Published online Jan 24, 2024. doi: 10.5306/wjco.v15.i1.32
Peer-review started: September 8, 2023
First decision: October 17, 2023
Revised: November 20, 2023
Accepted: December 19, 2023
Article in press: December 19, 2023
Published online: January 24, 2024
Processing time: 136 Days and 21.9 Hours
Glioma is one of the most common intracranial tumors, characterized by invasive growth and poor prognosis. Actin cytoskeletal rearrangement is an essential event of tumor cell migration. The actin dynamics-related protein scinderin (SCIN) has been reported to be closely related to tumor cell migration and invasion in several cancers.
To investigate the role and mechanism of SCIN in glioma.
The expression and clinical significance of SCIN in glioma were analyzed based on public databases. SCIN expression was examined using real-time quantitative polymerase chain reaction and Western blotting. Gene silencing was performed using short hairpin RNA transfection. Cell viability, migration, and invasion were assessed using cell counting kit 8 assay, wound healing, and Matrigel invasion assays, respectively. F-actin cytoskeleton organization was assessed using F-actin staining.
SCIN expression was significantly elevated in glioma, and high levels of SCIN were associated with advanced tumor grade and wild-type isocitrate dehydrogenase. Furthermore, SCIN-deficient cells exhibited decreased proliferation, migration, and invasion in U87 and U251 cells. Moreover, knockdown of SCIN inhibited the RhoA/focal adhesion kinase (FAK) signaling to promote F-actin depolymerization in U87 and U251 cells.
SCIN modulates the actin cytoskeleton via activating RhoA/FAK signaling, thereby promoting the migration and invasion of glioma cells. This study identified the cancer-promoting effect of SCIN and provided a potential therapeutic target for the treatment of glioma.
Core Tip: Actin dynamics-related protein scinderin (SCIN) was found to be significantly upregulated in glioma, and high SCIN expression was associated with advanced tumor grade and wild-type isocitrate dehydrogenase. Furthermore, silenced-SCIN cells exhibited decreased proliferation, migration, and invasion. Besides, knockdown of SCIN inhibited RhoA/focal adhesion kinase signaling to promote F-actin depolymerization in glioma cells.
- Citation: Lin X, Zhao Z, Sun SP, Liu W. Scinderin promotes glioma cell migration and invasion via remodeling actin cytoskeleton. World J Clin Oncol 2024; 15(1): 32-44
- URL: https://www.wjgnet.com/2218-4333/full/v15/i1/32.htm
- DOI: https://dx.doi.org/10.5306/wjco.v15.i1.32
Glioma is the most frequent and deadly tumor of the central nervous system, accounting for about 40%-60% of human intracranial tumors[1]. Based on the histologic types and malignancy grades, gliomas are classified into low-grade gliomas (LGGs) [World Health Organization (WHO) grade I-II] and high-grade gliomas (WHO III-IV grade)[2]. Low-grade gliomas are well-differentiated, while high-grade gliomas are poorly differentiated. The incidence rate of glioma in China ranged from 3 to 6 per 100000 people, most of which are grade III (anaplastic glioma) and IV gliomas [glioblastoma multiforme (GBM)]. Despite remarkable advances in microsurgery, radiotherapy, chemotherapy, and biotherapy, the prognosis of glioma patients remains unsatisfactory. Thus, exploring the molecular mechanism of glioma progression is of great significance for developing new prognostic indicators and therapeutic targets.
Actin dynamics-related protein scinderin (SCIN) (also known as adseverin) belongs to the gelsolin superfamily and is an important actin-severing protein[3]. SCIN regulates the reorganization of F-actin and participates in cell polarity, cell secretion, cell differentiation, and cell motility[4-6]. SCIN has been demonstrated to play diverse roles in chronic inflammation, coagulation processes, immune diseases, and tumors. The role of SCIN in tumors was first reported by Zunino in 2001[7]. They found that SCIN induced cell differentiation, maturation, and apoptosis by releasing platelet-like granules, and inhibited tumor cell formation and proliferation in megakaryoblastic leukemia. Since then, a growing body of studies have uncovered the biological roles of SCIN in various kinds of cancers[8-10]. For instance, in the cytotoxic lymphocyte-resistant mutants of non-small cell lung cancer, upregulated SCIN reduces the killing effect of cytotoxic T lymphocytes on tumor cells[11]. In a study of bladder cancer, SCIN was found to bind to mitochondrial voltage-dependent anion channel (VDAC) oligomers, thereby preventing VADC-induced mitochondrial apoptosis pathways. Besides, SCIN promotes the proliferation and metastasis of bladder, stomach, lung, and prostate cancers[12,13]. However, the biological role and molecular mechanism of SCIN in glioma remain unclear.
In this study, we analyzed the expression and clinical significance of SCIN in glioma based on public databases. Then, we utilized SCIN-specific short hairpin RNAs (shRNAs) to knock down SCIN expression in glioma cell lines and observed the effects of SCIN silencing on the proliferative, migrative, and invasive abilities of glioma cells. Furthermore, the effect of SCIN silencing on the cytoskeleton of glioma cells was also investigated. These experimental results will help us to further explore the mechanism of SCIN in glioma and lay a solid experimental foundation for glioma treatment.
Based on the Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/), we analyzed the difference in SCIN mRNA expression between 163 GBM samples or 518 LGG samples and 207 normal brain tissue samples and analyzed the relationship between SCIN expression and overall survival of glioma patients[14]. Based on the Chinese Glioma Genome Atlas (CGGA) database (http://www.cgga.org.cn/), we further analyzed the rela
U87 MG and U251 cell lines were provided by Procell (Wuhan, China). U87 MG cells were cultured in Minimum Essential Medium. U251 cells were cultured in Dulbecco's modified Eagle medium. All culture medium was supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Cells were maintained in an incubator containing 5% CO2 at 37 °C.
shRNAs targeting SCIN (sh-SCIN) were obtained from Shanghai Gene Pharmaceutical Co., LTD. (Shanghai, China). RNA double-stranded random sequences were used as the negative control for shRNA (sh-NC). Lipofectamine® 2000 (ThermoFisher Scientific, Waltham, MA, United States) was used to transfect these plasmids into U87 MG and U251 cells at a concentration of 50 nM. The transfected cells were screened with puromycin. The clones were then selected and cultured for further experiments.
