Published online Jan 24, 2024. doi: 10.5306/wjco.v15.i1.62
Peer-review started: October 23, 2023
First decision: November 23, 2023
Revised: December 12, 2023
Accepted: December 28, 2023
Article in press: December 28, 2023
Published online: January 24, 2024
Processing time: 91 Days and 12.6 Hours
Transcatheter arterial embolisation (TACE) is the primary treatment for intermediate-stage hepatocellular carcinoma (HCC) patients while some HCC cases have shown resistance to TACE.
To investigate the key genes and potential mechanisms correlated with TACE refractoriness in HCC.
The microarray datasets of TACE-treated HCC tissues, HCC and non-HCC tissues were collected by searching multiple public databases. The respective differentially expressed genes (DEGs) were attained via limma R package. Weighted gene co-expression network analysis was employed for identifying the significant modules related to TACE non-response. TACE refractoriness-related genes were obtained by intersecting up-regulated TACE-associated and HCC-associated DEGs together with the genes in significant modules related to TACE non-response. The key genes expression in the above two pairs of samples was compared respectively via Wilcoxon tests and standard mean differences model. The prognostic value of the key genes was evaluated by Kaplan-Meier curve. Multivariate analysis was utilised to investigate the independent prognostic factor in key genes. Single-cell RNA (scRNA) sequencing analysis was conducted to explore the cell types in HCC. TACE refractoriness-related genes activity was calculated via AUCell packages. The CellChat R package was used for the investigation of the cell–cell communication between the identified cell types.
HCC tissues of TACE non-responders (n = 66) and TACE responders (n = 81), HCC (n = 3941) and non-HCC (n = 3443) tissues were obtained. The five key genes, DLG associated protein 5 (DLGAP5), Kinesin family member 20A (KIF20A), Assembly factor for spindle microtubules (ASPM), Kinesin family member 11 (KIF11) and TPX2 microtubule nucleation factor (TPX2) in TACE refractoriness-related genes, were identified. The five key genes were all up-regulated in the TACE non-responders group and the HCC group. High expression of the five key genes predicted poor prognosis in HCC. Among the key genes, TPX2 was an independent prognostic factor. Four cell types, hepatocytes, embryonic stem cells, T cells and B cells, were identified in the HCC tissues. The TACE refractoriness-related genes expressed primarily in hepatocytes and embryonic stem cells. Hepatocytes, as the providers of ligands, had the strongest interaction with embryonic stem cells that provided receptors.
Five key genes (DLGAP5, KIF20A, ASPM, KIF11 and TPX2) were identified as promoting refractory TACE. Hepatocytes and embryonic stem cells were likely to boost TACE refractoriness.
Core Tip: This is a study that explored the key genes and mechanisms related to transcatheter arterial chemoembolisation (TACE) refractoriness in hepatocellular carcinoma. Through the combination of tissue microarrays and RNA-seq with single-cell RNA sequencing, the TACE refractoriness-related genes were identified and five key genes (DLGAP5, KIF20A, ASPM, KIF11 and TPX2) associated with TACE refractoriness were revealed. The TACE refractoriness-related genes were found to mainly express in hepatocytes and embryonic stem cells. Hepatocytes providing ligands had the strongest interaction with embryonic stem cells as receptors providers.
- Citation: Huang JZ, Li JD, Chen G, He RQ. Identification of the key genes and mechanisms associated with transcatheter arterial chemoembolisation refractoriness in hepatocellular carcinoma. World J Clin Oncol 2024; 15(1): 62-88
- URL: https://www.wjgnet.com/2218-4333/full/v15/i1/62.htm
- DOI: https://dx.doi.org/10.5306/wjco.v15.i1.62
Hepatocellular carcinoma (HCC) is a major primary liver tumour, accounting for more than 90% of primary liver tumour cases[1]. HCC is currently the fifth most common cancer worldwide and the second leading cause of cancer death in men, with the five-year survival rate of 18% second only to pancreatic cancer[1]. The complex pathogenesis of HCC and its various risk factors like chronic hepatitis B virus infection and alcohol abuse lead to the increasing incidence of HCC[2,3]. A retrospective study showed that the incidence of HCC increased markedly in America from 1975 to 2016[4]. It is worth noting that HCC is generally diagnosed in its advance stage[5]. The appearance of symptoms stage means the rapid progress of HCC with the poor therapeutic effect and slim survival[6].
Transcatheter arterial embolisation (TACE) is a strategy in which catheters, with the support of various embolisation drugs, are placed in the large arteries, typically the femoral arteries, to block the tumour’s blood supply[7,8]. The selection of the treatment is based on the Barcelona Clinic Liver Cancer staging system, which was created according to the number size of the HCC and the performance of the patient[9]. TACE is a standard therapy for intermediate-stage HCC patients[10]. Currently, surgical treatment, liver transplantation and ablation are available for early-HCC patients and contribute to a high survival rate[8,11]. However, metastatic disease may be a barrier to surgical therapy[12]. Local ablation is regarded as a safer alternative to surgery, but it is also not suitable for patients who have tumours in the subcapsular or domed position or near the main bile duct, large blood vessels or the intestine[13]. Patients who cannot benefit from the above treatments despite the early stage of the disease may be candidates for TACE[14]. Chemotherapy is universally considered the first choice for advanced HCC when there is a low pharmaceutical response or the formation of portal vein tumour thrombosis[15]. In clinical practice, multiple combinations of liver-directed therapies (LDT) pave the way for the curative treatment[16], and TACE is still the most widely used as an LDT for locally advanced HCC patients[17]. Therefore, TACE is an important method for treating HCC at all stages.
Unfortunately, some investigations have shown that a load of HCC patients did not respond to TACE, which reduces the efficacy of the systematic therapy significantly[18]. One recent study employed a cohort of HCC patients after TACE treatment, and a series of vitro experiments revealed that pyruvate kinase M1/2 was highly expressed, which may promote HCC tolerance to TACE through glycolysis dysregulation[19], and was associated with a low survival rate in TACE non-response HCC patients. Additionally, another study found that the low expression of miRNA-125b induced an HCC tolerance to TACE through a transcriptome analysis of 680 HCC patients[20]. However, HCC is featured in the high heterogeneity of genetic drivers leading to therapeutic susceptibility[21]. Therefore, individual genes heterogeneity requires the exploration of other potential factors through systematic measures.
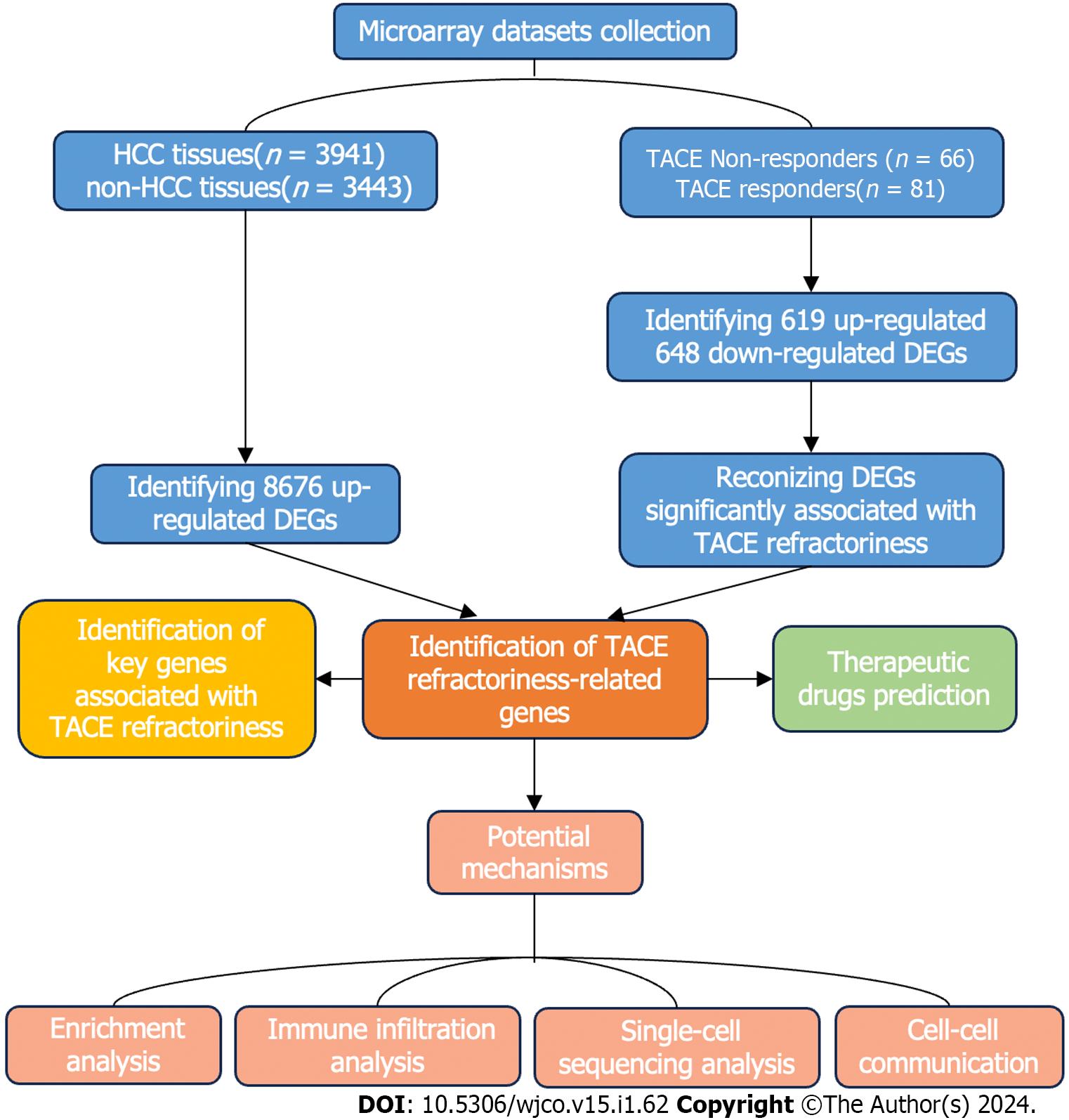
In this study, we aimed to identify the key genes of TACE refractoriness in HCC. Additionally, the potential mechanisms of TACE refractoriness in HCC were explored. Finally, potential therapeutic drugs against TACE refractoriness in HCC were discovered. The flow chart of this study is shown in Figure 1.
We retrieved the datasets including TACE non-responders and TACE responders from the Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), International Cancer Genome Consortium and ArrayExpress databases, with the query formulation 'Hepatocellular carcinoma AND Transcatheter arterial embolisation'. The datasets included were required to belong to Homo sapiens and contained HCC tissues from TACE non-responders and TACE responders who had not received any prior treatment. Simultaneously, we collected HCC and its control samples from the TCGA, GEO, Genotype-tissue Expression (GTEx) and ArrayExpress databases with keywords ‘hepatocellular carcinoma’ based on the following criteria: (1) The matrix must be Homo sapiens; (2) the data sets should include HCC and normal liver tissues or one of the above groups and can be merged; and (3) the collected samples should not be influenced by any chemicals or physical treatment. Subsequently, the profiles of all of the screened datasets downloaded from the above database were annotated according to the platform annotation files and then conducted by log2(x+1) conversion. To integrate the expression matrix from the same platforms or append the lack of normal liver tissue samples in some matrices, the R software (Version 3.6.2) was used to merge and normalise datasets with different sequencing and background through limma and surrogate variable analysis (sva) packages.
The limma package was applied to identify the differentially expressed genes (DEGs). The DEGs between TACE non-responders and TACE responders (TACE-associated DEGs) had to meet the requirement of P value < 0.05 and |log2 (Foldchange)| > 0.5. The DEGs between HCC and non-HCC tissues (HCC-associated DEGs) from the datasets were integrated by calculating the standardised mean difference (SMD). DEGs with P < 0.05 and |SMD| > 0 were recorded as the DEGs in HCC.
