran MA. ATR-mediated CD47 and PD-L1 up-regulation restricts radiotherapy-induced immune priming and abscopal responses in colorectal cancer. Sci Immunol. 2022;7:eabl9330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 126] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 38. | Pawlik TM, Keyomarsi K. Role of cell cycle in mediating sensitivity to radiotherapy. Int J Radiat Oncol Biol Phys. 2004;59:928-942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 669] [Cited by in RCA: 822] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 39. | Pauwels B, Wouters A, Peeters M, Vermorken JB, Lardon F. Role of cell cycle perturbations in the combination therapy of chemotherapeutic agents and radiation. Future Oncol. 2010;6:1485-1496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 1.6] [Reference Citation Analysis (3)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/