Cells were lysed by RIPA buffer (Solarbio, Beijing, China). The protein concentration was determined by the BCA Protein Assay Kit (Solarbio). Protein samples (20 μg) were loaded into 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and electrophoresis was performed at 100 V for 1-2 h. Then, the protein was transferred from the gel to the polyvinylidene fluoride membrane (Whatman, Maidstone, United Kingdom). After sealing the membrane with sealing buffer for 1 h, the membranes were incubated with primary antibody at 4 °C overnight. The membranes were then incubated with HRP-IgG antibody (1: 5000; Proteintech, Wuhan, China). Image J software was used to analyze the data. Anti-SCIN, anti-RhoA, and anti-GAPDH antibodies were obtained from Proteintech. Focal adhesion kinase (FAK), phospho-FAK, phospho-Cofilin (Ser3), Cofilin, and Talin antibodies were obtained from Cell Signaling Technology (Boston, MA, United States).
TRIzol reagent (ThermoFisher) was used to isolate total RNAs from cells. The purity of total RNAs was detected by agarose gel electrophoresis. The cDNA was synthesized using SuperScript® III Reverse Transcriptase (ThermoFisher). Polymerase chain reaction (PCR) amplification kit (ThermoFisher) was used for PCR amplification. SCIN primers were provided by Sangon Biotech (Shanghai, China), with sequences (forward primer, 5’-ACTGAGTGGCAGTTGCATTAT-3’, reverse primer, 5’-TGTGGGATGAATTGTTGGACCC-3’).
Cell proliferation was measured using Cell Counting Kit-8 Cell Proliferation and Cytotoxicity Assay Kit (Solarbio).
Cells were seeded into Petri dishes at 1 × 103 cells per well. After incubation at 37 °C and 5% CO2, the cells were saturated with humidity for 14 d. Cells were rinsed twice with phosphate buffered saline buffer. Cells were incubated with paraformaldehyde for 30 min, and stained with the crystal violet solution for 30 min. Finally, the clones were counted under a microscope.
The wound-healing experiment was carried out for migration ability. Glioma cells were plated on 6-well plates. Pipettes (200 μL) were used to draw a straight wound. After incubation in a serum-free medium for 24 h or 48 h, cell images were taken under a microscope.
Transwell chambers were precoated with Matrigel (Corning Costar, Cambridge, MA, United States). Transfected (1 × 103) cells in 100 μL serum-free medium were added to the upper Transwell chamber. The lower chamber was added with medium of 10% fetal bovine serum. After incubation for 6 h, the invading cells that adhered to the lower surface of the membrane were fixed and stained. Finally, the number of invading cells was counted under an inverted microscope (Olympus, Tokyo, Japan).