Weighted gene co-expression network analysis (WGCNA) is a systematic biological method for calculating the correlation among genes for finding gene clusters (modules) and connecting gene modules with external sample traits using eigengene network methodology[22]. In this study, we utilised WGCNA analysis to identify more reliable hub genes associated with TACE refractoriness in HCC.
WGCNA R package was utilised to construct the co-expression network using the expression profile of the TACE-associated DEGs in GSE104580. Firstly, Pearson's correlation matrix and average linkage methods were both used for all pair-wise genes. Secondly, an adjacency matrix was established to calculate the correlation value. The soft threshold was used to emphasise the correlation between two genes and eliminate weak correlations. Thirdly, adjacency was trans
Module eigengene was the first principal component of the module and was defined as the expression file of the whole genes in the module. The correlation coefficient between module eigengene and the clinical trait, namely the reactivity to TACE, was calculated to identify the clinically significant module. Module membership was regarded as the correlation between the genes and the module eigengene and was used to evaluate whether the genes belonged to the module or not. Additionally, gene significance was defined as the mediated P in the linear regression between expression and clinical traits. In addition, module significance was the average absolute gene significance of whole genes in the module. Finally, the modules with the top-two correlation coefficients between module eigengene and clinical status TACE non-responders were selected as the clinically significant modules for further analysis.
The up-regulated genes in TACE-associated DEGs, up-regulated HCC-associated DEGs and the genes in the clinically significant modules were intersected and their shared genes were considered as TACE refractoriness-related genes. TACE refractoriness-related genes were utilised for protein–protein interaction (PPI) network construction via STRING database. Then, the key genes were identified using the Maximal Clique Centrality (MCC) method of cytoHubba plugin in Cytoscape software (Version 3.7.2). MCC algorithm is a method of scoring each protein in the PPI network. A higher score means the protein is associated with more proteins in the network.
The mRNA expression level of the key genes between TACE non-responders and TACE responders was compared through Wilcoxon tests. Then, to verify that the key genes were differentially expressed in HCC, SMD model and a 95% confidence interval (95%CI) were carried out using Meta R packages and STATA 12.0 by integrating the datasets containing HCC and non-HCC tissues. A random-effect model or fixed-effect model was selected in the light of the I2 test and Chi-square test. The random-effect model was chosen when there was high heterogeneity (I2 > 50% or P < 0.05). Otherwise, the fixed-effect model was chosen. Sensibility analysis was conducted to detect the high heterogeneous datasets. The protein expression of key genes in HCC tissues was compared to that in non-HCC tissues by browsing The Human Protein Atlas (THPA) database. We then gathered the clinical information of the HCC tissues in the datasets. The clinical information was required to include survival status and survival time, and only the clinical information in TCGA datasets was eligible. To identify the independent prognostic factors in the key genes, multivariate analysis was used. For survival analysis, the optimum cut-off for division into high expression and low expression group was determined by running Survminer and Survival packages and the hazard ratio (HR) was calculated by Cox-proportional hazards regression model. Finally, the Kaplan–Meier was visualised using the Survival package.
TACE refractoriness-related genes were utilised in gene ontology (GO) annotation analysis, Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway using the clusterProfiler R package and Reactome pathway using Reactome pathway database. The terms with P < 0.05 and FDR < 0.25 were considered statistically significant. The top-five GO terms in respect to biological process (BP), cellular component (CC) and molecular function (MF) were investigated. Finally, the KEGG pathways and the top-ten Reactome pathways were visualised.
To further explore the cell types involved in TACE refractoriness, single-cell RNA sequencing (scRNA-seq) analysis was adopted. The scRNA-seq data were obtained from the GEO database with the search strategy 'ScRNA OR Single Cell AND Hepatocellular Carcinoma'. The samples in the scRNA-seq data had to be HCC samples that were not treated using any chemical or physical factors. The quality control process of the scRNA-seq data was performed with the Seurat R package. Cells with < 500 genes, > 6000 genes or > 20% mitochondrial genes were filtered out. Then, the gene expression matrix was normalised and scaled. Subsequently, the Harmony R package was employed for eliminating batch effect. A uniform manifold approximation and projection (UMAP) analysis was performed for dimensional reduction. The FindClusters function was able to classify the cells into different clusters with a resolution of 0.5, and the annotation of the cell clusters depended on the SingleR package. The activity of TACE refractoriness-related genes in each cell line was determined using the AUCell R package. Cells expressing more TACE refractoriness-related genes had higher area under-the-curve (AUC) values. The ‘AUCell_exploreThresholds’ function was used to determine the threshold for identifying TACE refractoriness-related genes’ active cells. The AUC score of each cell was then mapped to the UMAP plot embedding using the ggplot2 R package to visualise the active clusters.
CellChat, an open R package, was utilised for the analysis of intercellular communications using scRNA-seq data[23]. After identifying the cell types in the HCC tissues, the CellChat R package (Version 1.1.3) was employed to investigate the number of interactions, the communication strength and the over-expressed ligands and receptors between each identified cell type. The discovery of overexpressed ligand-receptor pairs was based on the data of ligand-receptor pairs related to ‘Secreted Signalling’ in datasets CellChatDB. The communication probability was calculated by means of the function ‘computeCommunProb’ in the CellChat R package. Subsequently, the cell–cell communication was predicted and aggregated. The number of interactions was visualised to show integrated communication.
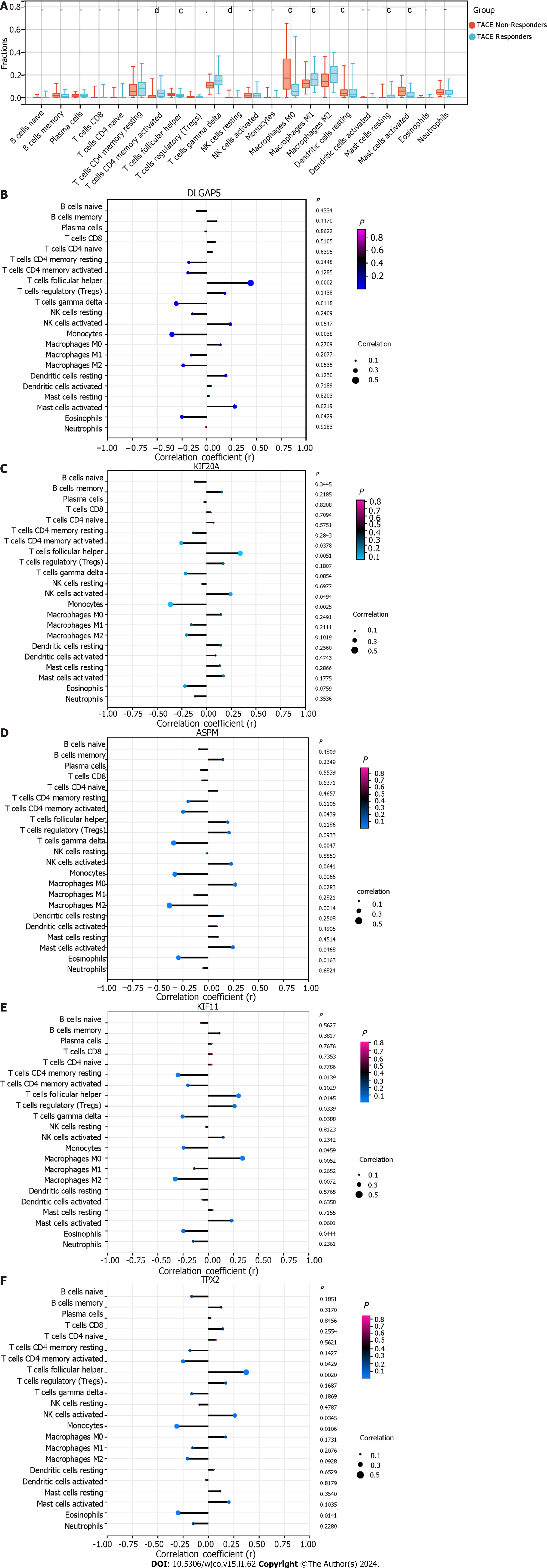
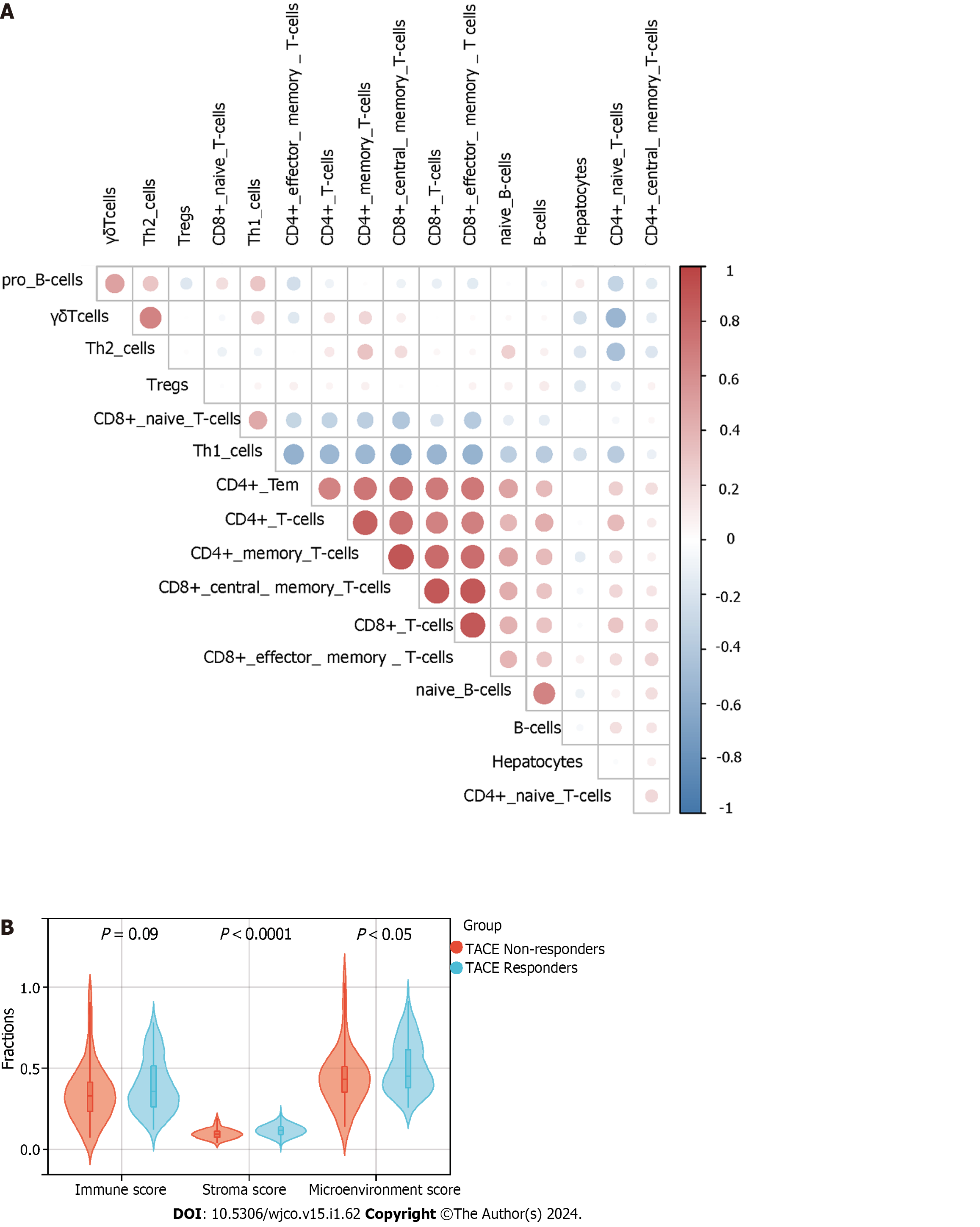
To elucidate the correlation between the TACE resistance and tumour microenvironment in the HCC, we conducted immune infiltration analysis based on CIBERSORT and xCell algorithm. The CIBERSORT algorithm is a deconvolution method used, in this case, to process marker gene expression values to estimate the proportion of various types of immune cells[24]. The xCell algorithm was performed for cell-type enrichment analysis from gene expression data of 64 immune and stroma cell types[25]. In this study, the CIBERSORT algorithm assessed the proportions of 22 kinds of immune cells between TACE non-responders and TACE responders. Meanwhile, the relationship between the expression of the five key genes and the 22 kinds of immune cells’ infiltration in the HCC samples from TACE non-responders was explored. The xCell algorithm evaluated the immune score, stromal score, microenvironment score and proportions of the cell types of the scRNA-seq analysis. Statistical significance required P < 0.05.
