INTRODUCTION
Germ-line mutations in BRCA1 and BRCA2 genes are the most well-known cause of hereditary cancer predisposition. BRCA1/2 pathogenic variants contribute to approximately 5%-10% and 15%-30% breast and ovarian cancer morbidity, respectively[1-6]. In addition, both mentioned genes are involved in the pathogenesis of a subset of stomach cancers, and the inheritance of BRCA2 inactive alleles is associated with an increased risk of prostate and pancreatic malignancies[7,8]. BRCA1/2-driven tumors tend to have particular clinical characteristics, being associated with younger age at onset and highly malignant phenotype[9,10]. Breast carcinomas (BCs) occurring in BRCA1 mutation carriers usually lack the expression of estrogen and progesterone receptors, and BRCA1/2-associated ovarian cancers (OCs) are characterized by serous high-grade histological appearance[10,11].
Breast and ovarian tumors arising in patients with BRCA1/2-associated hereditary cancer syndrome usually develop via somatic inactivation of the remaining allele of the involved gene. BRCA1 and BRCA2 play a key role in the maintenance of genomic integrity. Consequently, cancers lacking functional BRCA1 or BRCA2 proteins are deficient in DNA repair by homologous recombination (HR). Platinum compounds and poly (ADP-ribose) polymerase inhibitors (PARPi) induce massive DNA damage, which requires an HR-mediated repair. BRCA1/2-null cells are deficient for HR and consequently die upon the action of platinum salts or PARPi. This drug sensitivity is tumor-selective, as the normal cells of the patient retain one functional copy of BRCA1/2 gene and therefore remain capable of coping with the DNA damage[12-14].
As mentioned above, BRCA1/2-related hereditary tumors constitute a significant portion of OCs. In addition, many high-grade serous OCs have other causes of HR deficiency, e.g., somatic biallelic inactivation of BRCA1/2 genes or the presence of germ-line mutations in other members of DNA repair pathways[1,15]. This explains the high efficacy of platinum-based chemotherapeutic regimens in OC, which were developed empirically before the discovery of BRCA1/2 genes and constitute a standard-of-care for OC management. As expected, platinum therapy demonstrates increased efficacy in hereditary vs sporadic ovarian tumors[2,16]. In contrast to OC, BRCA1/2 deficiency is characteristic only for a minority of breast tumors; therefore, platinum compounds are not incorporated in the conventional treatment schemes for non-selected BC patients. Several trials demonstrated that platinum salts might outperform other chemotherapeutic agents when applied to BRCA1/2-driven BCs[17-19]. PARPi have been developed specifically for targeting tumors characterized by BRCA1/2 and/or HR deficiency. There are several PARPi approved for clinical use with slightly varying medical indications[20,21].
Although BRCA1/2-driven tumors have a clear-cut vulnerability, the use of platinum salts or PARPi does not usually result in a cure from metastatic disease. Platinum- and PARPi-exposed cancers eventually manage to escape from the action of BRCA1/2-specific therapy. Multiple preclinical and clinical studies have identified various BRCA1/2-restoring mechanisms or bypass pathways, which resume resistance to DNA damage in initially HR-deficient tumor cells. Recent investigations also provided evidence for an alternative scenario, where the emergence of the platinum-resistant tumor clone is attributed to a selection of pre-existing BRCA1-proficient cells; these therapy-resistant cells persist in small amounts in chemonaive tumors but are enriched in the residual lesion. This paper provides a brief overview of the mechanisms of acquired platinum and PARP-resistance in BRCA1/2-driven tumors.
