©The Author(s) 2026.
World J Diabetes. Jan 15, 2026; 17(1): 112885
Published online Jan 15, 2026. doi: 10.4239/wjd.v17.i1.112885
Published online Jan 15, 2026. doi: 10.4239/wjd.v17.i1.112885
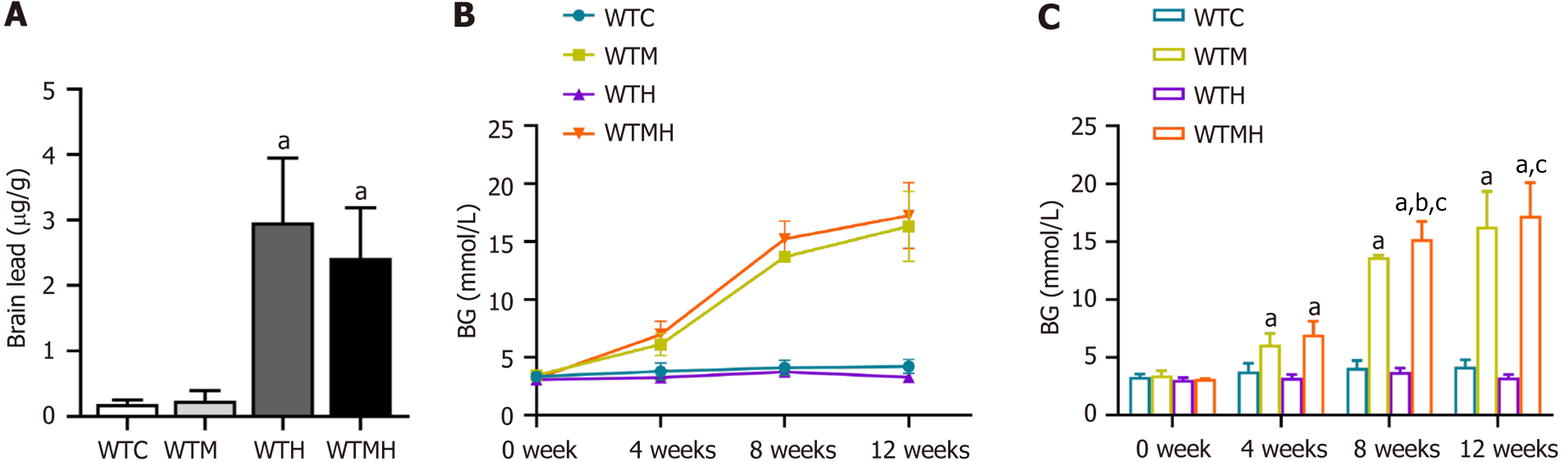
Figure 1 Lead and blood glucose levels in different mice groups.
A: Results of mouse brain levels; B and C: Blood glucose levels of mice in different groups at different time points. aP < 0.05 vs control group. bP < 0.05 vs diabetes group. cP < 0.05 vs lead exposure group. WTC: Control group; WTM: Diabetes group; WTH: Lead exposure group; WTMH: Diabetes with lead exposure group.