F-actin staining was performed using an F-actin Staining Kit (Phalloidin-Green; AmyJet Scientific, Wuhan, China). Cells were first inoculated on 96-well cover slides and grown to 70% confluence. Next, the cells were fixed with precooled 4% paraformaldehyde on ice for 20 min and permeabilized with 0.1% Triton X-100 for 10 min at room temperature. Then, after cells were incubated with 100 μL staining solution for 30 min, a 4’,6-diamidino-2-phenylindole solution (100 mL) was applied to stain the nuclei. The immediate observation was carried out under a fluorescence microscope (200 cells counted per field): excitation wavelength 488 nm, emission wavelength 530 nm (F-actin staining) or 350 nm excitation wavelength, 460 nm emission wavelength (nuclear staining).
SPSS 22.0 (IBM Corp., Armonk, NY, United States) was applied for statistical analysis. Data were expressed as mean ± SD. Two-tailed Student t-test and one-way analysis of variance (ANOVA) were used to analyze the statistical difference. Each experiment was replicated at least three times independently. P < 0.05 was considered to be statistically significant.
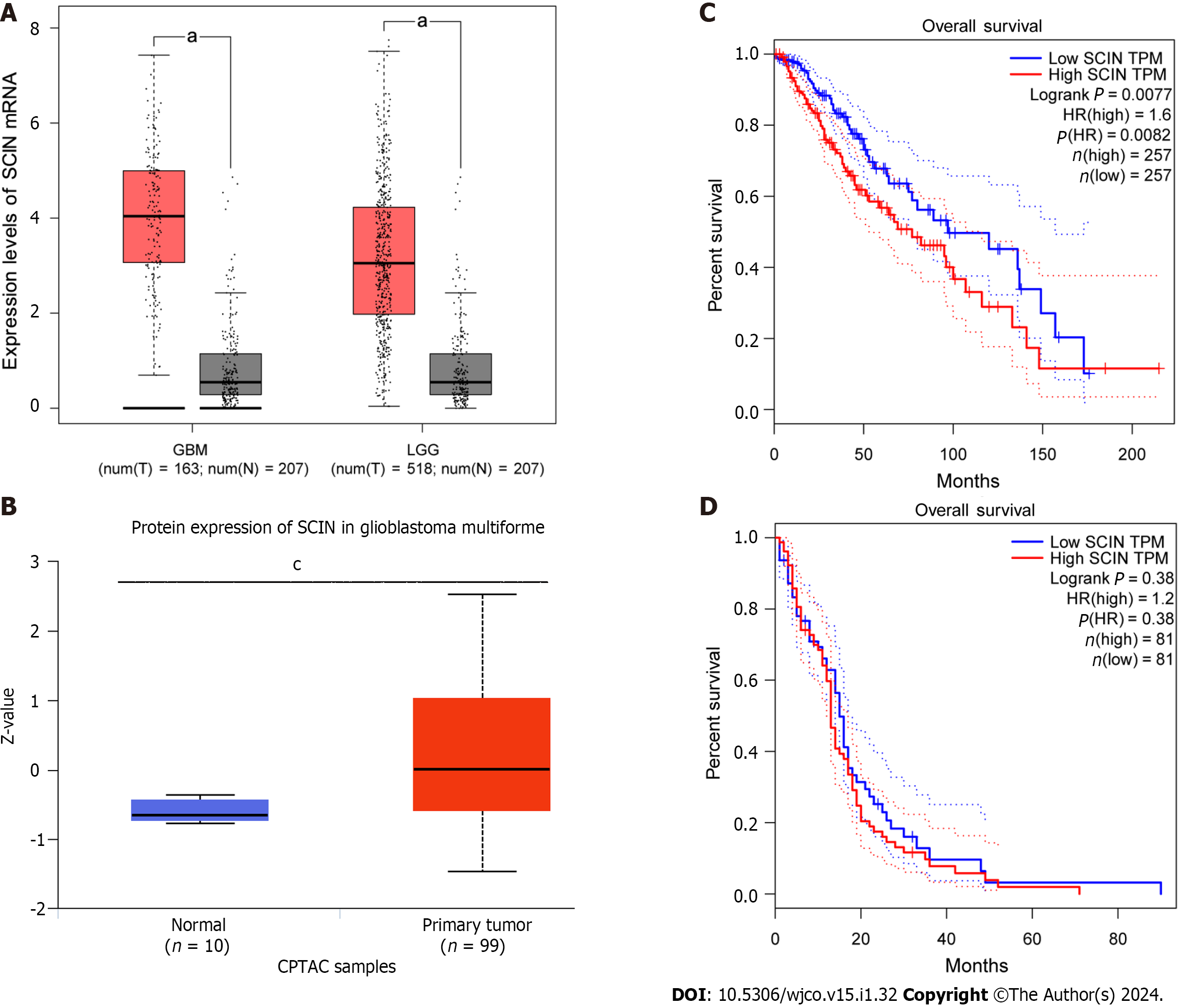
Based on the GEPIA database, we found that the SCIN abundance was remarkably higher in both LGG and GBM than that of normal tissues (Figure 1A). Consistently, the Clinical Proteomic Tumor Analysis Consortium data showed higher SCIN protein levels in primary GBM tissues than in normal tissues (Figure 1B). Furthermore, analysis results of the overall survival curve showed that high levels of SCIN predicted poor prognosis of LGG patients (Figure 1C). However, SCIN expression was not associated with the prognosis of patients with GBM (Figure 1D).
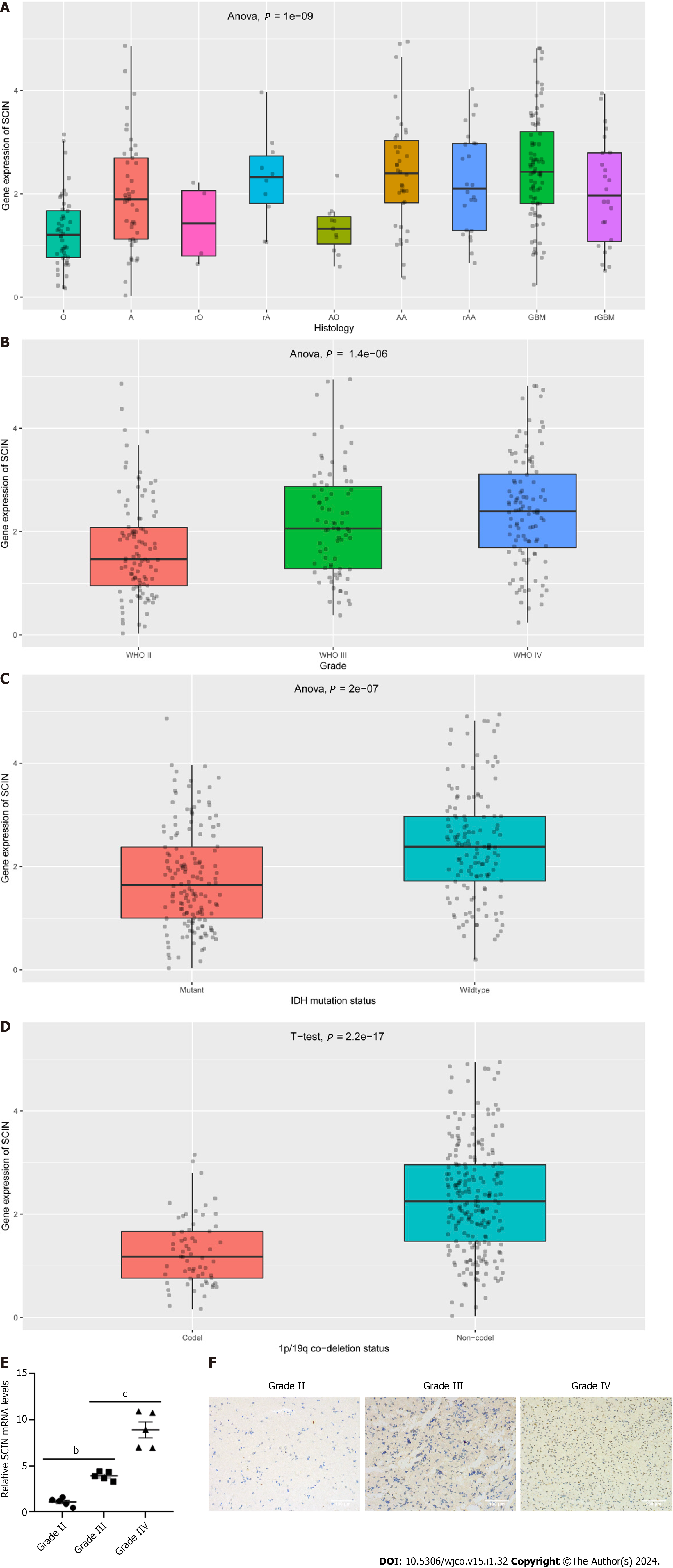
Then, we analyzed SCIN expression in gliomas with different characteristics based on the CGGA dataset. As displayed in Figure 2A, SCIN expression was higher in various types of gliomas than in normal tissues. Moreover, the expression of SCIN increased with advanced tumor grades (Figure 2B). Furthermore, a significant decrease in SCIN expression was observed in gliomas with IDH mutation and 1p/19q co-deletion (Figures 2C and D). Furthermore, we examined the expression of SCIN in glioma by qRT-PCR and Immunohistochemical staining. As shown in Figure 2E, the expression levels of SCIN mRNA were positively correlated with the tumor grades of glioma, and the highest expression was found in grade IV glioma. In line with these results, Immunohistochemical staining showed strong staining of SCIN in grade IV glioma tissues, whereas low and moderate staining was observed in grade I-III gliomas (Figure 2F).