RESTORATION OF BRCA1/2 FUNCTION BY SECONDARY SOMATIC MUTATIONS
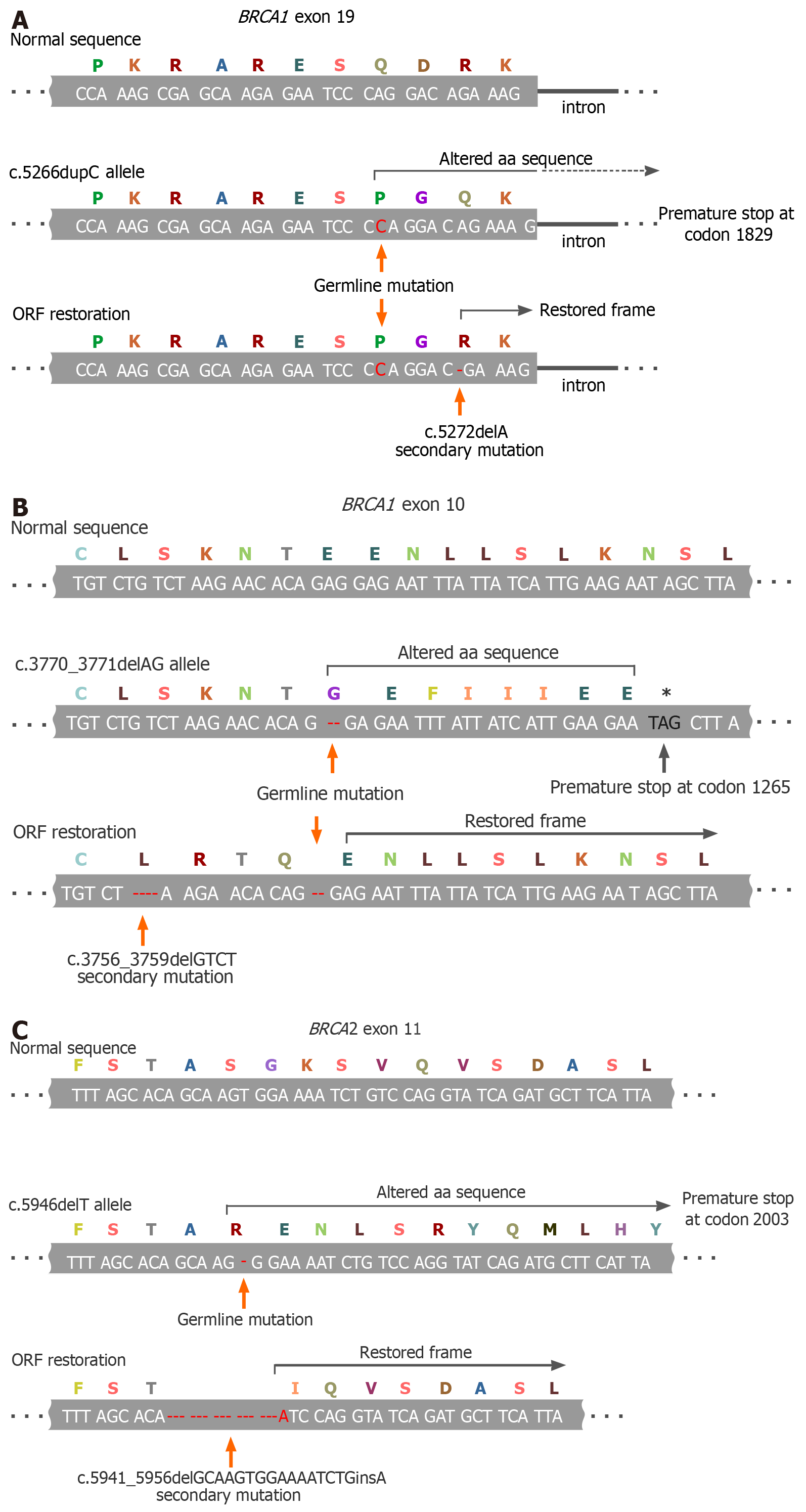
The vast majority of BRCA1/2 inherited pathogenic alleles are represented by small alterations in the nucleotide sequence, which cause a frameshift and emergence of premature stop-codons. The open reading frame (ORF) can be rescued by a nearby second mutation if it restores an original 3-letter genetic code, or by small deletion, which excises the pathogenic allele and reconstitutes the ORF, or by the true back mutation (Figure 1)[22-24]. A secondary ORF-restoring BRCA2 mutation was first described in an acute myeloid leukemia cell line obtained from a patient with Fanconi anemia[25]. The discovery of PARPi and the recognition of BRCA1/2-specific action of platinum compounds stimulated intense investigations of the mechanisms of tumor resistance to these drugs. A series of studies revealed that the emergence of secondary BRCA1/2 mutations is the most reproducible hallmark of the acquisition of a drug-resistant phenotype. Indeed, the reversion mutations have been repeatedly observed in the experiments with cell lines, patient-derived xenografts (PDX) and clinical samples[26,27].
Figure 1 Examples of BRCA1/2 open-reading frame restoration in BRCA1/2-mutated tumors during systemic therapy.