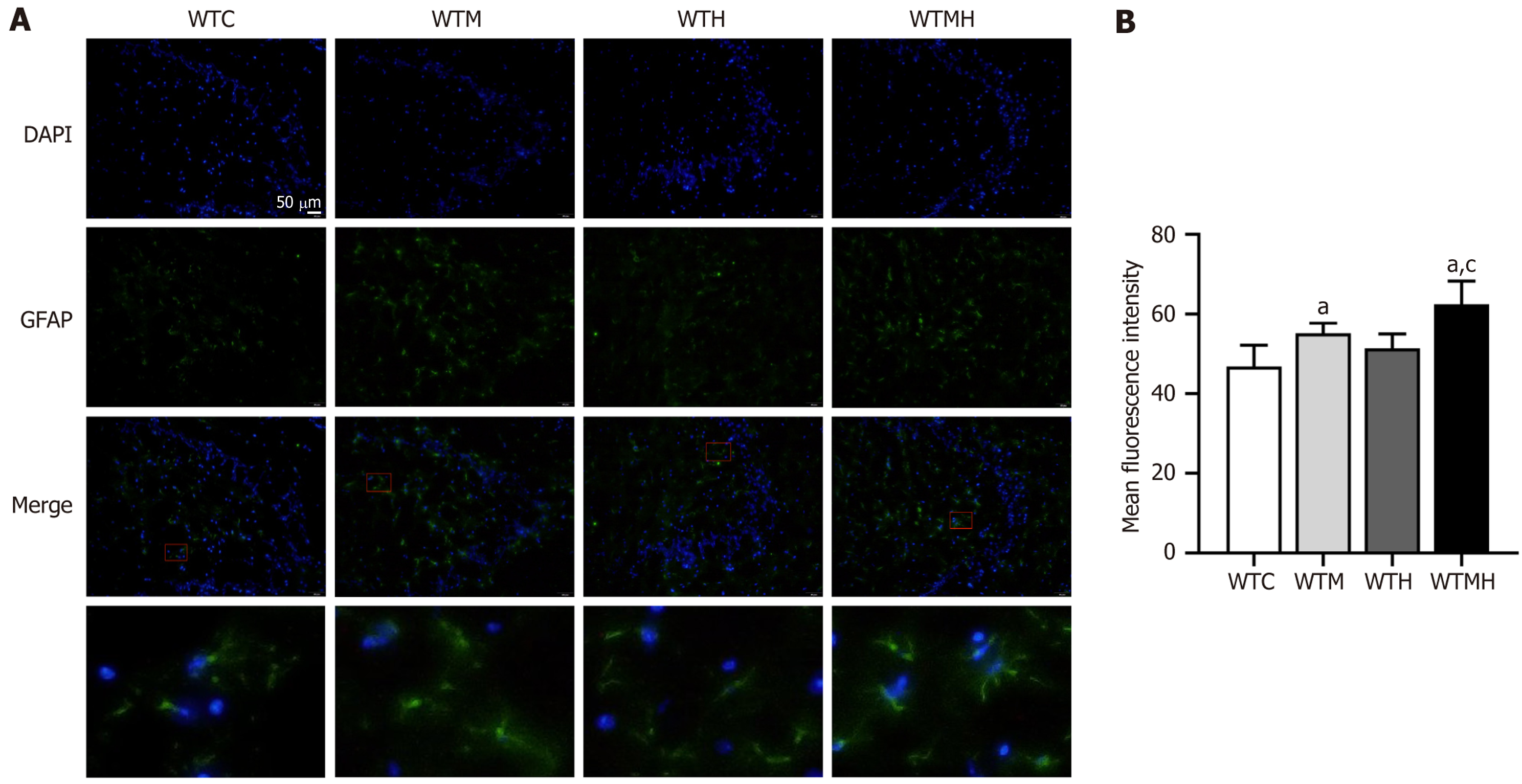
Figure 2 Immunofluorescence results.
A: Expression of GFAP in the hippocampal tissue of each group of mice; B: Statistical analysis of average fluorescence intensity. aP < 0.05 vs control group. cP < 0.05 vs lead exposure group. WTC: Control group; WTM: Diabetes group; WTH: Lead exposure group; WTMH: Diabetes with lead exposure group.
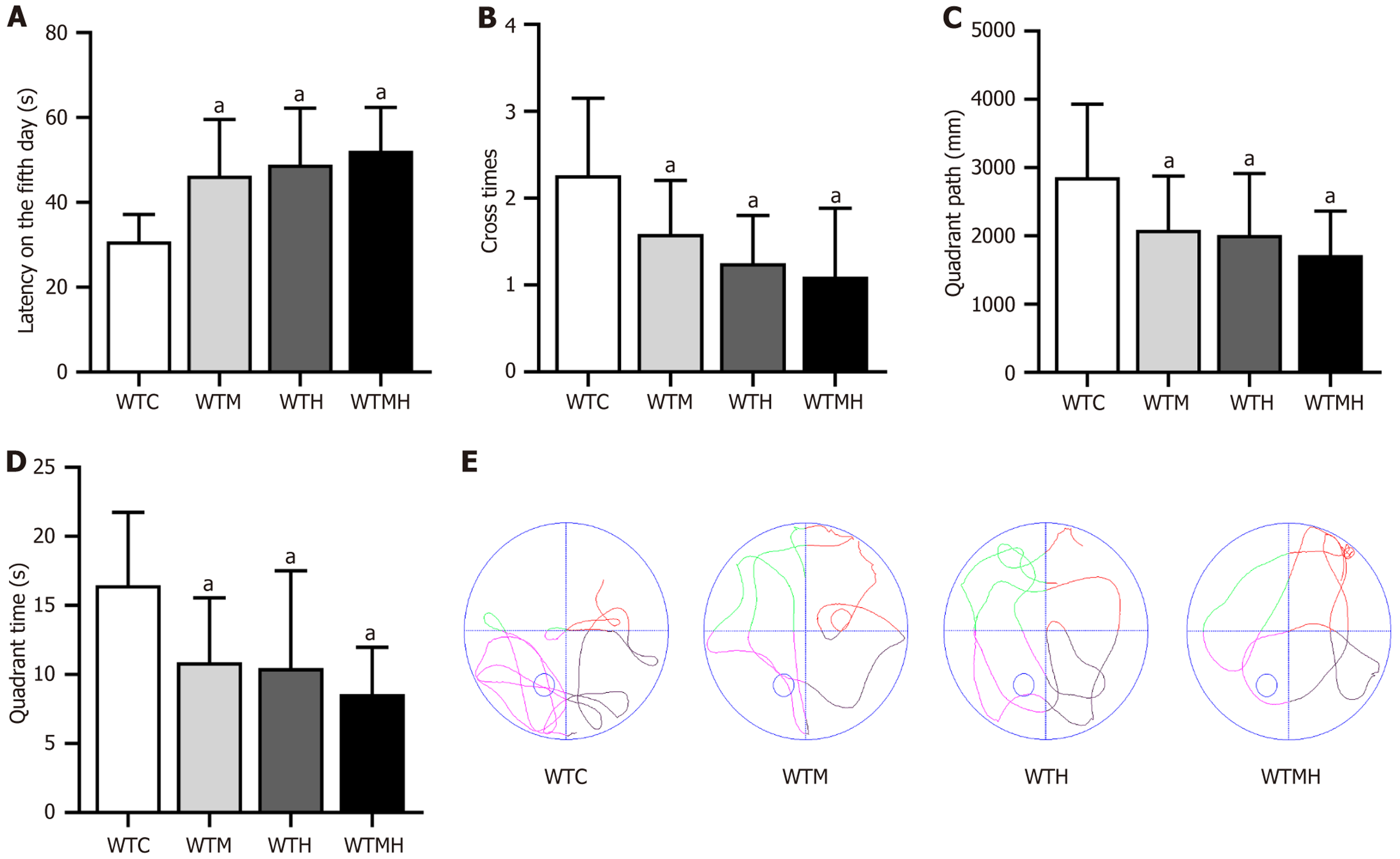
Figure 3 Results of the Morris water maze experiment in mice.
A: Escape latency period of each group of mice on the fifth day; B: Platform crossing frequency of each group of mice; C: Target quadrant time of each group of mice; D: Target quadrant swim distance of each group of mice; E: Representative figure of swimming trajectories of each group of mice in the spatial exploration test. Sample size: WTC: 20, WTM: 20, WTH: 20, WTMH: 20. aP < 0.05 vs control group. WTC: Control group; WTM: Diabetes group; WTH: Lead exposure group; WTMH: Diabetes with lead exposure group.