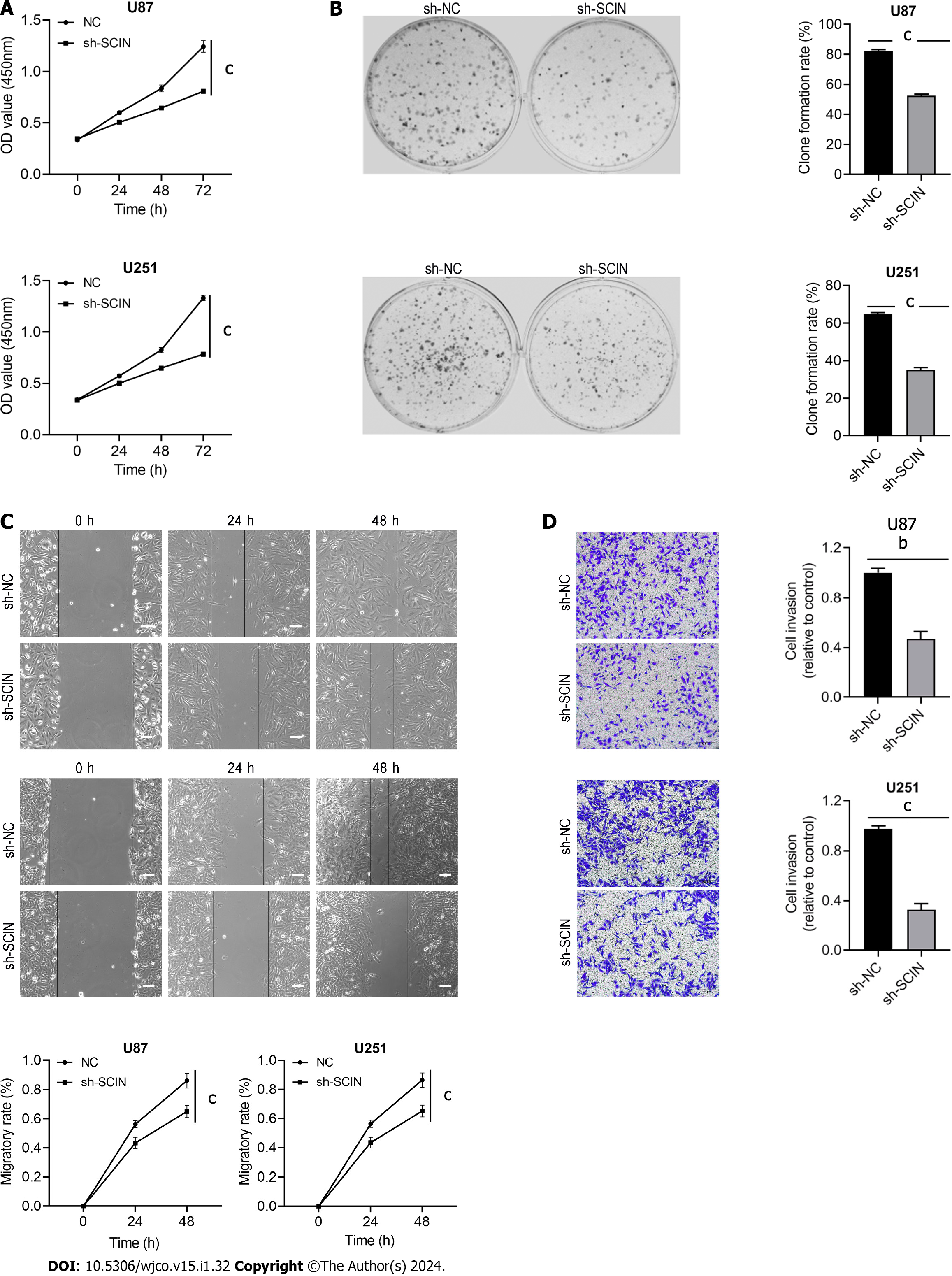
We constructed three shRNAs targeting SCIN and found that sh-SCIN#3 showed the strongest inhibitory effect on SCIN expression in both U87 and U251 cells (Supplementary Figure 1A and B). Furthermore, cell viability was most significantly inhibited by sh-SCIN#3, compared to the other two shRNAs (Supplementary Figure 1C). Thus, sh-SCIN#3 was used in the further experiments. CCK8 assay indicated that cell proliferation was decreased after silencing endogenous SCIN (Figure 3A), which was also confirmed by colony formation assay in U87 and U251 cells (Figure 3B). Moreover, the wound-healing assay showed that the SCIN-deficient cells migrated into the scratching area at a significantly slower rate than those in the sh-NC groups (Figure 3C). Besides, the knockdown of SCIN could inhibit cell invasive ability (Figure 3D). Therefore, these results suggested that SCIN had a promoting effect on migration and invasion in glioma cells.