A: Secondary 1-bp deletion occurring downstream to the germline mutation (BRCA1 c.5266dupC; described in[22]); B: Secondary 4-bp deletion located upstream to the germline mutation (BRCA1 c.3770_3771delAG; an example from[23]); C: Secondary in-frame deletion/insertion excising the mutation-containing gene fragment (BRCA2 c.5946delT; an example from[24]). ORF: Open-reading frame.
For the time being, the development of secondary ORF-restoring mutations in BRCA1/2 genes is the only clinically proven mechanism of the tumor adaptation to the therapy, which is relevant both to platinum compounds and PARPi, characteristic both for BRCA1 and BRCA2 genes, and has been convincingly validated in patient samples. The true incidence of secondary BRCA1/2 mutations is difficult to presently define due to various selection biases and technical limitations of available molecular genetic assays: They appear to be found in approximately a quarter of PARPi/platinum-resistant tumors, although some studies provide even higher estimates. Importantly, many reports describe the emergence of multiple distinct BRCA1/2 ORF-restoring mutations in independent drug-resistant clones obtained from the same patient, thus providing evidence for the functional convergence of tumor adaptation pathways[26,27].
Some data suggest that the genetic reversion is somewhat more characteristic for BRCA2- than for BRCA1-driven tumors. Distinct pathogenic variants of BRCA1 and BRCA2 may differ in their ability to be rescued by the second mutation: It is hypothesized that the genetic reversion is more acceptable for non-conservative regions of the above genes, which are more or less dispensable for their function. Indeed, in all cases, except genuine back mutations, the involved region of BRCA1 and BRCA2 genes undergoes subtle alterations (i.e. the deletion of a few coding nucleotides or the change of the sequence for a few amino acids); therefore, highly conserved parts of these genes may not tolerate this mechanism of genetic adaptation[14,26,27].
In addition to secondary mutations affecting the coding sequence of BRCA1/2 genes, there are functionally similar events resulting in the production of hypomorphic but still functional protein. For example, loss of exon 11 is compatible with the participation of BRCA1 in HR; consequently, alternative splicing resulting in the BRCA1 exon 11 skipping may contribute to the acquired drug resistance[28]. The mutation located in the N-terminal portion of the BRCA1 gene can be bypassed by the production of a hypomorphic protein, whose translation starts after the frameshift[29]. Another mechanism of the partial rescue of BRCA1 function involves gene rearrangements, which terminate BRCA1 translation before the mutation-containing BCRT domain, consequently preventing the proteasomal degradation of BRCA1. These truncated versions of BRCA1 are capable of maintaining HR and mediate PARPi resistance[30]. Upregulation of HSP90 may stabilize some BRCA1-mutant proteins and thus support their function[31]. Amplification of mutated BRCA2 was shown to compensate for partial loss of BRCA2 function and rendered PARPi resistance in cell line experiments[32].
Some BRCA1/2 germ-line pathogenic alleles are represented by so-called large gene rearrangements (LGRs), which may involve deletions of multiple exons. By definition, these tumors cannot be repaired by the second ORF-restoring mutation. One would expect that these tumors are likely to demonstrate a more pronounced and prolonged response to BRCA1/2-specific therapy. BRCA1/2 LGRs are not specifically considered in the studies on tumor drug sensitivity. However, there are case reports supporting exceptional responsiveness of BRCA1/2 LGR-associated tumors to PARPi[33].
BYPASS MECHANISMS
There are two key mechanisms of the repair of DNA double-strand breaks (DSB). Accurate correction of DNA sequence can be achieved exclusively by HR. In the absence of functional HR, error-prone non-homologous end-joining (NHEJ) becomes a prevailing mechanism of DSB repair. The choice between HR and NHEJ is mediated by the balance between their regulators, BRCA1 and 53BP1. When BRCA1 is inactivated by mutation, NHEJ-driven DNA repair prevails. This results in the accumulation of multiple DNA lesions and eventual cell death. BRCA1-deficient cells may adapt to the platinum or PARPi pressure by down-regulation of 53BP1. As a result of consequent NHEJ suppression, tumor cells re-activate HR and eventually become resistant to the drug exposure[31,34,35]. Down-regulation of 53BP1 has been observed in some clinical samples that failed platinum-based or PARPi therapy[31,36,37]. In vitro studies revealed several other proteins whose loss also contributes to the switch from NHEJ to HR or to other bypass pathways. Noticeably, the involvement of 53BP1 exemplifies the differences between BRCA1- and BRCA2-mutated tumors, as the loss of 53BP1 or related proteins is relevant only for the treatment escape of BRCA1-deficient cancers[34]. Preclinical studies also identified HR-independent platinum/ PARPi resistance mechanisms, which involve stabilization of replication forks[38].
