©The Author(s) 2025.
World J Diabetes. Nov 15, 2025; 16(11): 110428
Published online Nov 15, 2025. doi: 10.4239/wjd.v16.i11.110428
Published online Nov 15, 2025. doi: 10.4239/wjd.v16.i11.110428
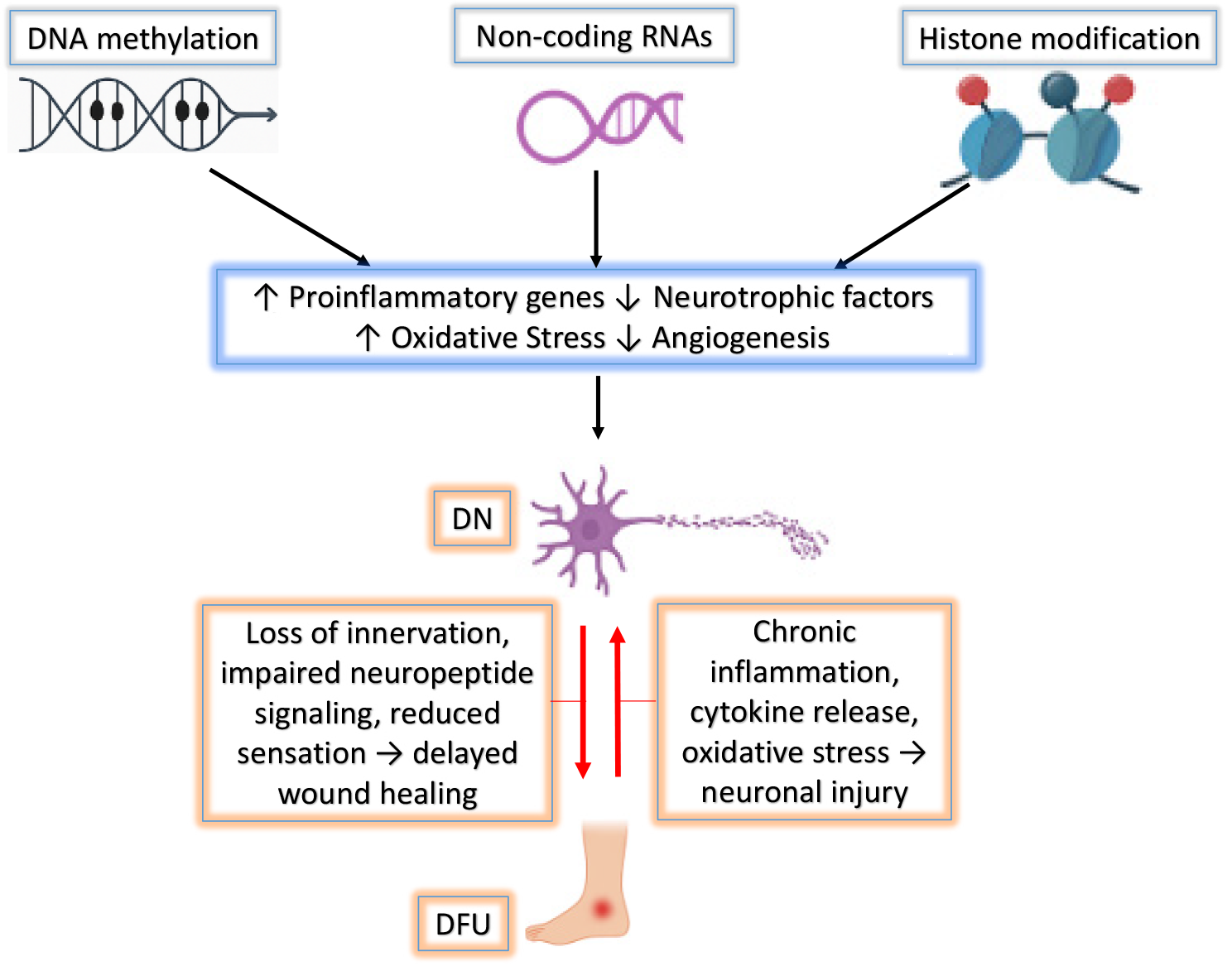
Figure 1 Integrated epigenetic network in diabetic complications.
This schematic illustrates the interplay between major epigenetic modifications (DNA methylation, histone modifications, and non-coding RNAs) in driving chronic inflammation, oxidative stress, impaired neurovascular repair, and tissue degeneration. These processes contribute to the development and persistence of diabetic neuropathy (DN) and diabetic foot ulcers (DFUs) by disrupting key protective pathways and promoting pathological gene expression.