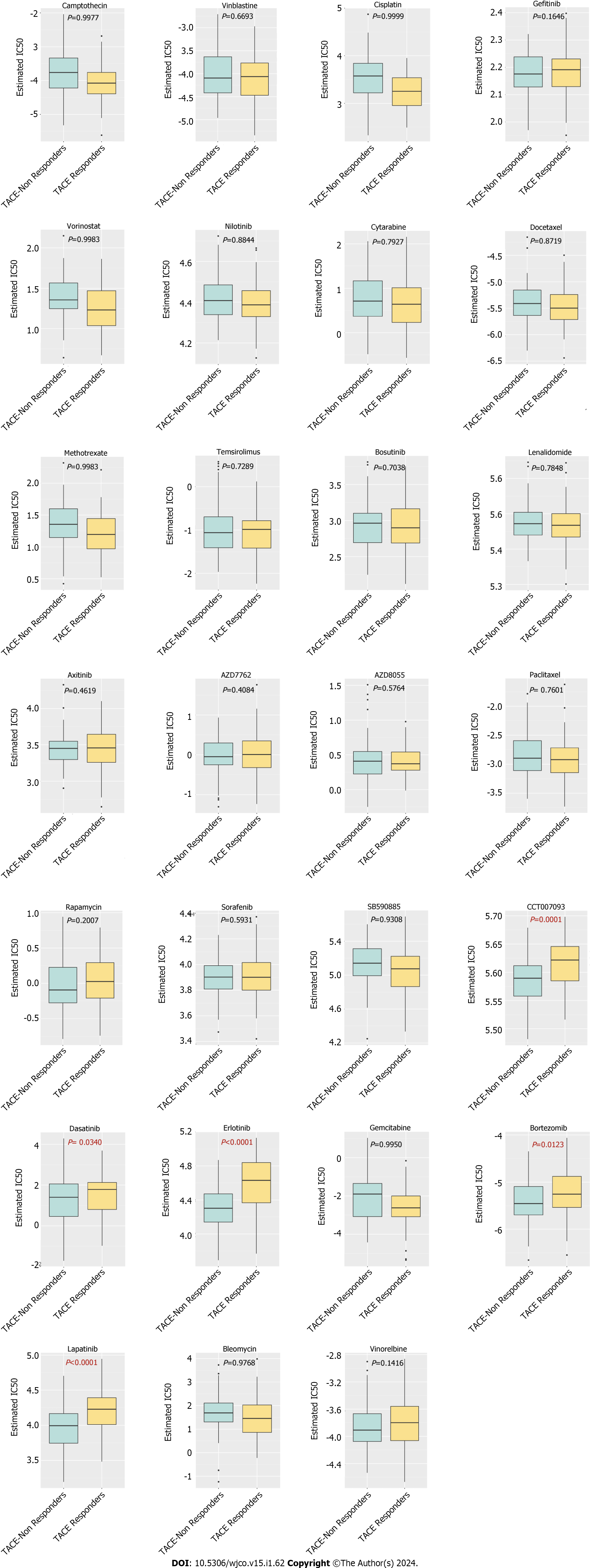
The pRRophetic R package was adopted to predict the half maximal inhibitory concentration (IC50) of chemotherapeutic agents antagonising HCC in the Genomics of Drug Sensitivity in Cancer database. IC50 represented the effectiveness of each kind of chemotherapeutic agent inhibiting specific biological or biochemical functions. The comparison of the IC50s of the TACE non-responders group and the TACE responders group was tested by Wilcoxon signed-rank test.
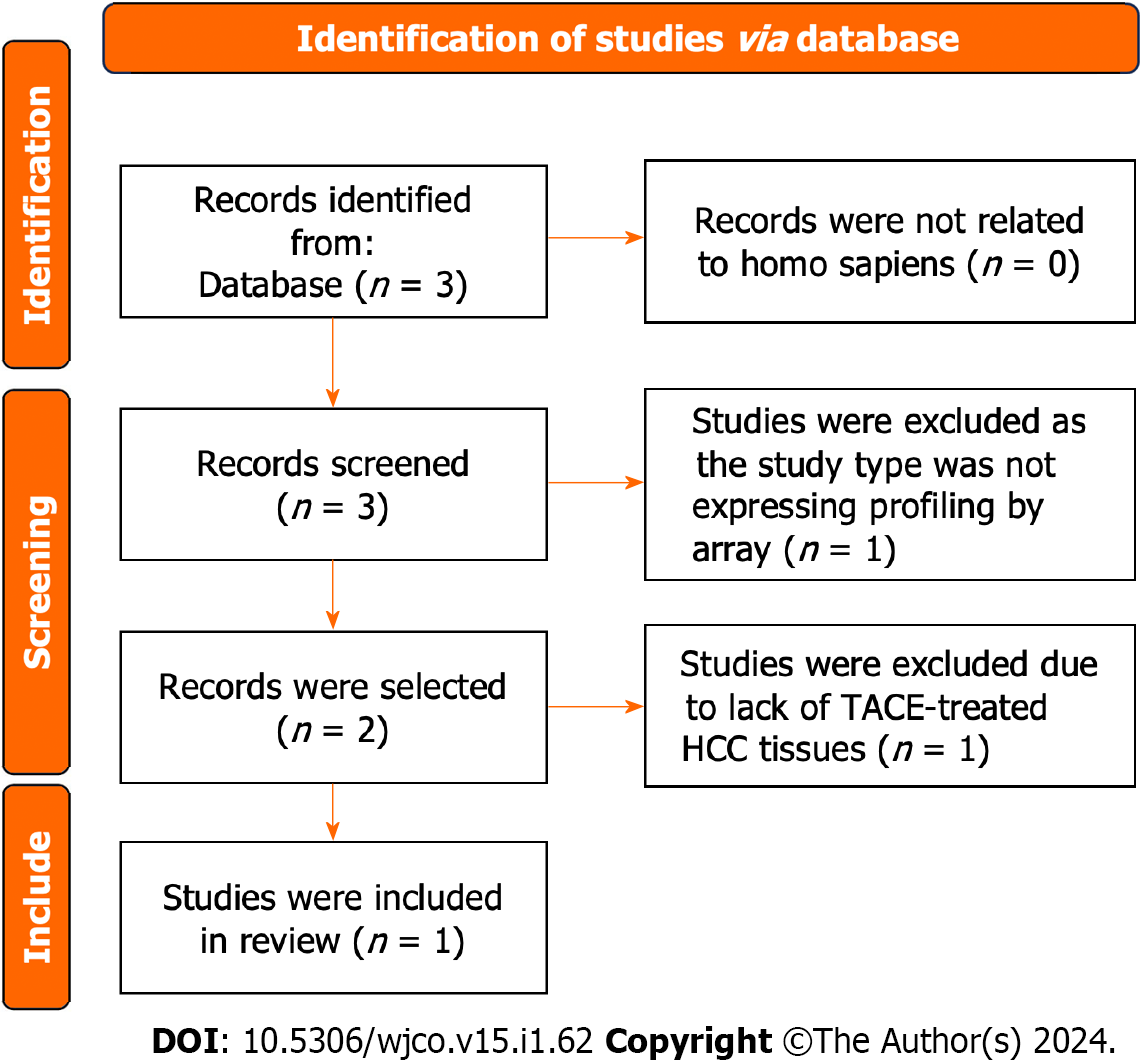
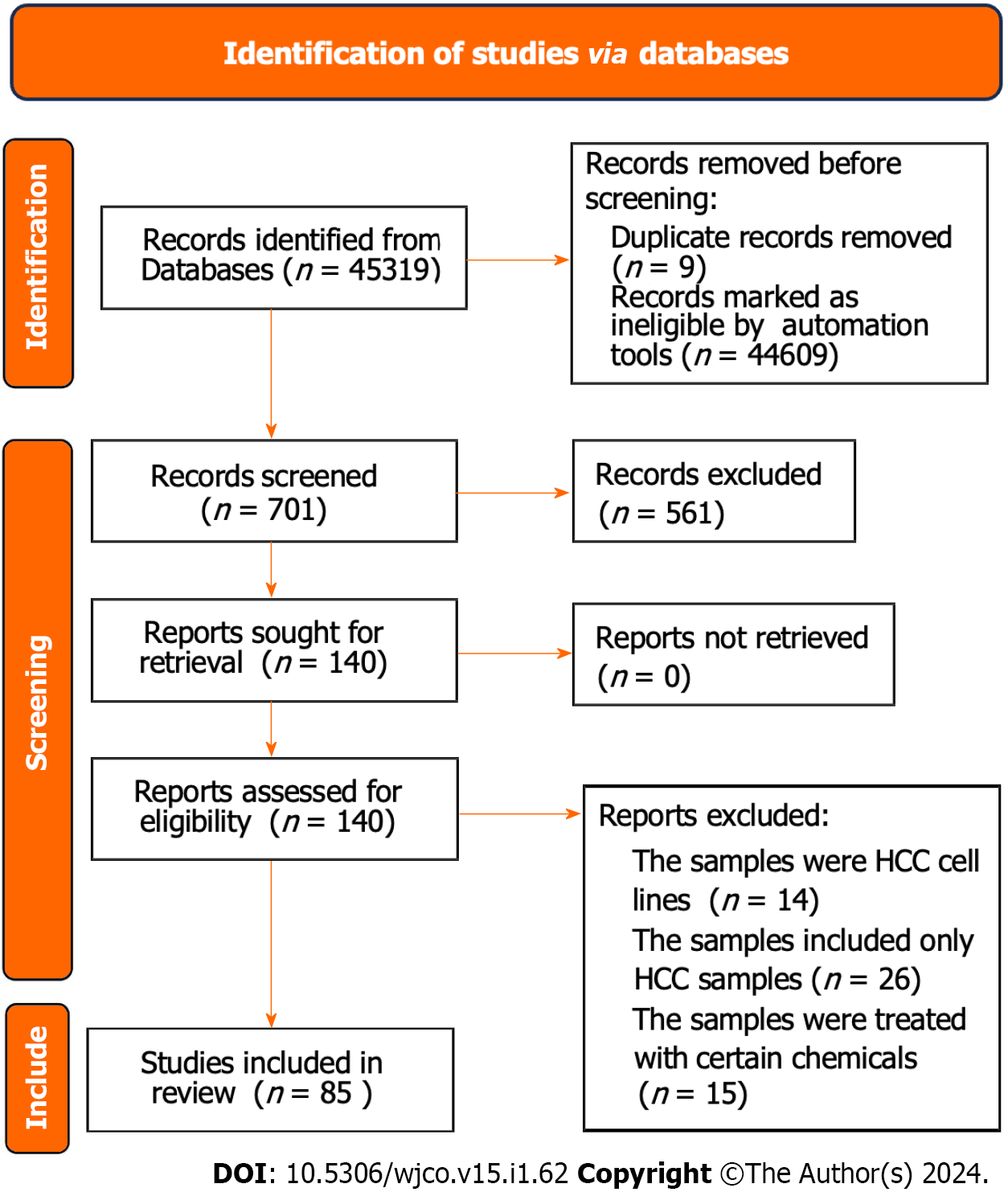
The gene expression matrix GSE104580 including HCC samples of 66 TACE non-responders and 81 TACE- responders was obtained from the GEO database. The screening procedure can be seen in Figure 2. Additionally, a total of 3941 HCC and 3443 non-HCC samples were obtained from the TCGA, GEO, GTEx and ArrayExpress databases. The 85 studies that included HCC and non-HCC tissues were collected according to the procedure in Figure 3. The detailed information of the 84 datasets (datasets from TCGA and GTEx databases have been merged as one dataset) are listed in Supple
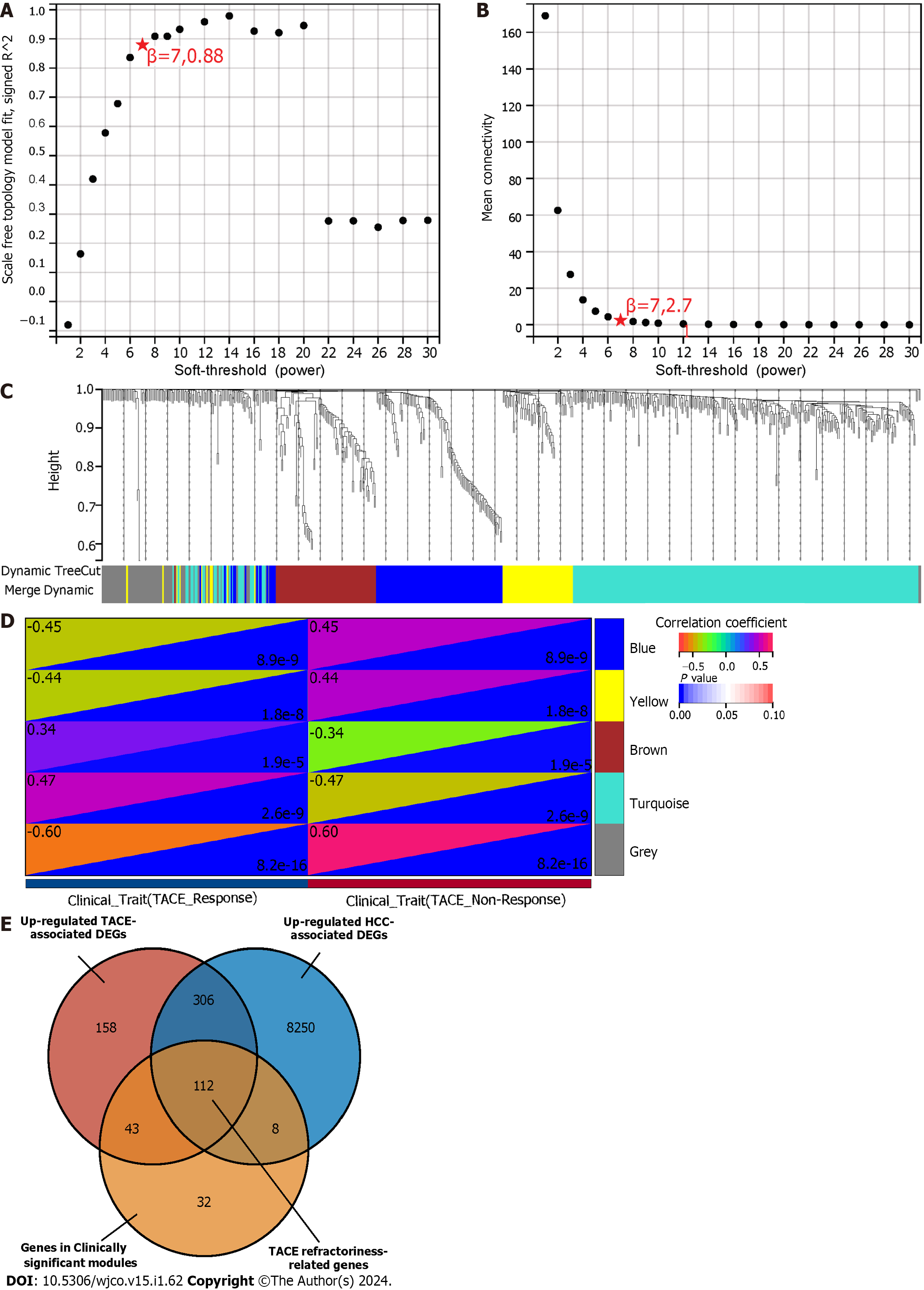
The expression file of 1267 TACE-associated DEGs and the clinical traits in GSE104580 were used for the WGCNA analysis. The soft-threshold β = 7 with the highest scan-free topology model fit (R2 = 0.88) and the mean connectivity lower than 100 was set to construct a scan-free network (Figure 5A and B). Subsequently, the adjacency matrix was built and turned into a TOM to establish a gene-clustering dendrogram (Figure 5C). As shown in Figure 5C, each colour in the merge-dynamic row corresponds to a gene module after they were merged based on the cut line 0.25, and, finally, a total of five modules were identified.