BRCA1/2-NON-RELATED MECHANISMS OF ACQUIRED RESISTANCE TO PLATINUM COMPOUNDS AND PARPi
The above-described mechanisms of acquired therapy resistance are more or less specific for the BRCA1/2-associated action of platinum salts and PARPi. There are also general mechanisms for the adaptation of tumor cells to the therapy, which are indirectly related to the targeted biological pathway and may involve activation of the drug efflux, down-deregulation of apoptosis, preservation of tumor cancer stem cells (CSCs) and epithelial-mesenchymal transition (EMT)[39]. Up-regulation of ABCB1 (MDR1) transporter has been implicated in multidrug resistance. Therapy-resistant ovarian and breast carcinomas are characterized by gene fusions, which result in increased expression of the ABCB1 gene[40]. The translational implications of these observations are not immediately clear: Drug transporters are involved in multiple physiological processes and are characterized by significant redundancy, so their targeting may be associated with significant adverse events and insufficient clinical efficacy[41]. There are reports demonstrating the selection of CSCs upon PARPi exposure[42]. The role of EMT in the development of PARPi resistance has been shown in preclinical studies involving BRCA2-deficient cells[43].
SELECTION OF PRE-EXISTING BRCA1-PROFICIENT CELLS DURING PLATINUM-BASED THERAPY
A significant portion of OC patients present with the inoperable disease; therefore, they undergo neoadjuvant (first-line) therapy aimed to reduce the tumor burden and permit surgical excision of the remaining cancer lumps. BRCA1-driven cancers are particularly sensitive to systemic platinum-based treatment; hence, this category of OC is usually amenable to complete surgical debulking. Despite that presumably efficient platinum-based therapy is administered again after the surgery, it apparently cannot eliminate the residual cancer cells, given that almost all OC treated by this scheme eventually relapse[44].
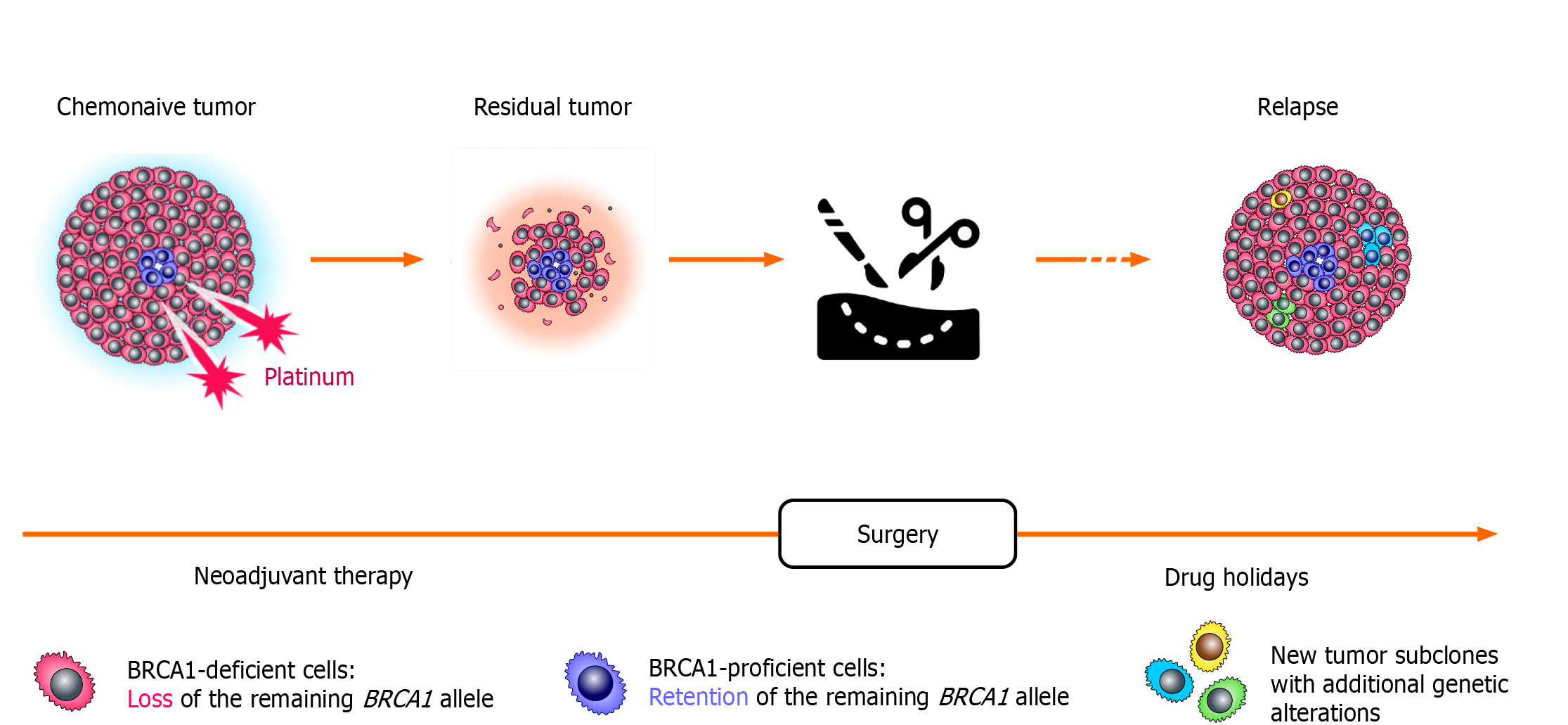
Comparison of tumor specimens obtained before the start of the treatment and after a few weeks of neoadjuvant therapy revealed surprising findings (Figure 2). While chemonaive BRCA1-associated OCs are characterized by somatic loss of heterozygosity (LOH) of the remaining allele, the residual tumors obtained after a few weeks of neoadjuvant therapy often show the retention of the wild-type BRCA1 copy. This “restoration of heterozygosity” occurs due to the selection of preexisting BRCA1-proficient cells, which persist in small amounts in chemonaive tumors; these isolated tumor cells with retained BRCA1 function can be visualized by various imaging techniques. Importantly, both primary cancers and residual tumor masses were shown to retain the same mutation in the