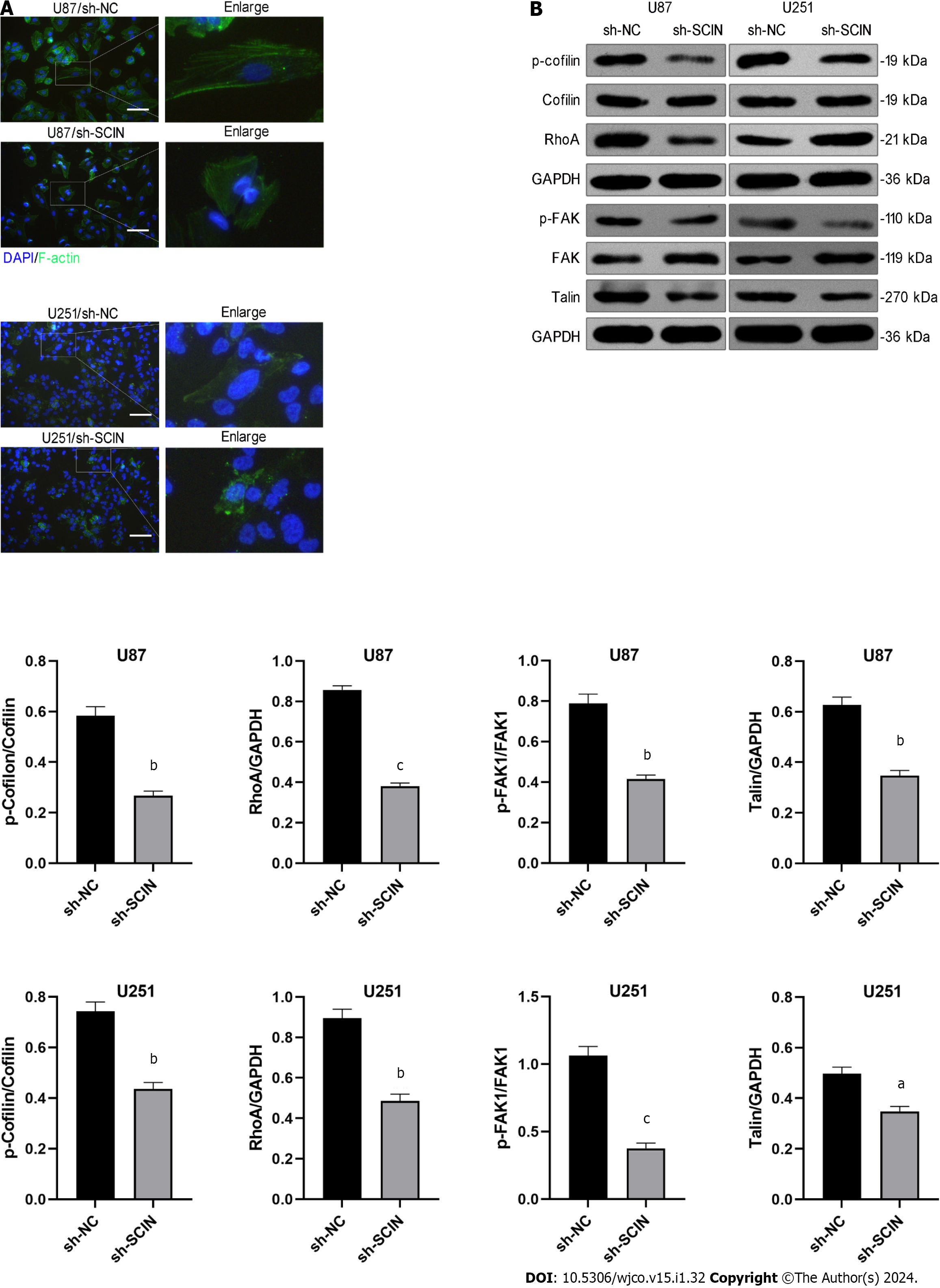
SCIN is an actin severing and capping protein and controls actin organization. Therefore, we investigated the effect of SCIN on F-actin polymerization in glioma cells. Immunofluorescence with F-actin staining indicated the actin stress fibers were clustered and arranged in the negative control cells, while the SCIN-deficient cells showed significant morphological changes, showing sparse disorder of actin stress fibers and less dendrite-like structures (Figure 4A), suggesting that the mobility activity of the cells was weakened. Consistently, western blot results demonstrated that knockdown of SCIN suppressed RhoA, Talin, and phosphorylated cofilin and FAK levels in U87 and U251 cells (Figure 4B), indicating that silenced SCIN inhibited the activation of RhoA/FAK signaling pathway. Notably, the RhoA/FAK pathway is a well-known F-actin polymerization-related signaling pathway, and its inactivation indicated the weakness of F-actin polymerization[16]. This phenomenon indicates that SCIN regulates F-actin polymerization via RhoA/FAK signaling.