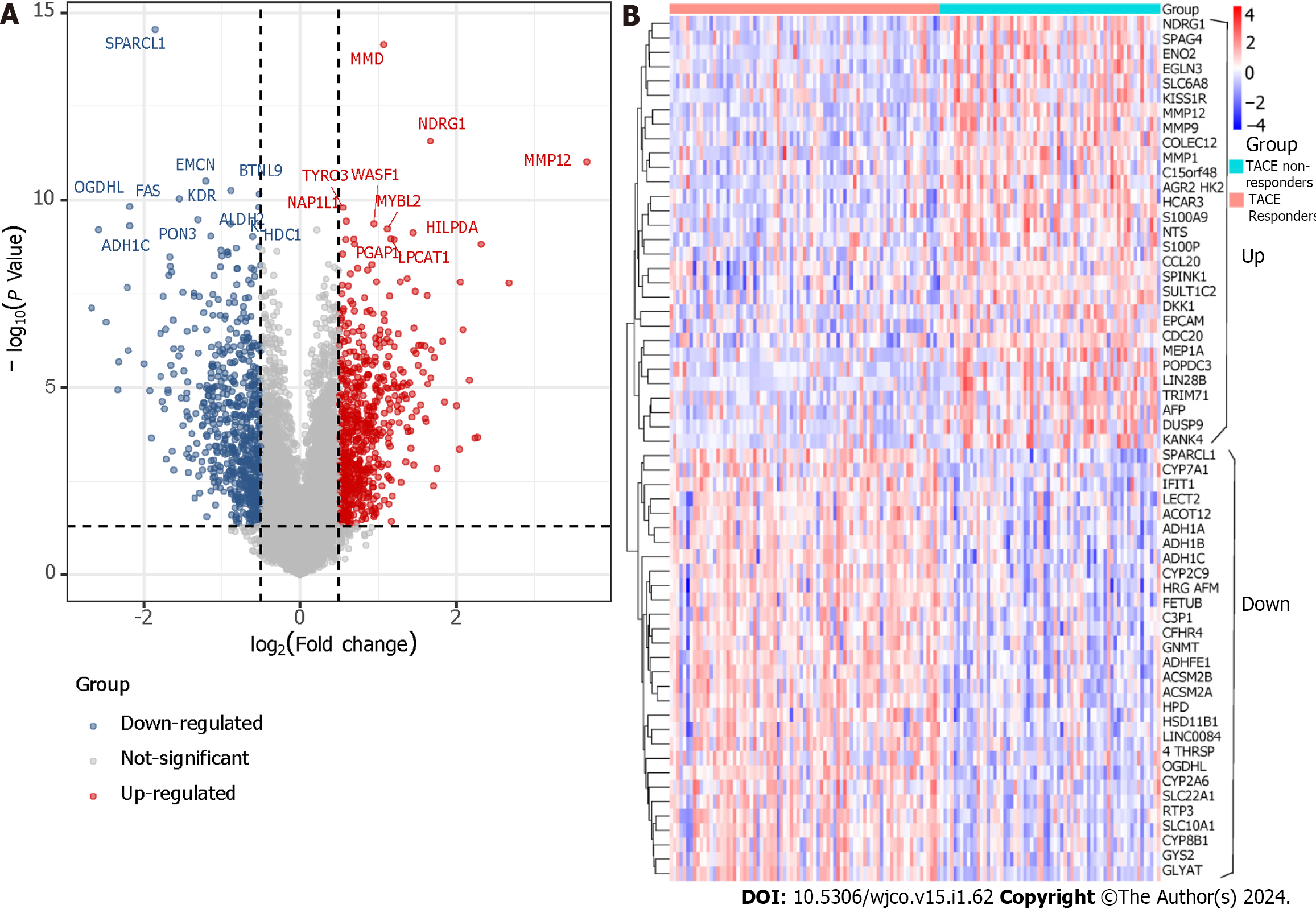
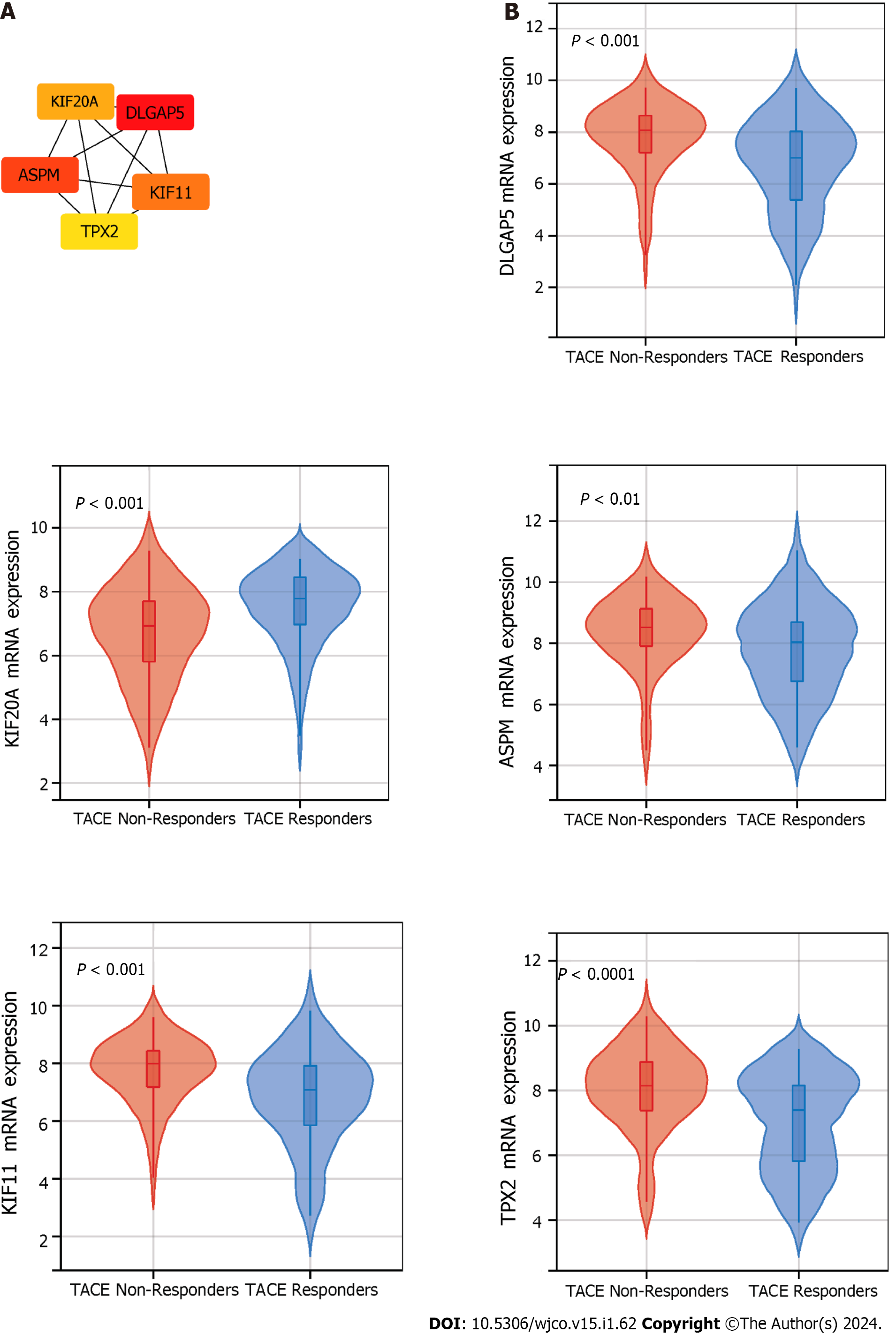
The clinical traits, namely reactivity to TACE in GSE104580, were combined with the five identified modules to screen for the clinically significant module(s) (Figure 5D). Finally, the blue and grey modules were identified as the clinically significant modules due to having the highest correlation coefficient with the clinical trait TACE non-responders. A total of 195 genes were in the blue and grey modules. After intersecting 619 up-regulated TACE-associated DEGs, 8676 up-regulated HCC-associated DEGs and 195 genes in the clinically significant modules, a total of 112 shared genes were identified as TACE refractoriness-related genes for further analysis (Figure 5E). According to the scoring results from the MCC algorithm, the top-five genes had relatively high scores, while the rest had extremely low scores. To ensure reliability and accuracy, we selected the top-five genes as the key genes among the TACE refractoriness-related genes. The five key genes, DLG associated protein 5 (DLGAP5), Kinesin family member 20A (KIF20A), Assembly factor for spindle microtubules (ASPM), Kinesin family member 11 (KIF11) and TPX2 microtubule nucleation factor (TPX2), were identified using the MCC methods in the CytoHubb plugin contained in Cytoscape software (Figure 6A).