TP53 gene. Loss of BRCA1 in normal cells triggers apoptosis, while cells with inactive TP53 may survive BRCA1 deficiency. It appears that TP53 mutation must be acquired in the very initial stages of the tumor evolution, while BRCA1 LOH, being a key event in the pathogenesis of BRCA-driven cancers, can emerge and be tolerated only after TP53 inactivation. The persistence of isolated BRCA1-proficient cells within a gross tumor mass is common for BRCA1-driven cancers, as the “restoration” of BRCA1 heterozygosity is observed approximately in two-thirds of BRCA1-associated OCs[45].
Figure 2 Selection of pre-existing BRCA1-proficient cells during platinum-based therapy.
Loss of the remaining BRCA1 allele is observed in the majority of cells forming the tumor; however, even chemonaive BRCA1-driven ovarian cancers contain a small fraction of transformed cells with retained BRCA1 heterozygosity. These cells are platinum-resistant and rapidly repopulate tumor mass during neoadjuvant therapy for ovarian cancer. During platinum-free interval, which occurs after the completion of the adjuvant therapy, these BRCA1-proficient cells become outcompeted by cells carrying BRCA1 LOH. Therefore, ovarian cancer relapses resemble primary tumors with regard to the BRCA1 status, as they demonstrate again the BRCA1 deficiency and the sensitivity to platinum compounds.
Intriguingly, the relapse OC tissues obtained from the same patients after therapy holidays show BRCA1 LOH again, thus providing a mechanistic explanation for the platinum sensitivity of recurrent BRCA1-associated cancers (Figure 2). Exome sequencing revealed that only TP53 mutation is stably maintained throughout the natural history of BRCA1-driven cancers, while the profiles of somatic point mutations and chromosome number alterations show some variations between chemonaive, post-neoadjuvant and recurrent tumor specimens. Overall, it appears that BRCA1-driven tumors present an ecosystem: While the gross majority of tumor mass is BRCA1-deficient, there are apparently some biological reasons to maintain the persistence of small amounts of BRCA1-proficient cells. In the absence of external hazards, BRCA1-deficient cells clearly outcompete cells with retained BRCA1 function. However, these cells are sensitive to platinum exposure and perhaps to some other kinds of unfavorable environment, so the maintenance of the reservoir of invulnerable (BRCA1-proficient) cells is important for warranting tumor plasticity. Upon drug pressure, BRCA-proficient cells take advantage and increase their relative fraction in residual tumor mass; however, they again lose the competition after the cessation of the systemic treatment[46]. The above observations fit very well with the concept of tumor “stem cells” as a cause of acquired drug resistance.