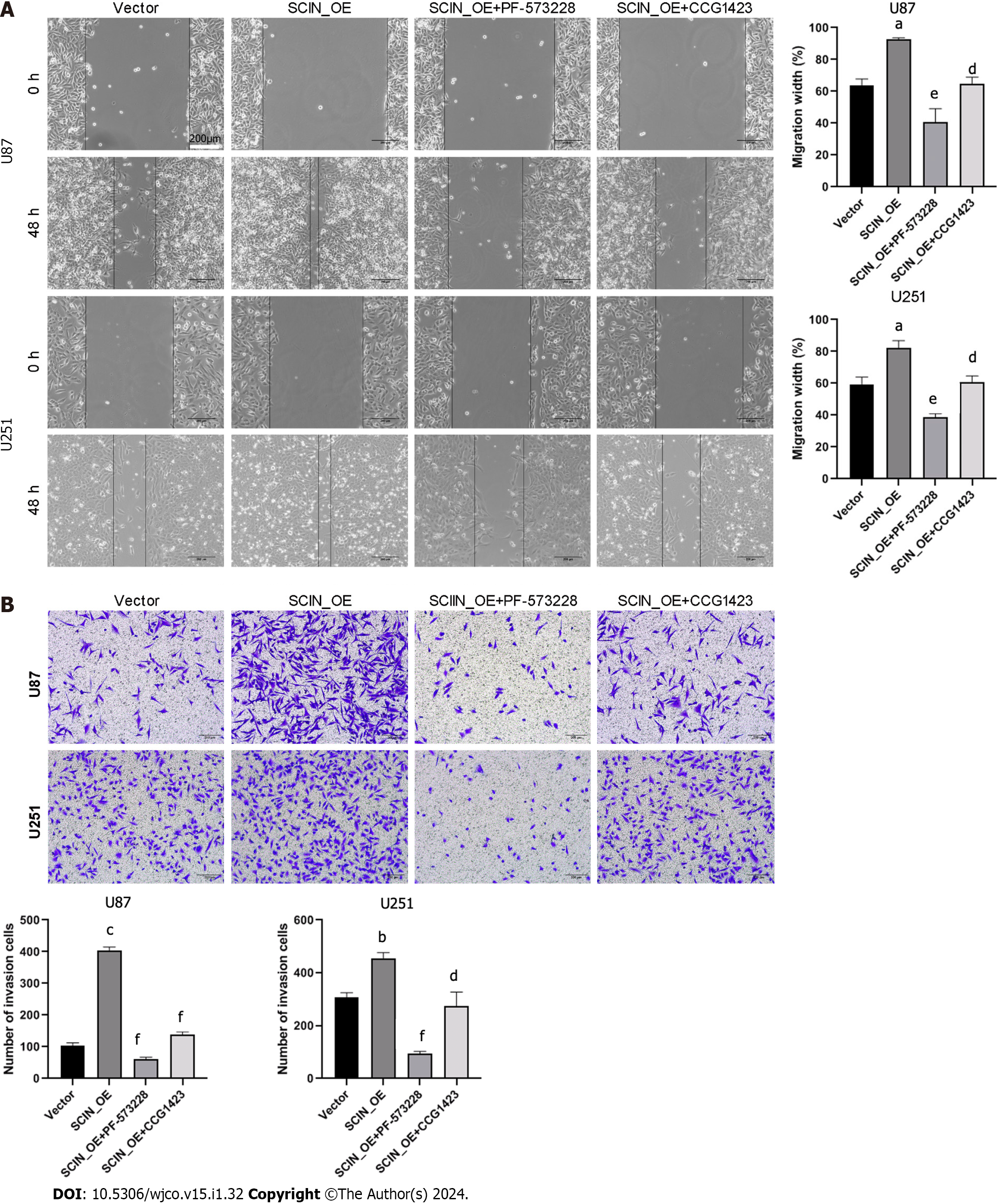
To shed light on the role of RhoA/FAK signaling in SCIN-mediated glioma cell migration and invasion, a selective FAK inhibitor, PF-573228, and a RhoA inhibitor, CCG1423, were used in SCIN-overexpressed U87 and U251 cells. The wound healing assays demonstrated that SCIN overexpression promoted the motility of the cells, which was inhibited after treatment with PF-573228 or CCG1423 (Figure 5A). The Transwell assays revealed that either PF-573228 or CCG1423 treatment reversed the excessive cell invasion induced by SCIN overexpression (Figure 5B). These data indicated that SCIN promotes malignant behaviors in glioma cells via RhoA/FAK signaling pathway.
Glioma is one of the most common brain tumors with rapid progression and dismal prognosis. The occurrence and development of glioma is a complex process involving multiple factors, levels, and genes. In current study, we found that SCIN expression was upregulated in glioma tissues and that high levels of SCIN were associated with high tumor grade and poor prognosis. The depletion of SCIN inhibited the proliferation, invasion, and migration of glioma cells. Mechanistically, SCIN affected cytoskeleton remodeling and inhibited the formation of lamellipodia via RhoA/FAK signaling pathway. This study identifies the cancer-promoting role of SCIN and provides a potential therapeutic target of SCIN for glioma treatment.
Numerous studies revealed aberrant expression of SCIN in several human cancers[17]. It was reported that SCIN was upregulated in gastric cancer tissues and increased SCIN expression was related to metastasis and poor overall survival[18]. Overexpression of SCIN was an independent predictor of poor prognosis in colorectal cancer patients[8]. Consistent with these findings, we found that the expression of SCIN was upregulated in LGG and GBM, and the overexpression of SCIN correlated with a poor prognosis in LGG patients. Further analysis of data from the CGGA database showed that higher levels of SCIN correlated with advanced tumor grade, while lower levels of SCIN were associated with IDH mutation and 1p/19q co-deletion in glioma. Currently, IDH mutation and 1p/19q co-deletion are considered to be good prognostic factors for patients with glioma[19]. Thus, these results support that SCIN is a potential prognostic biomarker for glioma.
Further, SCIN has been reported to participate in a variety of cellular biological processes in human cancer cells. Some studies have shown that SCIN promotes proliferation, inhibits apoptosis, and regulates the cell cycle in several cancers, including prostate, breast, lung, and hepatocellular cancers[10,20,21]. For example, SCIN is identified as a functional apoptosis regulator in hepatocellular carcinoma (HCC). Overexpression of SCIN inhibited apoptotic death and promoted xenografted HCC cell growth, while SCIN knockdown enhanced the chemosensitivity of HCC cells and suppressed tumor growth in vivo[22]. Other studies revealed the positive role of SCIN in cell migration, invasion, and metastasis[18,23]. Herein, CCK8 and colony-forming assays were used to verify that SCIN silencing suppressed cell proliferative ability in glioma cells. Meanwhile, wound healing and Transwell invasion assays revealed that SCIN silencing repressed cell migratory and invasive capabilities of glioma cells. These findings were consistent with previous reports, confirming the carcinogenic activity of SCIN in glioma cells.