To ascertain the key genes’ differential expression in the HCC samples of the TACE non-responders and TACE responders, the mRNA expression of DLGAP5, KIF20A, ASPM, KIF11 and TPX2 was analysed. The results showed that the five key genes were all more highly expressed in the TACE non-responders group compared with TACE responders group (Figure 6B).
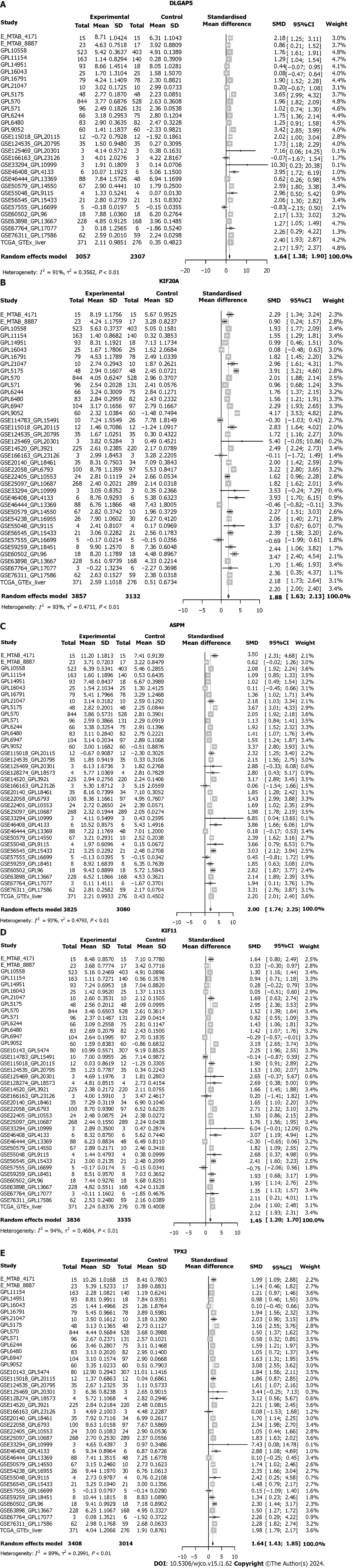
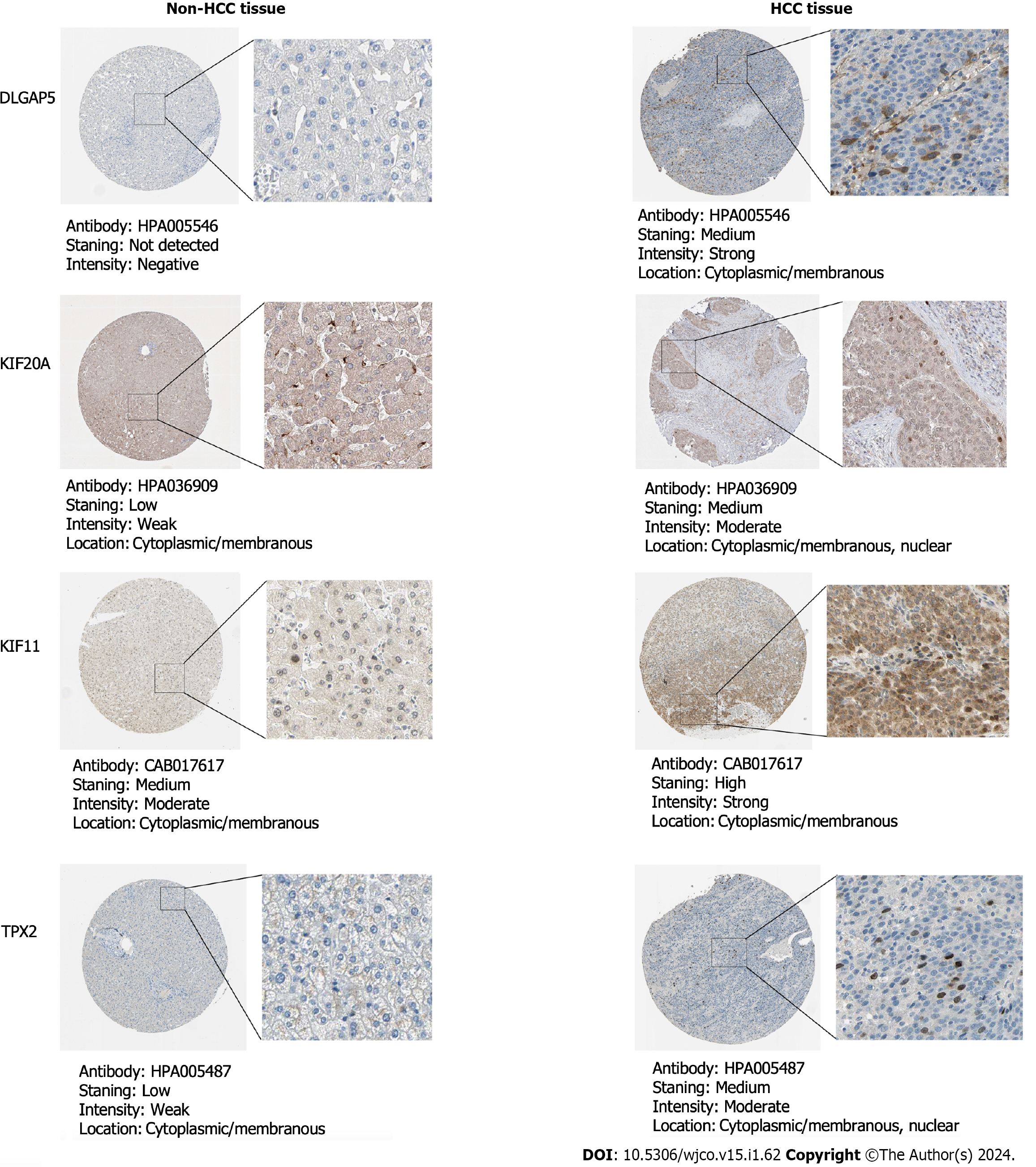
After integrating all enrolled data including HCC and non-HCC tissues, we found that the SMD and 95%CI of the five key genes under random effects were as follows: DLGAP5 (SMD: 1.54, 95%CI: 1.38–1.90), KIF20A (SMD: 1.88, 95%CI: 1.63–2.13), ASPM (SMD: 2.00, 95%CI: 1.74–2.25), KIF11 (SMD: 1.45, 95%CI: 1.20–1.70) and TPX2 (SMD: 1.64, 95%CI: 1.43–1.85) (Figure 7). These indicated that the expression level of the five key genes was remarkably higher in the HCC than in the non-HCC tissues. The protein expression of key genes DLGAP5, KIF20A, KIF11 and TPX2 in the HCC tissues was higher than in the non-HCC tissues (Figure 8). The IHC resource for protein expression of ASPM was lack in THPA database.
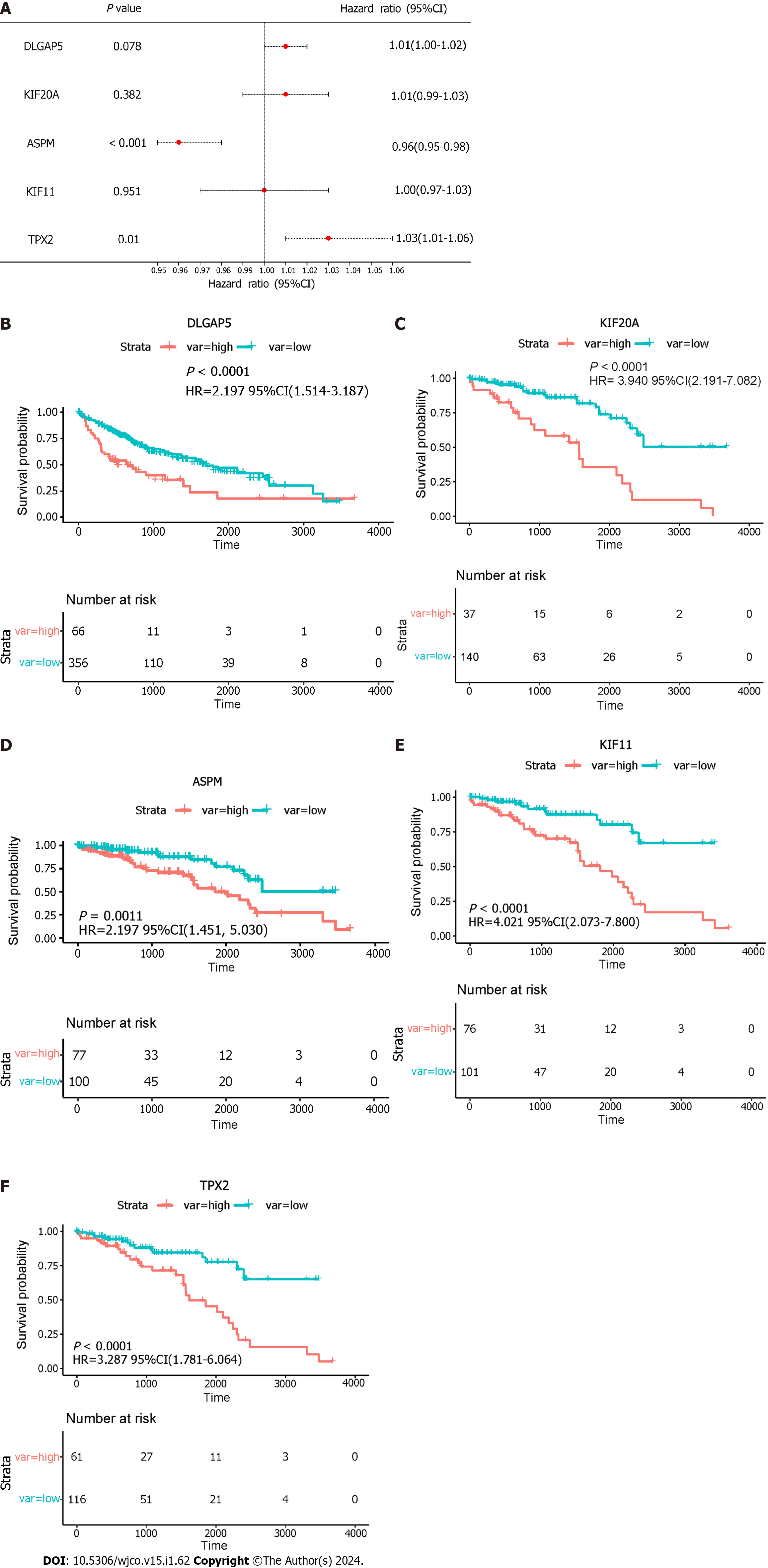
TPX2 was considered an independent prognostic factor in HCC via multivariate analysis (Figure 9A). The results showed that a high expression of the key genes was correlated with a low overall survival. To verify the key genes participating in HCC poor prognosis, the survival curves are shown in Figure 9B–F. In addition, HR was as follows: DLGAP5 (HR: 2.197, P < 0.0001), KIF20A (HR: 3.940, P < 0.0001), ASPM (HR: 2.702, P = 0.0011), KIF11 (HR: 4.021, P < 0.0001) and TPX2 (HR: 3.287, P < 0.0001).