The platinum-induced selection of pre-existing BRCA-proficient cells has been demonstrated only for the BRCA1 gene, while similarly designed studies have not been performed yet for BRCA2-associated tumors. It is not self-explanatory that the same phenomenon is applicable to BRCA2-driven cancers. Indeed, although both BRCA1 and BRCA2 proteins are involved in the response to DNA damage, they have essential dissimilarities in their structure and function[14]. Consequently, they demonstrate differences regarding the spectrum of associated tumors, with prostate and pancreatic cancer been strongly linked to BRCA2 but not to BRCA1 heterozygosity[7]. Breast carcinomas arising in BRCA1 germ-line mutation carriers are usually triple-negative with regard to the receptor status (ER, PgR and HER2), while BRCA2 pathogenic alleles are generally associated with the development of tumors expressing steroid hormone receptors[10,11]. BRCA1 but not BRCA2 is essential for taxane-mediated cell death, so the resistance to taxanes is characteristic for BRCA1- but not for BRCA2-deficient cells[12]. The emergence of ORF-restoring secondary mutations in heavily pretreated tumors appears to be somewhat more common for BRCA2 than for BRCA1 gene[26,27]. BRCA1 deficiency is lethal for normal cells; therefore, the development of cancers in BRCA1 germ-line mutation carriers always involves mutation-driven inactivation of the TP53 gene, which results in down-regulation of apoptosis and provides the ground for the survival of BRCA1-null cells. In contrast, BRCA2 inactivation is compatible with cell viability, so BRCA2-associated tumors often have wild-type TP53 status[47]. While the persistence of BRCA1-proficient cells in chemonaive BRCA1-driven tumors is essential for the adaptation of OC to platinum-based therapy, it is unclear how this intratumoral heterogeneity supports the maintenance of tumor mass in “natural” conditions. This intratumoral heterogeneity may not necessarily be characteristic for the cancers arising in BRCA2 germ-line mutation carriers. Further studies are needed to reveal whether the persistence of isolated HR-proficient “stem” cells is relevant for BRCA2-driven tumors or sporadic OCs with BRCAness phenotype.
CONTROVERSIAL AND UNRESOLVED ISSUES
Platinum compounds and PARPi converge in their mechanisms with regard to targeting HR-deficient cells; however, there are also some differences in their action. For example, platinum salts appear to target tumors with deficient nucleotide excision repair[34]. Consequently, while secondary BRCA1/2 mutations or other HR-restoring events are likely to result in cross-resistance between platinum and PARPi, other modes of tumor adaptation to the therapy may be more drug-specific. Platinum is commonly used for the treatment of ovarian cancer, and the clinical trials demonstrated that the use of PARPi results in significantly better outcomes in platinum-sensitive vs platinum-resistant disease[48,49]. Similarly, the advantage of talazoparib was more pronounced in BRCA1/2-driven breast cancer patients who did not receive prior cisplatin or carboplatin[50]. However, some presumably platinum-resistant ovarian tumors still demonstrate some sensitivity to PARPi[48,49]. On the other hand, PARPi therapy may result, for example, in the emergence of mutations in the PARP1 gene, which alter PARP1 trapping to DNA but are unlikely to affect tumor sensitivity to drugs other than PARPi[51]. It needs to be stressed that in the clinical setting, the platinum sensitivity of ovarian cancer is usually defined not by the actual tumor response to carboplatin or cisplatin but by the time interval exceeding 6 mo since the last platinum exposure. It is not impossible that some tumors may actually restore HR deficiency within a shorter period of time, so their response to PARPi could be explained by conventional PARPi-associated biological mechanisms. While the use of PARPi after chemotherapy has been evaluated in many clinical trials[49], we are unaware of a systematic analysis of chemotherapy response in PARPi-resistant tumors.
BRCA1 and BRCA2 germ-line mutations are usually viewed as equivalent in all clinical trials involving DNA damaging treatments. Although this approach is generally well justified, some differences between these two genes need to be acknowledged. Preclinical experiments have demonstrated mechanisms for therapy escape that are relevant for BRCA1- but not for BRCA2-driven tumors[34]. The spectrum of associated cancers is somewhat different for these two genes; for example, the analysis of PARPi-resistant prostate malignancies is almost entirely limited to BRCA2 mutation carriers, as BRCA1 plays a negligible role in the predisposition to this disease[26].