Cytoskeleton constituents, including F-actin, maintain epithelial integrity and their disruption is a major cause of cancer progression[24]. SCIN, as an important regulator of F-actin organization, regulates actin filament dynamics[6]. Previous studies showed that the dysfunction of SCIN promoted cytoskeleton remodeling, resulting in changes in cellular behaviors. High levels of SCIN were observed in gastric cancer and silenced SCIN suppressed metastasis of gastric cancer cells, and decreased filopodium formation[18]. SCIN is involved in subcortical actin remodeling and promotes the formation of cell extensions and collagen degradation in MCF7 cells, thereby affecting matrix invasion and metastasis[25]. In this study, aggregated and arranged actin stress fibers were observed in the glioma cells, while the knockdown of SCIN caused the formation of sparse and disordered actin stress fibers. Accordingly, we suggest that SCIN may play a key role in F-actin polymerization. The reduction of actin stress fibers indicates that cell migration is inhibited, which explains the phenomenon of inhibited cell migration caused by SCIN loss at the subcellular levels.
FAK and RhoA have been shown to play critical roles in the F-actin reorganization, leading to tumor invasion[26]. The role of RhoA in regulating actin-filament formation has been well described[27]. RhoA promotes F-actin formation in various cancer cells[28,29]. FAK serves as a scaffolding protein for the binding sites of multiple oncogenic tyrosine kinases and regulates diverse cellular processes, including adhesion, migration, invasion, and metastasis[30]. Our investigation showed that SCIN silencing inhibited the expression levels of RhoA, p-cofilin, p-FAK, and Talin. To further clarify whether the FAK/RhoA signaling axis may be involved in SCIN-mediated migration and invasive activity of glioma cells, PF-573228 or CCG1423 was used to inhibit the FAK or RhoA activity in SCIN-overexpressed glioma cells. Expectedly, PF-573228 or CCG1423 suppressed the migration and invasiveness of glioma cells. Collectively, SCIN has the potential to promote malignant behaviors and F-actin polymerization in human glioma cells, and the underlying mechanism is related to the activation of the RhoA/FAK signaling axis.
In summary, SCIN promotes cell proliferation, migration, and invasion of glioma cells through remodeling the actin cytoskeleton. Our work illustrates a novel mechanism of SCIN-mediated glioma progression and suggests the possibility that SCIN might be a potential therapeutic target for glioma treatment.
Glioma is one of the most common intracranial tumors, characterized by invasive growth and poor prognosis. Actin cytoskeletal rearrangement is an essential event of tumor cell migration. The actin dynamics-related protein scinderin (SCIN) has been reported to be closely related to tumor cell mobility and invasion in several cancers.
The biological role and molecular mechanism of SCIN in glioma remain unclear.
This study aims to investigate the role and mechanism of SCIN in glioma.
The expression and clinical significance of SCIN were analyzed in glioma based on public databases. Then, we utilized SCIN-specific short hairpin RNAs to knock down SCIN expression in glioma cell lines and observed the effects of SCIN silencing on the proliferative, migrative, and invasive abilities of glioma cells. Furthermore, the effect of SCIN silencing on the cytoskeleton of glioma cells was also investigated.
SCIN expression was significantly elevated in glioma, and high levels of SCIN were associated with advanced tumor grade and wild-type dehydrogenase. SCIN-deficient cells exhibited repressed proliferation, migration, and invasion in U87 and U251 cells. The knockdown of SCIN promotes F-actin depolymerization in U87 and U251 cells via inhibiting RhoA/FAK signaling.
Our work illustrates a novel mechanism of SCIN-mediated glioma progression and suggests the possibility that SCIN might be a potential therapeutic target for glioma treatment.
To explore SCIN as a biomarker for glioma diagnosis in more clinical samples. To investigate the potential anticancer value of SCIN as an intervention target in vivo.
| 1. | Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2955] [Cited by in RCA: 3202] [Article Influence: 177.9] [Reference Citation Analysis (0)] |
| 2. | Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, Soffietti R, von Deimling A, Ellison DW. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 2021;23:1231-1251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4523] [Cited by in RCA: 7660] [Article Influence: 1532.0] [Reference Citation Analysis (1)] |
| 3. | Lejen T, Pene TD, Rosé SD, Trifaró JM. The role of different Scinderin domains in the control of F-actin cytoskeleton during exocytosis. Ann N Y Acad Sci. 2002;971:248-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 4. | Wang K, Kong F, Qiu Y, Chen T, Fu J, Jin X, Su Y, Gu Y, Hu Z, Li J. Autophagy regulation and protein kinase activity of PIK3C3 controls sertoli cell polarity through its negative regulation on SCIN (scinderin). Autophagy. 2023;19:2934-2957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 5. | Wang X, Shelton SD, Bordieanu B, Frank AR, Yi Y, Venigalla SSK, Gu Z, Lenser NP, Glogauer M, Chandel NS, Zhao H, Zhao Z, McFadden DG, Mishra P. Scinderin promotes fusion of electron transport chain dysfunctional muscle stem cells with myofibers. Nat Aging. 2022;2:155-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 6. | Zhang J, Yu Q, Jiang D, Yu K, Yu W, Chi Z, Chen S, Li M, Yang D, Wang Z, Xu T, Guo X, Zhang K, Fang H, Ye Q, He Y, Zhang X, Wang D. Epithelial Gasdermin D shapes the host-microbial interface by driving mucus layer formation. Sci Immunol. 2022;7:eabk2092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 87] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 7. | Zunino R, Li Q, Rosé SD, Romero-Benítez MM, Lejen T, Brandan NC, Trifaró JM. Expression of scinderin in megakaryoblastic leukemia cells induces differentiation, maturation, and apoptosis with release of plateletlike particles and inhibits proliferation and tumorigenesis. Blood. 