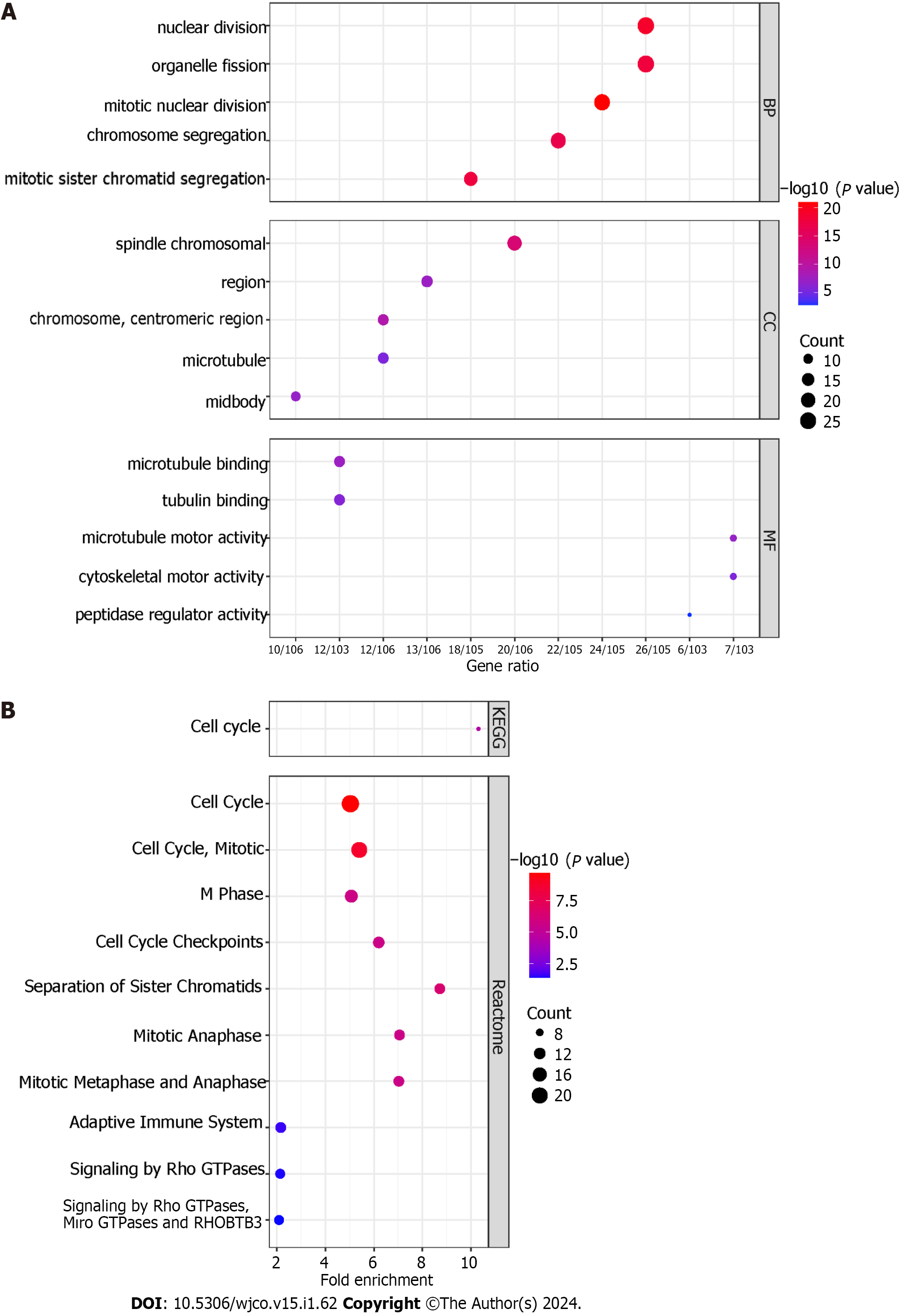
For BP, the 112 TACE refractoriness-related genes were mainly enriched in nuclear division, organelle fission, mitotic nuclear division, chromosome segregation and mitotic sister chromatid segregation. CC involved spindle, chromosomal region, chromosome, centromeric region, microtubule and midbody. As for MF, these TACE refractoriness-related genes were associated with microtubule binding, tubulin binding, microtubule motor activity, cytoskeletal motor activity and peptidase regulator activity (Figure 10A). With regards to the pathways, these TACE refractoriness-related genes were mainly active in cell cycle pathways in the KEGG pathway enrichment analysis, and the relevant reactome pathways are shown in Figure 10B.
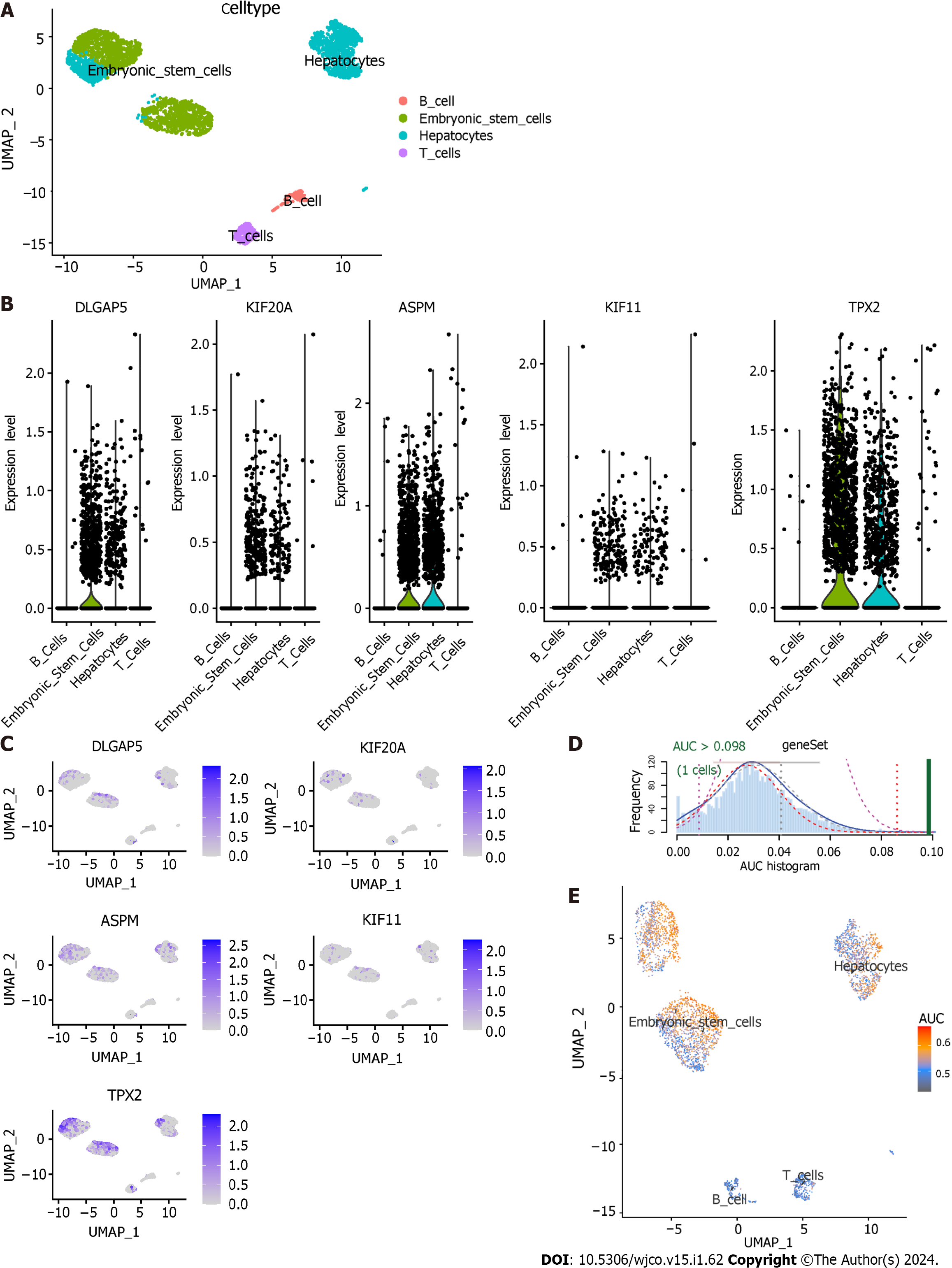
The dataset GSE103867 satisfied our requirements and was obtained for scRNA analysis. After quality control, a total of 3363 cells from HCC samples were identified. Four cell types were identified: hepatocytes, embryonic stem cells, T cells and B cells (Figure 11A). The expression level of five key genes in the four cell types is shown in Figures 11B and C. The AUCell R package was used for determining the TACE refractoriness-related activity in each cell line (Figure 11D). TACE refractoriness-related genes were mainly active in hepatocytes and embryonic stem cells (Figure 11E).
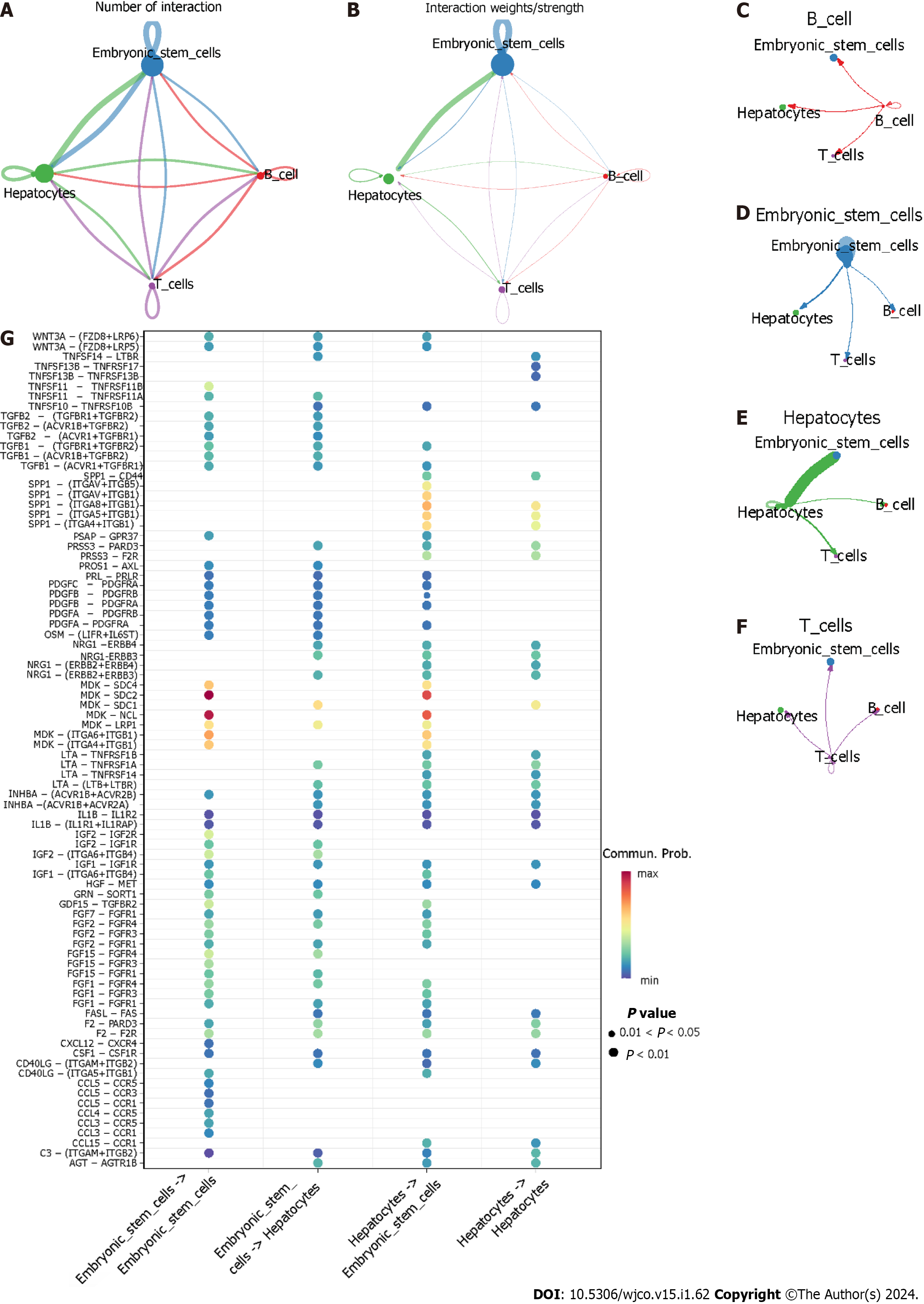
As shown in Figure 12A–F, hepatocytes as providers of ligands and embryonic stem cells as the providers of receptors had the highest interaction strength. When hepatocytes expressed ligands and embryonic stem cells provided the corresponding receptors, some ligand-receptor pairs like MDK-SDC2 and MDK-NCL had the greatest communication probability of accomplishing cell–cell communication (Figure 12G).