The regimens of administration of platinum salts and PARPi significantly differ. Cisplatin or carboplatin are usually administered in several cycles, so there are peak drug concentrations and significant intervals between chemotherapy infusions. In contrast to this intermittent drug administration of platinum drugs, PARPi are used at a continuous dose for a prolonged period of time. It is very likely that the mode of drug administration may influence the pathways of tumor adaptation to therapeutic intervention. Furthermore, published clinical experiments included very heterogeneous groups of patients with regard to the duration of prior treatment. It appears that the majority of secondary BRCA1/2 mutations were detected mainly in heavily pretreated patients, while the initial cycles of chemotherapy rarely resulted in the genetic reversion of the BRCA1/2 sequence[26,27,46,52].
Cell and animal experiments cannot fully recapitulate the complexity of intratumoral heterogeneity, tumor microenvironment, interplay with the immune system, drug dosing, etc., characteristic for a clinical setting. The investigation of biological material obtained from cancer patients is challenging, particularly when it comes to the analysis of acquired therapy resistance. Tumor re-biopsy, by definition, requires sound clinical and ethical justification; therefore, some studies relied on circulating tumor DNA (ctDNA). Liquid biopsy is capable, in theory, to uncover the entire spectrum of subclonal secondary mutations, although it may underestimate the frequency of back mutations and does not account for the proportion of BRCA1/2-restored cells within a tumor mass. Current technologies for gene sequencing, which are utilized for the detection of secondary mutations, may miss some large deletions of genetic material[26]. It is highly desirable to continue the collection of platinum- and PARPi-resistant tumor samples from cancer patients, to subject these specimens to comprehensive molecular profiling, and to monitor the response of these tumors to subsequent treatment modalities. This effort may identify gene-response correlation and help to guide the clinical management of BRCA1/2-related cancers after the failure of the standard therapy.
CONCLUSION
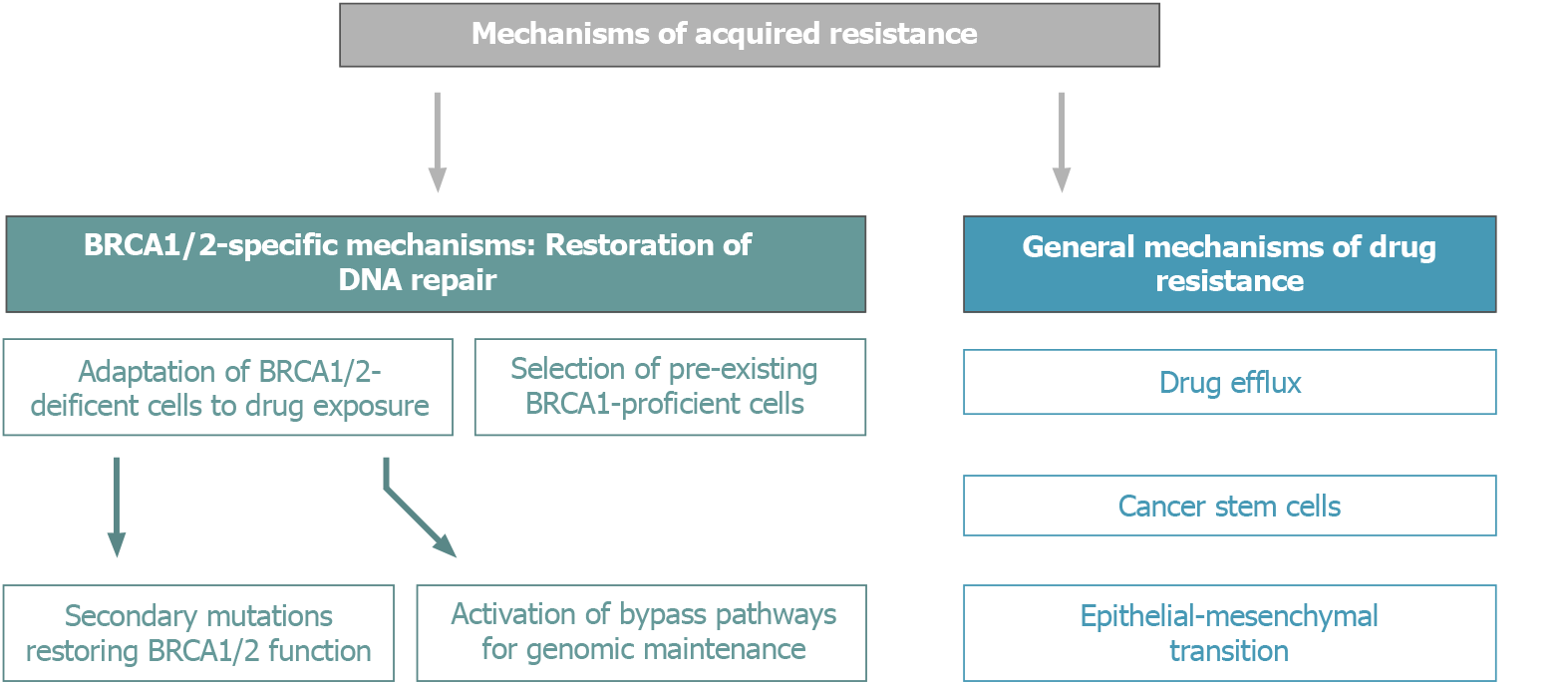
BRCA1/2-driven tumors have a number of in-built mechanisms of adaptation to conventional schemes of platinum-based therapy and PARPi (Figure 3). Nowadays, an increasing number of OC patients are subjected to long-term PARPi maintenance therapy, which certainly affects the biological and clinical properties of recurrent tumors. It is somewhat surprising that the available medical research literature does not put an emphasis on the potential treatment options for tumors arising on the background of continuous PARPi exposure, despite that multiple lines of preclinical and clinical data suggest the involvement of cross-resistance mechanisms[34].