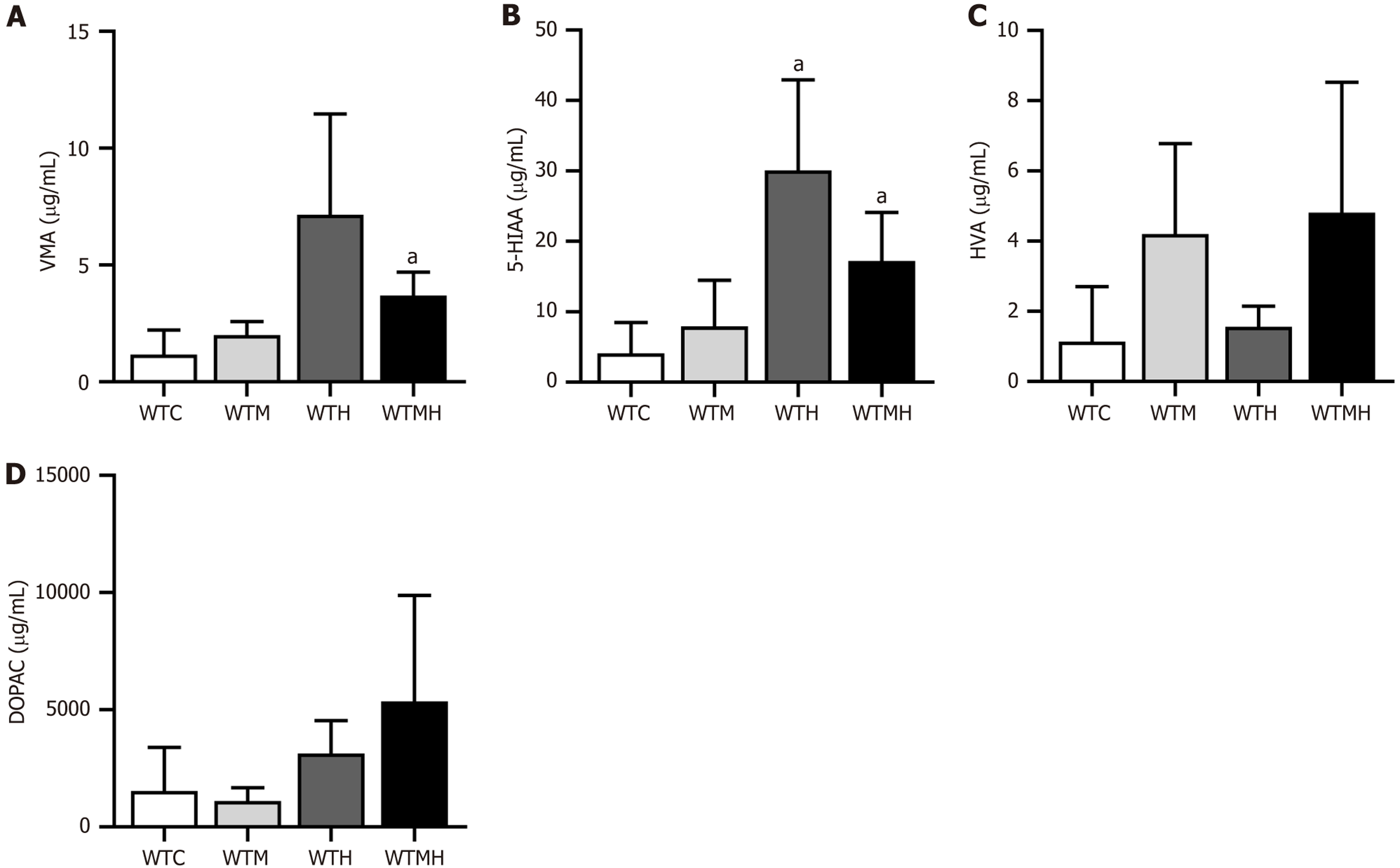
Figure 4 High performance liquid chromatography analysis of urinary neurotransmitters in mice.
A: Vanillylmandelic acid; B: 5-Hydroxy
Figure 5 Hippocampal proteomics results in mice from different groups.
A: Differentially expressed proteins between control group (WTC) and diabetes group in proteomic analysis; B: Differentially expressed proteins between WTC and diabetes with lead exposure group in proteomic analysis; C and D: Top 10 differentially expressed genes in Gene Ontology biological process enrichment analysis; E and F: Top 10 differentially expressed genes in Kyoto Encyclopedia of Genes and Genomes enrichment analysis.
Figure 6 Detection of gut microbiota in different groups of mice and correlation analysis between gut microbiota and proteomics.
A: Analysis of Chao1, observed species indices, Faith’s PD index, and Simpson diversity index; B: Microbial Venn diagram analysis of intergroup differences; C: Analysis of metabolic pathways of differential microbial communities; D: Correlation analysis between differential microbiota and top 60 differential proteins between control group and diabetes group; E: Correlation analysis between differential microbiota and top 60 differential proteins between control group and diabetes with lead exposure group.