TACE non-responders had a higher level of infiltration of follicular helper T cells, macrophages M0 and active mast cells than TACE responders. While TACE non-responders were associated with a lower level of infiltration of active memory CD4+ T cells, macrophages M1 and M2 macrophages (Figure 13A). The analysis of the connection between the expression of key genes and the infiltration of 22 kinds of immune cell in CIBERSORT algorithm was performed (Figure 13B–F). The correlation among hepatocytes, phenotypes of T cells and phenotypes of B cells was computed using the xCell algorithm (Figure 14A). TACE non-responders were calculated as having lower stroma and microenvironment scores (Figure 14B).
There were 27 kinds of drugs used in the treatment of HCC in the pRRophetic R package. The IC50 values of the 27 chemotherapeutics were compared between TACE non-responders and TACE responders. Five chemotherapeutics (CCT007093, Dasatinib, Erlotinib, Bortezomib and Lapatinib) had the potential for reversing TACE non-response (Figure 15).
In this study, we have achieved the following targets: (1) Five key genes (DLGAP5, KIF20A, ASPM, KIF11 and TPX2) were identified among TACE refractoriness-related genes and all overexpressed in the TACE non-responders group and the HCC group; (2) a higher expression of five key genes predicted a worse overall survival (OS) probability in HCC, and TPX2 was considered to be an independent prognostic factor in HCC; (3) cell cycle pathway was likely to play a role in resisting TACE therapy with enrichment analysis using TACE refractoriness-related genes; (4) four cell types, hepatocytes, embryonic stem cells, T cells and B cells, were identified in HCC samples via scRNA sequencing analysis, and hepatocytes and embryonic stem cells were seen as possibly promoting TACE refractoriness via AUCell and CellChat analysis; (5) the immune environment landscapes of the TACE non-response group and the TACE response group were revealed; (6) some compounds, like CCT007093, Dasatinib, Erlotinib, Bortezomib and Lapatinib, might be effective for the TACE non-response group according to drug-sensitivity analysis.
Some advanced imaging techniques have been reported as predicting the therapeutic response of TACE in HCC through the detection of liver hemodynamics, water molecule diffusion capacity, metabolic changes and blood-oxygen level[26]. One study found that the blood-oxygen level significantly decreased in the cancerous region of 30 HCC patients after receiving TACE but saw no notable changes in 30 healthy volunteers[27]. TACE creates a hypoxic microenvironment due to arterial embolisation, which contributes to a surge in the vascular endothelial growth factor, p53, which enlarges tumour size and decreases the survival rate[28,29]. Cheng et al[30] found that TPX2, as one of the hypoxia-related genes, was obviously and clearly expressed in a high-risk HCC group and participated in TACE refractoriness. Our study has identified TPX2 as a key gene for possibly inducing TACE refractoriness.
On the basis of some studies, autophagy may be a significant mechanism mediating the therapeutic efficacy of TACE in HCC. Autophagy protects the cancer cells from abominable conditions such as hypoxia, starvation and cell apoptosis induced by chemotherapy and enhances the cancer cells’ resistance to hypoxic and chemotherapy[27]. Mao et al[29] demonstrated that apoptosis-stimulating p53 protein 2 (ASPP2) was low-expression in recurrent HCC patients after TACE treatment and confirmed that ASPP2, as a co-expression factor of the cancer suppressor p53, was responsible for decreasing the autophagy maker Beclin-1. Gao et al[31] concluded that TACE in combination with the autophagic inhibitor chloroquine shown to be more efficient at curing tumours in rabbit liver than TACE alone.
Further investigation of research showed that MicroRNA (miRNA) also has a substantial effect on the low therapeutic efficacy of TACE in HCC. Wei et al[20] found that down-regulated miR-125b was related to the recurrence of HCC after TACE treatment. They concluded that the low miR-125b expression attenuated the HIF1α translation to activate the HIF1α/pAKT loop and block the autocrine HIF1α/platelet-derived growth factor β (PDGFβ)/pAKT/HIF1α loop of HIF1α translation by targeting PDGFβ. Tumour-initiating cells (T-ICs) or cancer stem cells (CSCs) are a phenotype of cancer cells with a self-renewal and tumorigenesis faculty[32]. T-ICs and CSCs are connected to cancer proliferation and anti-cancer therapy resistance[33]. Clinical cohort analysis indicates that HCC patients with high miR-186 expression benefit from TACE[34]. Yao et al[34] found that a miR-186 knockdown led to the expansion of CSCs while a high expression of miR-186 inhibited the process. Additionally, other miRNA, like miR-26a, miR-107 and miR-106, were differentially expressed between TACE non-responders and TACE responders, and their differential expression was correlated with OS and progression-free survival[35].
Consistent with the description above, the engagement of T-IC and CSCs is another critical mechanism in the resistance of TACE in HCC. Zeng et al[36] stated that the CSC makers like epithelial cell adhesion molecule had a higher expression in TACE-treated specimens than non-TACE specimens, which indicated that HCC with abundant CSCs phenotype had a high risk of recurrence after TACE treatment. Subsequently, Xiang et al[37] explained that a low expression of Src-homology 2 domain-containing phosphatase 2 (Shp2) was favourable for a high therapeutic effect from TACE. As for the mechanism, they found that Shp2 appeared in sorted epithelial cell adhesion molecule-positive or clusters of differentiation 133-positive liver CSCs and in CSC-enriched hepatoma spheroids from patients, and that a high expression of Shp2 promoted the proliferation of liver CSCs by enhancing an accumulation of β-catenin[37]. Some transcription factors of embryonic stem cells had the ability to regulate the CSCs in pancreatic cancer[38]. Zhou et al[39] identified some potential methylation-related genes that become enriched in the embryonic stem cell pathway to characterise HCC, which indicated embryonic stem cells might play a role in HCC progression. The derivatives of embryonic stem cells were able to generate hepatocytes[40]. Hepatocytes were found to create a profound metabolic rewiring when preparing to proliferate in HCC[41]. However, there was little evidence to show the function of hepatocytes and embryonic stem cells in TACE refractoriness.
The metabolic process is considered to play a part in the occurrence of TACE refractoriness in HCC. Aberrant glutamine metabolism (GM) is involved in tumorigenesis and poor prognosis[42,43]. Ying et al[43] employed 41 GM-associated genes to construct a transcriptome-based approach named ‘GM score’ and validated it using two independent HCC cohorts. The results showed that a high GM score had a positive correlation with a slim OS of HCC patients after TACE treatment. Therefore, it is plausible that the metabolic process and immune infiltration level may be connected to TACE refractoriness in HCC.
In this study, five key genes have been identified as being highly correlated with TACE refractoriness in HCC, and its potential mechanisms have been discussed. However, some limitations exist in this study. Firstly, although the five key genes (DLGAP5, KIF20A, ASPM, KIF11 and TPX2) were verified as being associated with TACE refractoriness, the clinical applicability of these genes requires more cases as confirmation due to the limited number of TACE-treated patients in this study and the heterogeneity in the diagnosis. Secondly, due to insufficient samples of HCC cells and stroma treated by TACE, we have no scope for the further validation of the five key genes. Thirdly, the results of the AUCell analysis that TACE refractoriness-related genes were mainly active in hepatocytes and embryonic stem cells require further verification due to only one cell reaching the AUC threshold of TACE refractoriness-related genes list. Lastly, the potential mechanisms identified also need further proof through additional vivo and vitro experiments.
Five key genes (DLGAP5, KIF20A, ASPM, KIF11 and TPX2) were all up-regulated to facilitate TACE refractoriness. Hepatocytes and embryonic stem cells had intimate intercellular communication and were likely to boost TACE refractoriness. CCT007093, Dasatinib, Erlotinib, Bortezomib and Lapatinib possibly played a curable role in TACE non-responders.
Transcatheter arterial embolisation (TACE) is a primary therapeutic strategy for hepatocellular carcinoma (HCC) patients in the intermediate and advanced stages. In China, TACE refractoriness is defined as the intrahepatic target lesion that remains in a disease progression state after receiving standardised and refined TACE treatment for three or more times consecutively.
It is essential to identify biomarkers for predicting TACE refractoriness and to explore the potential mechanisms of TACE refractoriness.
The purpose of our study is to identify the key genes associated with TACE refractoriness and investigate the potential mechanisms of TACE refractoriness.
The gene expression profile was obtained from the public databases. Weighted gene co-expression network analysis and the cytoHubba plugin were utilised to identify the key genes in TACE refractoriness. Multivariate Cox regression and Kaplan–Meier were employed. ScRNA analysis was used for exploring the potential mechanisms of TACE refractoriness.
Five key genes (DLGAP5, KIF20A, ASPM, KIF11, and TPX2) were all up-regulated in TACE non-responders, which predicted poor prognosis. TPX2 is recognised as an independent prognostic factor. TACE refractoriness-related genes were mainly active in hepatocytes and embryonic stem cells. Hepatocytes and embryonic stem cells showed strong cellular interactions in HCC.
Five key genes (DLGAP5, KIF20A, ASPM, KIF11, and TPX2) were identified as being associated with TACE refractoriness. Hepatocytes and embryonic stem cells probably promoted TACE refractoriness.
More vivo and vitro experiments are essential to elaborate and verify the significance of the key genes and the potential mechanisms involved in TACE refractoriness.