Figure 3 Mechanisms of acquired resistance of BRCA1/2-driven tumors to platinum compounds and PARP inhibitors.
Genome profiling of drug-resistant tumors obtained from BRCA1/2 mutation carriers has not identified recurrent actionable molecular lesions[46]. However, despite the restoration of HR proficiency or the emergence of bypass pathways, these tumors continue to contain the genomic scar of BRCAness, i.e. the existence of multiple genomic rearrangements. These genetic lesions may underlie an increased antigenicity of BRCA1/2-driven tumors. Interestingly, second mutations, which are the cause of drug resistance, are often associated with the emergence of additional antigenic epitopes[26]. The feasibility of the use of immune therapy against platinum/PARPi-resistant OCs has not been evaluated systematically, although case series support the promise of this option[53].
The best approach would be to implement treatment that would prevent the appearance of drug-resistant clones. There is a number of ongoing trials evaluating the efficacy of combinations of PARPi with other drugs[34,54]. Several studies demonstrated the potentially curative impact of high-dose chemotherapy for BRCA1/2 mutation carriers; however, the use of this treatment is associated with excessive adverse effects[55]. Neoadjuvant combination of cisplatin and mitomycin C resulted in complete pathological responses, i.e. in the elimination of all detectable cancer cells, in some BRCA1/2-driven OCs[44].
There are several recent breakthroughs in the management of BRCA1/2-driven tumors, which resulted in significant improvement of disease outcomes. Continued understanding of the mechanisms of platinum/PARPi resistance inspired the development of a multitude of novel therapeutic approaches, which are likely to contribute to further advances in cancer treatment. BRCA1/2-associated carcinomas have well-defined vulnerabilities and are characterized by pronounced drug sensitivity. They are similar in this respect to germ-cell tumors and some hematological malignancies, which are generally curable by already available therapeutic tools. There are reasonable chances that cure rates for BRCA1/2-associated malignancies will significantly increase in the near future.
ACKNOWLEDGEMENTS
We appreciate greatly the contribution of Dr. Ekatherina Kuligina (N.N. Petrov Institute of Oncology) in the preparation of Figures. We are cordially thankful to Dr. Barbara Vona (University of Tuebingen) for critical reading of this manuscript.
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/Licenses/by-nc/4.0/
Manuscript source: Invited manuscript
Corresponding Author's Membership in Professional Societies: European Society for Medical Oncology, No. 6102.
Specialty type: Oncology
Country/Territory of origin: Russia
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Zheng Z S-Editor: Liu M L-Editor: Filipodia P-Editor: Yuan YY