Published online Feb 15, 2026. doi: 10.4251/wjgo.v18.i2.116345
Revised: November 26, 2025
Accepted: December 10, 2025
Published online: February 15, 2026
Processing time: 86 Days and 17 Hours
Esophageal squamous cell carcinoma (ESCC) remains one of the most lethal malignancies worldwide, with pronounced geographic and ethnic disparities in incidence and outcomes. Rapid advances in genome-wide and sequencing technologies have revealed population-specific mutation spectra and risk loci, high
Core Tip: Ethnic genomic differences in esophageal squamous cell carcinoma (ESCC) reveal distinct genetic risk loci, mutational signatures, and gene-environment interactions that shape disease susceptibility across populations. Integrating multi-ethnic, multi-omics data is crucial for developing personalized strategies for prevention, diagnosis, and treatment, addressing disparities in ESCC outcomes. Ethnic genomic differences in ESCC reveal distinct genetic risk loci, mutational signatures, and gene-environment interactions that shape disease susceptibility across populations. Integrating multi-ethnic, multi-omics data is crucial for developing personalized strategies for prevention, diagnosis, and treatment, addressing disparities in ESCC outcomes.
- Citation: Li WM, Jiao Y, Liu SQ, Wang CX, He M. Ethnic genomic diversity in esophageal squamous cell carcinoma. World J Gastrointest Oncol 2026; 18(2): 116345
- URL: https://www.wjgnet.com/1948-5204/full/v18/i2/116345.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v18.i2.116345
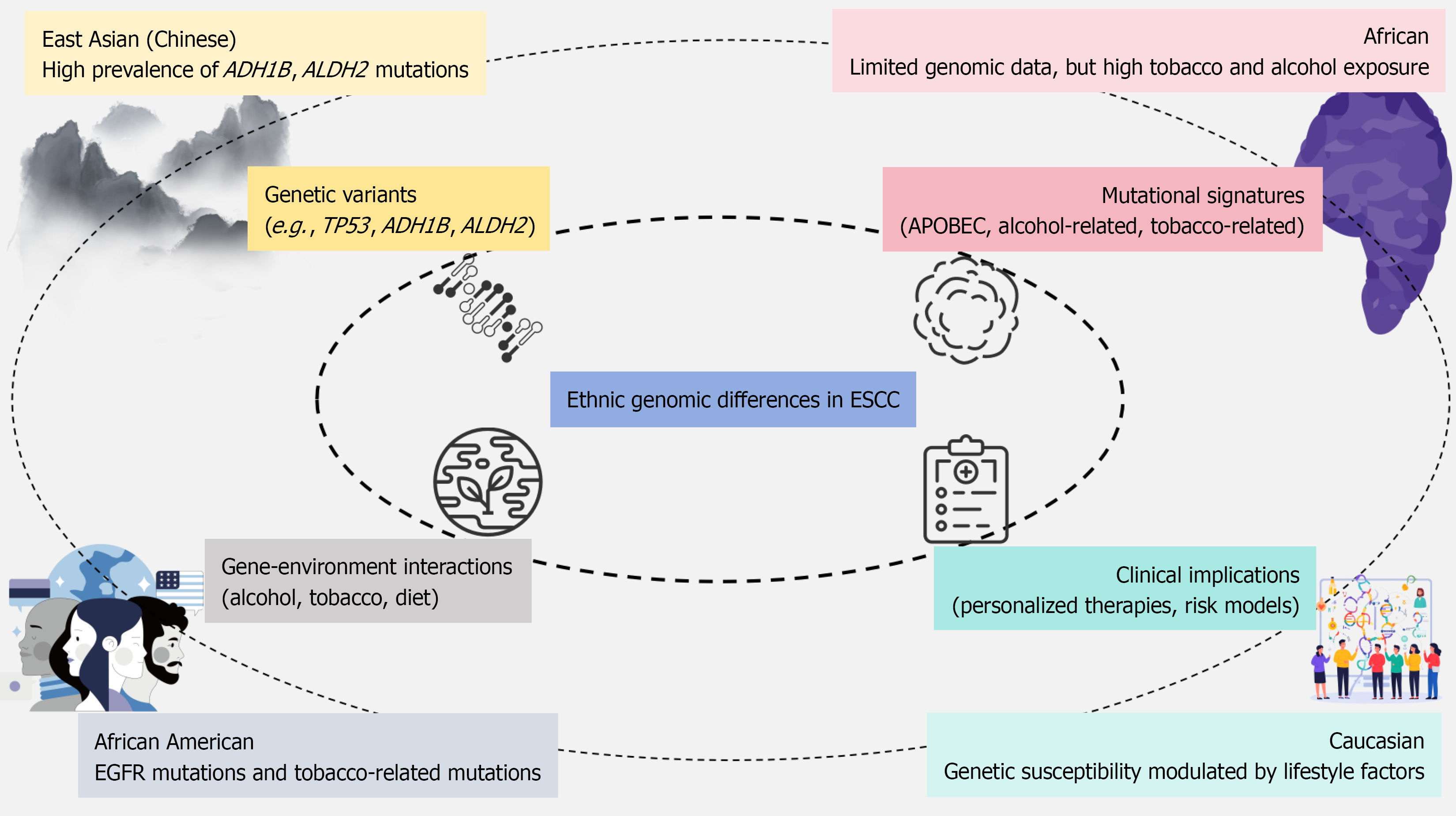
Esophageal squamous cell carcinoma (ESCC) remains one of the deadliest malignancies globally, exhibiting marked ethnic and geographic disparities in incidence and outcomes. In particular, the disease burden is disproportionately high in East Asia and sub-Saharan Africa[1,2]. Despite the advances in treatment options, the prognosis for ESCC remains poor, with authoritative epidemiological reports indicating a five-year survival rate commonly below 25% in high-incidence regions[3-5]. Moreover, recent studies have identified survival disparities among different ethnic groups, with certain African and African American populations experiencing comparatively poorer outcomes than some East Asian cohorts, further highlighting the importance of understanding ethnicity-related factors influencing prognosis[6-8]. This grim survival rate is compounded by the distinct genetic landscapes across different ethnic groups, which influence both the risk and progression of the disease[9,10]. The growing understanding of genetic factors influencing ESCC risk, such as mutations in genes like TP53, ADH1B, and ALDH2, underscores the importance of tailoring prevention and treatment strategies to the genetic backgrounds of diverse populations[11,12]. Recent technological advancements, such as genome-wide association studies and next-generation sequencing, have elucidated these ethnic-specific variations, revealing a complex interplay between inherited susceptibility and environmental exposures[2,12]. These major ethnic genomic patterns are illustrated in Figure 1.