The authors thank Guangxi Key Laboratory of Medical Pathology for their technical support.
| 1. | Chen X, Lu Y, Shi X, Han G, Zhang L, Ni C, Zhao J, Gao Y, Wang X. Epidemiological and Clinical Characteristics of Five Rare Pathological Subtypes of Hepatocellular Carcinoma. Front Oncol. 2022;12:864106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 2. | Zhang X, Zou H, Chen Y, Zhang H, Tian R, Meng J, Zhu Y, Guo H, Dai E, Zhu B, Liu Z, Jin Y, Li Y, Feng L, Zhuang H, Pan CQ, Li J, Duan Z. The effects of increased dose of hepatitis B vaccine on mother-to-child transmission and immune response for infants born to mothers with chronic hepatitis B infection: a prospective, multicenter, large-sample cohort study. BMC Med. 2021;19:148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 3. | Gufler S, Seeboeck R, Schatz C, Haybaeck J. The Translational Bridge between Inflammation and Hepatocarcinogenesis. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 4. | Yao J, Liang X, Liu Y, Li S, Zheng M. Trends in Incidence and Prognostic Factors of Two Subtypes of Primary Liver Cancers: A Surveillance, Epidemiology, and End Results-Based Population Study. Cancer Control. 2022;29:10732748211051548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 5. | Yang C, Zhang H, Zhang L, Zhu AX, Bernards R, Qin W, Wang C. Evolving therapeutic landscape of advanced hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2023;20:203-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 492] [Reference Citation Analysis (0)] |
| 6. | Chidambaranathan-Reghupaty S, Fisher PB, Sarkar D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv Cancer Res. 2021;149:1-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 330] [Cited by in RCA: 635] [Article Influence: 105.8] [Reference Citation Analysis (0)] |
| 7. | Kim H, Choi B, Mouli SK, Choi H, Harris KR, Kulik LM, Lewandowski RJ, Kim DH. Preclinical Development and Validation of Translational Temperature Sensitive Iodized Oil Emulsion Mediated Transcatheter Arterial Chemo-Immuno-Embolization for the Treatment of Hepatocellular Carcinoma. Adv Healthc Mater. 2023;12:e2300906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 8. | Yang Y, Lin K, Liu L, Qian Y, Yang Y, Yuan S, Zhu P, Huang J, Liu F, Gu F, Fu S, Jiang B, Liu H, Pan Z, Lau WY, Zhou W. Impact of preoperative TACE on incidences of microvascular invasion and long-term post-hepatectomy survival in hepatocellular carcinoma patients: A propensity score matching analysis. Cancer Med. 2021;10:2100-2111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 9. | Khan AR, Wei X, Xu X. Portal Vein Tumor Thrombosis and Hepatocellular Carcinoma - The Changing Tides. J Hepatocell Carcinoma. 2021;8:1089-1115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 81] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 10. | Hatanaka T, Yata Y, Naganuma A, Kakizaki S. Treatment Strategy for Intermediate-Stage Hepatocellular Carcinoma: Transarterial Chemoembolization, Systemic Therapy, and Conversion Therapy. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 38] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 11. | Wang P, Song X, Cao D, Cui K, Wang J, Utpatel K, Shang R, Wang H, Che L, Evert M, Zhao K, Calvisi DF, Chen X. Oncogene-dependent function of BRG1 in hepatocarcinogenesis. Cell Death Dis. 2020;11:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 12. | Yang S, Pang L, Dai W, Wu S, Ren T, Duan Y, Zheng Y, Bi S, Zhang X, Kong J. Role of Forkhead Box O Proteins in Hepatocellular Carcinoma Biology and Progression (Review). Front Oncol. 2021;11:667730. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 13. | Reig M, Darnell A, Forner A, Rimola J, Ayuso C, Bruix J. Systemic therapy for hepatocellular carcinoma: the issue of treatment stage migration and registration of progression using the BCLC-refined RECIST. Semin Liver Dis. 2014;34:444-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 115] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 14. | Liu B, Zhang Y, Chen H, Li W, Tsochatzis E. The combination of transcatheter arterial chemoembolisation (TACE) and thermal ablation versus TACE alone for hepatocellular carcinoma. Cochrane Database Syst Rev. 2022;1:CD013345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 15. | Kosaka Y, Kimura T, Kawaoka T, Ogawa Y, Amioka K, Naruto K, Yoshikawa Y, Kikukawa C, Suehiro Y, Yamaoka K, Ando Y, Uchikawa S, Morio K, Nakahara T, Murakami E, Takahashi S, Tsuge M, Hiramatsu A, Imamura M, Chosa K, Awai K, Nagata Y, Chayama K, Aikata H. Hepatic Arterial Infusion Chemotherapy Combined with Radiation Therapy for Advanced Hepatocellular Carcinoma with Tumor Thrombosis of the Main Trunk or Bilobar of the Portal Vein. Liver Cancer. 2021;10:151-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 16. | Núñez KG, Sandow T, Fort D, Hibino M, Wright P, Cohen AJ, Thevenot PT. PD-1 expression in hepatocellular carcinoma predicts liver-directed therapy response and bridge-to-transplant survival. Cancer Immunol Immunother. 2022;71:1453-1465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 17. | Zhao P, Zhao J, Deng Y, Zeng G, Jiang Y, Liao L, Zhang S, Tao Q, Liu Z, Tang X, Tu X, Jiang L, Zhang H, Zheng Y. Application of iron/barium ferrite/carbon-coated iron nanocrystal composites in transcatheter arterial chemoembolization of hepatocellular carcinoma. J Colloid Interface Sci. 2021;601:30-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 18. | Fako V, Martin SP, Pomyen Y, Budhu A, Chaisaingmongkol J, Franck S, Lee JM, Ng IO, Cheung TT, Wei X, Liu N, Ji J, Zhao L, Liu Z, Jia HL, Tang ZY, Qin LX, Kloeckner R, Marquardt J, Greten T, Wang XW. Gene signature predictive of hepatocellular carcinoma patient response to transarterial chemoembolization. Int J Biol Sci. 2019;15:2654-2663. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 19. | Martin SP, Fako V, Dang H, Dominguez DA, Khatib S, Ma L, Wang H, Zheng W, Wang XW. PKM2 inhibition may reverse therapeutic resistance to transarterial chemoembolization in hepatocellular carcinoma. J Exp Clin Cancer Res. 2020;39:99. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 80] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 20. | Wei X, Zhao L, Ren R, Ji F, Xue S, Zhang J, Liu Z, Ma Z, Wang XW, Wong L, Liu N, Shi J, Guo X, Roessler S, Zheng X, Ji J. MiR-125b Loss Activated HIF1α/pAKT Loop, Leading to Transarterial Chemoembolization Resistance in Hepatocellular Carcinoma. Hepatology. 2021;73:1381-1398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 60] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 21. | Myojin Y, Kodama T, Maesaka K, Motooka D, Sato Y, Tanaka S, Abe Y, Ohkawa K, Mita E, Hayashi Y, Hikita H, Sakamori R, Tatsumi T, Taguchi A, Eguchi H, Takehara T. ST6GAL1 Is a Novel Serum Biomarker for Lenvatinib-Susceptible FGF19-Driven Hepatocellular Carcinoma. Clin Cancer Res. 2021;27:1150-1161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 22. | Li X, Duan Y, Hao Y. Identification of super enhancer-associated key genes for prognosis of germinal center B-cell type diffuse large B-cell lymphoma by integrated analysis. BMC Med Genomics. 2021;14:69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 23. | Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, Myung P, Plikus MV, Nie Q. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12:1088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3052] [Cited by in RCA: 5144] [Article Influence: 1028.8] [Reference Citation Analysis (0)] |
| 24. | Huang J, Chen T, Wang J, Wang Z, Huang S. Weighted gene co-expression network analysis and CIBERSORT screening of key genes related to m6A methylation in Hirschsprung's disease. Front Genet. 2023;14:1183467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 25. | Teng L, Shen L, Zhao W, Wang C, Feng S, Wang Y, Bi Y, Rong S, Shushakova N, Haller H, Chen J, Jiang H. SLAMF8 Participates in Acute Renal Transplant Rejection via TLR4 Pathway on Pro-Inflammatory Macrophages. Front Immunol. 2022;13:846695. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 26. | Yang K, Zhang XM, Yang L, Xu H, Peng J. Advanced imaging techniques in the therapeutic response of transarterial chemoembolization for hepatocellular carcinoma. World J Gastroenterol. 2016;22:4835-4847. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 22] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 27. | Su H, Yang F, Fu R, Li X, French R, Mose E, Pu X, Trinh B, Kumar A, Liu J, Antonucci L, Todoric J, Liu Y, Hu Y, Diaz-Meco MT, Moscat J, Metallo CM, Lowy AM, Sun B, Karin M. Cancer cells escape autophagy inhibition via NRF2-induced macropinocytosis. Cancer Cell. 2021;39:678-693.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 148] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 28. | Lin ZH, Jiang JR, Ma XK, Chen J, Li HP, Li X, Wu XY, Huang MS, Lin Q. Prognostic value of serum HIF-1α change following transarterial chemoembolization in hepatocellular carcinoma. Clin Exp Med. 2021;21:109-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 29. | Mao J, Tan Z, Pan X, Meng F. ASPP2 expression predicts the prognosis of patients with hepatocellular carcinoma after transcatheter arterial chemoembolization. Exp Ther Med. 2021;21:397. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 30. | Cheng X, Li J, Feng L, Feng S, Wu X, Li Y. The role of hypoxia-related genes in TACE-refractory hepatocellular carcinoma: Exploration of prognosis, immunological characteristics and drug resistance based on onco-multi-OMICS approach. Front Pharmacol. 2022;13:1011033. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 31. | Gao L, Song JR, Zhang JW, Zhao X, Zhao QD, Sun K, Deng WJ, Li R, Lv G, Cheng HY, Wei LX. Chloroquine promotes the anticancer effect of TACE in a rabbit VX2 Liver tumor model. Int J Biol Sci. 2013;9:322-330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 32. | Sadasivam S, Subramanian R. A perspective on challenges and opportunities in characterizing oral cancer stem cells. Front Biosci (Landmark Ed). 2020;25:1011-1021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 33. | Han J, Won M, Kim JH, Jung E, Min K, Jangili P, Kim JS. Cancer stem cell-targeted bio-imaging and chemotherapeutic perspective. Chem Soc Rev. 2020;49:7856-7878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 153] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 34. | Yao H, Yang Z, Lou Y, Huang J, Yang P, Jiang W, Chen S. miR-186 Inhibits Liver Cancer Stem Cells Expansion via Targeting PTPN11. Front Oncol. 2021;11:632976. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 35. | Ali HEA, Emam AA, Zeeneldin AA, Srour R, Tabashy R, El-Desouky ED, Abd Elmageed ZY, Abdel-Wahab AA. Circulating miR-26a, miR-106b, miR-107 and miR-133b stratify hepatocellular carcinoma patients according to their response to transarterial chemoembolization. Clin Biochem. 2019;65:45-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 36. | Zeng Z, Ren J, O'Neil M, Zhao J, Bridges B, Cox J, Abdulkarim B, Schmitt TM, Kumer SC, Weinman SA. Impact of stem cell marker expression on recurrence of TACE-treated hepatocellular carcinoma post liver transplantation. BMC Cancer. 2012;12:584. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 37. | Xiang D, Cheng Z, Liu H, Wang X, Han T, Sun W, Li X, Yang W, Chen C, Xia M, Liu N, Yin S, Jin G, Lee T, Dong L, Hu H, Wang H, Ding J. Shp2 promotes liver cancer stem cell expansion by augmenting β-catenin signaling and predicts chemotherapeutic response of patients. Hepatology. 2017;65:1566-1580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 126] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 38. | Herreros-Villanueva M, Bujanda L, Billadeau DD, Zhang JS. Embryonic stem cell factors and pancreatic cancer. World J Gastroenterol. 2014;20:2247-2254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 65] [Cited by in RCA: 71] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 39. | Zhou S, Li M, Ostrow D, Ruble D, Mascarenhas L, Pawel B, Buckley JD, Triche TJ. Potential methylation-regulated genes and pathways in hepatocellular neoplasm, not otherwise specified. Front Oncol. 2022;12:952325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 40. | Mennen RH, Oldenburger MM, Piersma AH. Endoderm and mesoderm derivatives in embryonic stem cell differentiation and their use in developmental toxicity testing. Reprod Toxicol. 2022;107:44-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 41. | Hall Z, Chiarugi D, Charidemou E, Leslie J, Scott E, Pellegrinet L, Allison M, Mocciaro G, Anstee QM, Evan GI, Hoare M, Vidal-Puig A, Oakley F, Vacca M, Griffin JL. Lipid Remodeling in Hepatocyte Proliferation and Hepatocellular Carcinoma. Hepatology. 2021;73:1028-1044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 116] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 42. | Li B, Cao Y, Meng G, Qian L, Xu T, Yan C, Luo O, Wang S, Wei J, Ding Y, Yu D. Targeting glutaminase 1 attenuates stemness properties in hepatocellular carcinoma by increasing reactive oxygen species and suppressing Wnt/beta-catenin pathway. EBioMedicine. 2019;39:239-254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 175] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 43. | Ying L, Cheng M, Lu Y, Tao Q, Chen X, Shen B, Xiong F, Hu Z, Wang D, Li X. Glutamine Metabolism Scoring Predicts Prognosis and Therapeutic Resistance in Hepatocellular Carcinoma. Pathol Oncol Res. 2021;27:1610075. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Biochemistry and molecular biology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Lykoudis PM, United Kingdom S-Editor: Liu JH L-Editor: A P-Editor: Zhang XD