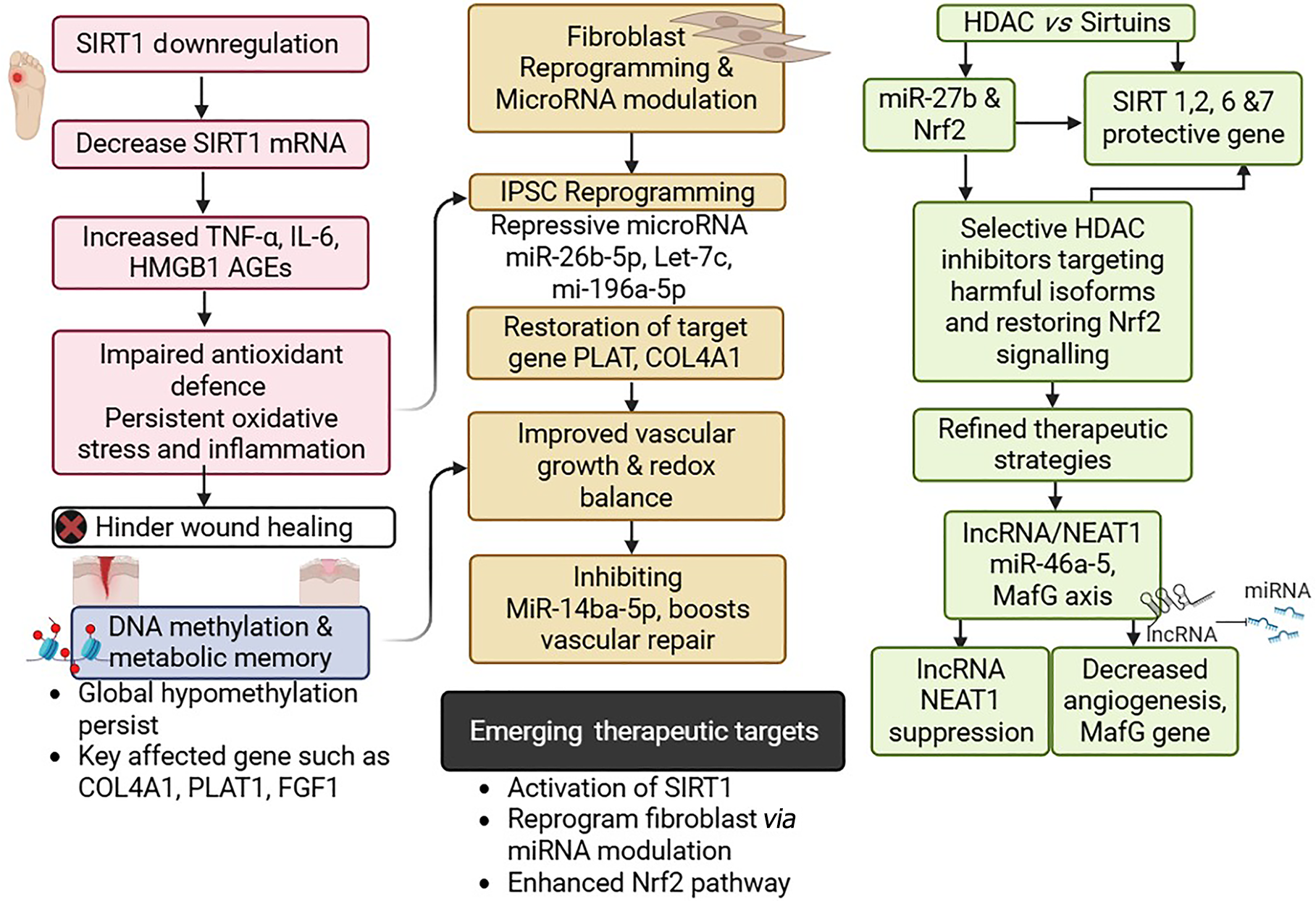
Figure 2 Epigenetic regulations in diabetic foot ulcer pathogenesis and healing.
SIRT1 downregulation leads to elevated pro-inflammatory mediators and oxidative stress, which impairs wound healing. Persistent DNA hypomethylation, despite normoglycemia, sustains metabolic memory and represses key reparative genes (e.g., collagen type IV alpha 1 [COL4A1], plasminogen activator, tissue type 1 [PLAT1], and fibroblast growth factor 1 [FGF1]). Reprogramming fibroblasts through modulation of microRNAs (e.g., miR-26b-5p, let-7c, miR-196a-5p) restores pro-repair gene expression, improves redox balance, and promotes vascular repair. Additionally, targeting the histone deacetylase (HDAC)–sirtuin axis via selective HDAC inhibition and enhancing Nrf2 signaling refines therapeutic strategies. Suppression of long non-coding RNA (lncRNA) nuclear enriched abundant transcript 1 (NEAT1) and modulation of the miR-46a-5–MafG axis further improve angiogenesis. Together, these insights identify promising therapeutic avenues.