2001;98:2210-2219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 52] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 8. | Lin Q, Li J, Zhu D, Niu Z, Pan X, Xu P, Ji M, Wei Y, Xu J. Aberrant Scinderin Expression Correlates With Liver Metastasis and Poor Prognosis in Colorectal Cancer. Front Pharmacol. 2019;10:1183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 9. | Jian W, Zhang X, Wang J, Liu Y, Hu C, Wang X, Liu R. Scinderin-knockdown inhibits proliferation and promotes apoptosis in human breast carcinoma cells. Oncol Lett. 2018;16:3207-3214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 10. | Liu H, Shi D, Liu T, Yu Z, Zhou C. Lentivirus-mediated silencing of SCIN inhibits proliferation of human lung carcinoma cells. Gene. 2015;554:32-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 11. | Abouzahr S, Bismuth G, Gaudin C, Caroll O, Van Endert P, Jalil A, Dausset J, Vergnon I, Richon C, Kauffmann A, Galon J, Raposo G, Mami-Chouaib F, Chouaib S. Identification of target actin content and polymerization status as a mechanism of tumor resistance after cytolytic T lymphocyte pressure. Proc Natl Acad Sci U S A. 2006;103:1428-1433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 53] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 12. | Zhou B, Chen TW, Jiang YB, Wei XB, Lu CD, Li JJ, Xie D, Cheng SQ. Scinderin suppresses cell proliferation and predicts the poor prognosis of hepatocellular carcinoma. Oncol Lett. 2020;19:2011-2020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 13. | Tavabe Ghavami TS, Irani S, Mirfakhrai R, Shirkoohi R. Differential expression of Scinderin and Gelsolin in gastric cancer and comparison with clinical and morphological characteristics. EXCLI J. 2020;19:750-761. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 14. | Tang Z, Li C, Kang B, Gao G, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5550] [Cited by in RCA: 7444] [Article Influence: 827.1] [Reference Citation Analysis (1)] |
| 15. | Zhao Z, Zhang KN, Wang Q, Li G, Zeng F, Zhang Y, Wu F, Chai R, Wang Z, Zhang C, Zhang W, Bao Z, Jiang T. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genomics Proteomics Bioinformatics. 2021;19:1-12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 707] [Article Influence: 141.4] [Reference Citation Analysis (0)] |
| 16. | Tang J, Kang Y, Huang L, Wu L, Peng Y. TIMP1 preserves the blood-brain barrier through interacting with CD63/integrin β 1 complex and regulating downstream FAK/RhoA signaling. Acta Pharm Sin B. 2020;10:987-1003. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 17. | Zhang ZH, Zhang W, Zhou JD, Zhang TJ, Ma JC, Xu ZJ, Lian XY, Wu DH, Wen XM, Deng ZQ, Lin J, Qian J. Decreased SCIN expression, associated with promoter methylation, is a valuable predictor for prognosis in acute myeloid leukemia. Mol Carcinog. 2018;57:735-744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 18. | Liu JJ, Liu JY, Chen J, Wu YX, Yan P, Ji CD, Wang YX, Xiang DF, Zhang X, Zhang P, Cui YH, Wang JM, Bian XW, Qian F. Scinderin promotes the invasion and metastasis of gastric cancer cells and predicts the outcome of patients. Cancer Lett. 2016;376:110-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 19. | Yang Z, Ling F, Ruan S, Hu J, Tang M, Sun X, Long W. Clinical and Prognostic Implications of 1p/19q, IDH, BRAF, MGMT Promoter, and TERT Promoter Alterations, and Expression of Ki-67 and p53 in Human Gliomas. Cancer Manag Res. 2021;13:8755-8765. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 20. | Wang D, Sun SQ, Yu YH, Wu WZ, Yang SL, Tan JM. Suppression of SCIN inhibits human prostate cancer cell proliferation and induces G0/G1 phase arrest. Int J Oncol. 2014;44:161-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 21. | Lai X, Su W, Zhao H, Yang S, Zeng T, Wu W, Wang D. Loss of scinderin decreased expression of epidermal growth factor receptor and promoted apoptosis of castration-resistant prostate cancer cells. FEBS Open Bio. 2018;8:743-750. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 22. | Qiao X, Zhou Y, Xie W, Wang Y, Zhang Y, Tian T, Dou J, Yang X, Shen S, Hu J, Qiao S, Wu Y. Scinderin is a novel transcriptional target of BRMS1 involved in regulation of hepatocellular carcinoma cell apoptosis. Am J Cancer Res. 2018;8:1008-1018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 23. | Chen XM, Guo JM, Chen P, Mao LG, Feng WY, Le DH, Li KQ. Suppression of scinderin modulates epithelialmesenchymal transition markers in highly metastatic gastric cancer cell line SGC7901. Mol Med Rep. 2014;10:2327-2333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 24. | Best M, Gale ME, Wells CM. PAK-dependent regulation of actin dynamics in breast cancer cells. Int J Biochem Cell Biol. 2022;146:106207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 25. | Tanic J, Wang Y, Lee W, Coelho NM, Glogauer M, McCulloch CA. Adseverin modulates morphology and invasive function of MCF7 cells. Biochim Biophys Acta Mol Basis Dis. 2019;1865:2716-2725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 26. | Schaks M, Giannone G, Rottner K. Actin dynamics in cell migration. Essays Biochem. 2019;63:483-495. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 224] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 27. | Kim JG, Islam R, Cho JY, Jeong H, Cap KC, Park Y, Hossain AJ, Park JB. Regulation of RhoA GTPase and various transcription factors in the RhoA pathway. J Cell Physiol. 2018;233:6381-6392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 103] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 28. | Denk-Lobnig M, Totz JF, Heer NC, Dunkel J, Martin AC. Combinatorial patterns of graded RhoA activation and uniform F-actin depletion promote tissue curvature. Development. 2021;148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 29. | Ma TJ, Zhang ZW, Lu YL, Zhang YY, Tao DC, Liu YQ, Ma YX. CLOCK and BMAL1 stabilize and activate RHOA to promote F-actin formation in cancer cells. Exp Mol Med. 2018;50:1-15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 30. | Hong KO, Ahn CH, Yang IH, Han JM, Shin JA, Cho SD, Hong SD. Norcantharidin Suppresses YD-15 Cell Invasion Through Inhibition of FAK/Paxillin and F-Actin Reorganization. Molecules. 2019;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Clinical neurology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: El-Shishtawy MM, Egypt; Ma Z, United States S-Editor: Liu JH L-Editor: Filipodia P-Editor: Zhang XD