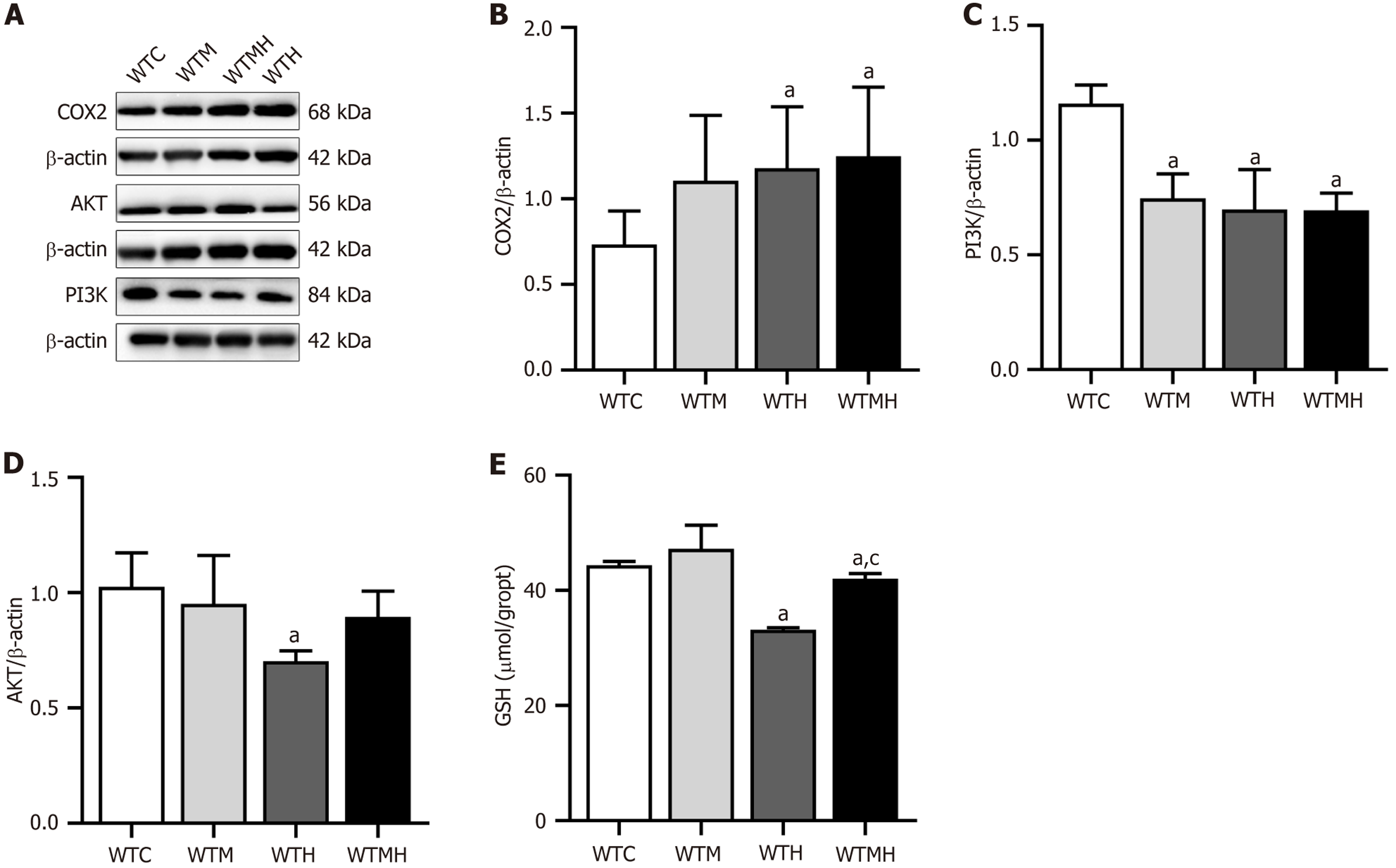
Figure 7 Western blot and glutathione detection in hippocampal tissue of mice.
A: Protein bands of each group of mice; B: Expression of OX2 protein; C: Expression of PI3K protein; D: Expression of AKT protein, E: Expression of glutathione in the brain of each group of mice. aP < 0.05 vs control group. cP < 0.05 vs lead exposure group. WTC: Control group; WTM: Diabetes group; WTH: Lead exposure group; WTMH: Diabetes with lead exposure group.
- Citation: Ding WJ, Liu CQ, Tang XY, Shang ZB, Liang X, Tao T, Liu RX, Jiang QY, Qiu YF, Sun Y. Role of gut microbiota in lead-induced neural damage in diabetic mice. World J Diabetes 2026; 17(1): 112885
- URL: https://www.wjgnet.com/1948-9358/full/v17/i1/112885.htm
- DOI: https://dx.doi.org/10.4239/wjd.v17.i1.112885