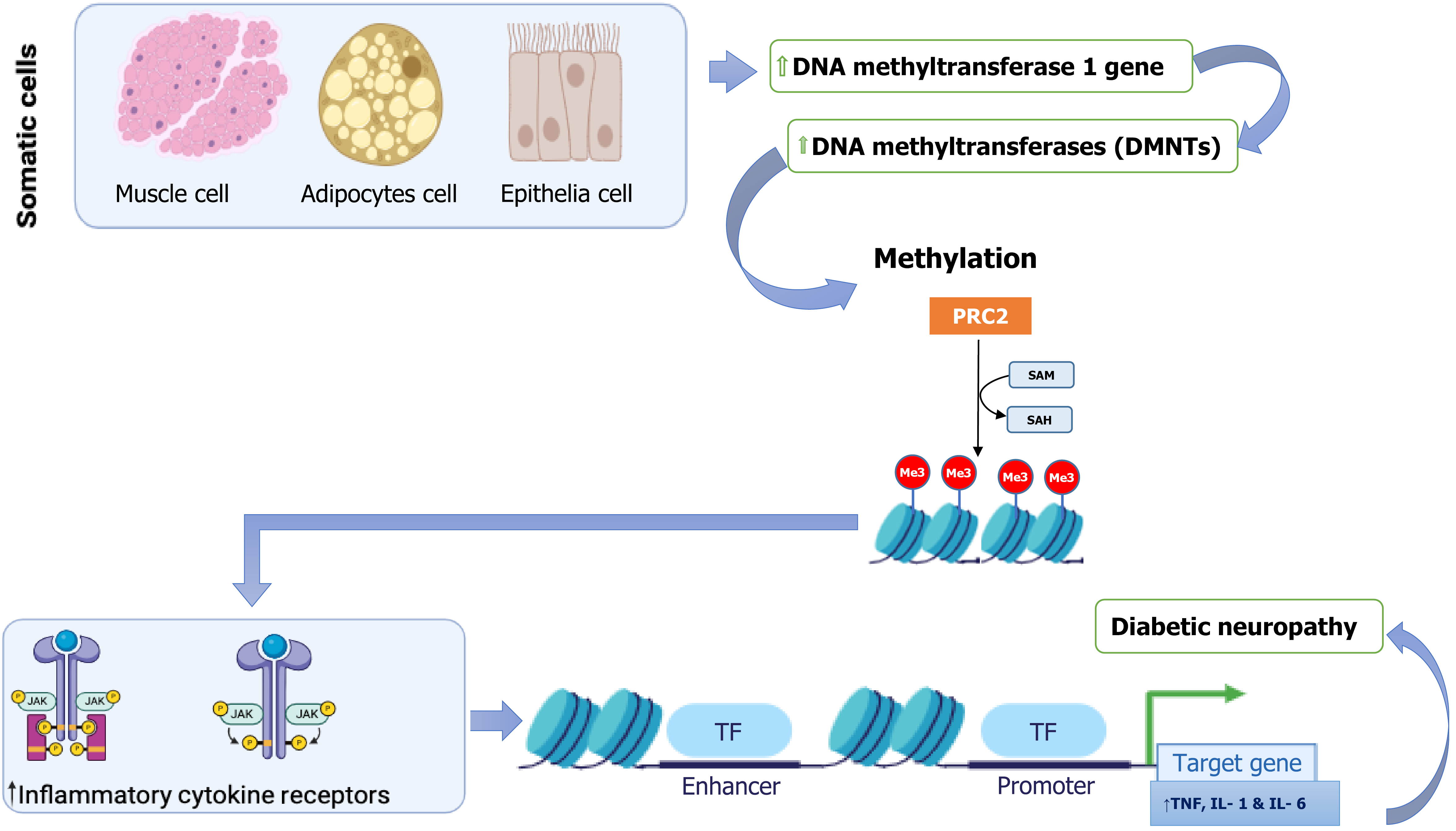
Figure 3 Impact of DNA methylation on inflammatory responses in diabetic neuropathy.
This schematic illustrates the role of aberrant DNA methylation in promoting inflammatory signaling pathways implicated in diabetic neuropathy. In somatic cells (muscle, adipocytes, epithelial), hyperactivation of DNA methylation machinery, including DNA-methyltransferase 1 (DNMT1) and other DNMTs, leads to increased chromatin methylation. The polycomb repressive complex 2 (PRC2) and S-adenosylmethionine (SAM) facilitate histone methylation (Me3) at key regulatory loci, repressing transcriptional activity by RNA polymerase II (RNAPII). This epigenetic repression alters immune signaling pathways, particularly by upregulating inflammatory cytokine receptors (e.g., Janus kinase/signal transducer and activator of transcription signaling), which in turn enhances transcription of pro-inflammatory cytokine genes, including tumor necrosis factor (TNF), interleukin 1 (IL-1), and IL-6. The persistent expression of these inflammatory mediators contributes to chronic neuroinflammation and the progression of diabetic neuropathy.
- Citation: Sanusi KO, Asiwe JN, Sulaimon FA, Bashar F, Yusuf SK, Abdulkadir HO. Diabetic neuropathy and wound healing: An update on epigenetic crosstalk. World J Diabetes 2025; 16(11): 110428
- URL: https://www.wjgnet.com/1948-9358/full/v16/i11/110428.htm
- DOI: https://dx.doi.org/10.4239/wjd.v16.i11.110428