Numerous studies have identified shared and distinct genetic risk loci associated with ESCC across ethnic populations. For instance, common susceptibility genes such as TP53, ADH1B, ALDH2, and PIK3CA have been implicated in ESCC risk across diverse groups, with certain alleles showing different prevalence and impacts based on ethnicity[2,11]. For example, the ALDH2 variant exhibits a high allele frequency of approximately 25%-40% in East Asian populations but is nearly absent in African and Caucasian groups, while ADH1B Arg47His shows a frequency above 60% in East Asians compared with < 5% in Africans and Caucasians[13,14]. Genetic variants in alcohol-metabolizing enzymes, especially ADH1B and ALDH2, have been particularly significant in East Asian populations, where these polymorphisms significantly modify the risk of ESCC in those with heavy alcohol consumption[15-17]. Meta-analytic data further demonstrate that carriers of the ALDH2 risk allele who consume alcohol have a markedly elevated risk [odds ratio (OR = 4.0-6.0)], whereas the corresponding OR in non-East Asian cohorts is substantially lower due to minimal allele prevalence[18-20]. These findings emphasize the critical role of gene-environment interactions in determining the risk of ESCC, where genetic predispositions interact with environmental factors such as alcohol and tobacco use[15,21]. However, studies conducted in African cohorts remain limited, and while loci such as CHEK2 have been identified in both African and Chinese populations, there is still insufficient data to fully understand the genetic predisposition to ESCC in these underrepresented groups[22,23]. A comparative overview of these population-specific variations is summarized in Table 1.
| Study population | Genetic variants | Mutational signatures | Gene-environment interactions | Clinical implications |
| East Asian (Chinese) | TP53, ADH1B, ALDH2, PIK3CA, PLCE1 | APOBEC, alcohol-related signature | Strong interaction with alcohol and tobacco | High risk due to alcohol metabolism variants, targeted therapies |
| African American | TP53, EGFR, CHEK2, BRCA2 | APOBEC, tobacco-related signature | Tobacco and alcohol significantly influence risk | Poor prognosis linked to EGFR mutations and tobacco use |
| African (sub-Saharan) | CHEK2, ADH1B, ALDH2 | Tobacco-related signatures | Limited data on gene-environment interactions | Need for more genetic studies and tailored interventions |
| Caucasian | TP53, GSTP1, PIK3CA, EGFR | Alcohol-related mutational signature | Tobacco use and genetic interaction with alcohol | Risk assessment models considering both genetic and lifestyle factors |
| Kazakh | ADH1B, ALDH2, TP53 | APOBEC, alcohol-related signature | Tobacco, alcohol, and dietary habits | Gene-environment modeling crucial for prevention strategies |
Mutational signature profiling has been a key focus in understanding the genomic landscape of ESCC, revealing a variety of mutations that differ across populations. The APOBEC-related mutational signature, commonly associated with tobacco and alcohol exposure, has been frequently observed in ESCC tumors, particularly in East Asian populations, highlighting the role of environmental carcinogens in shaping the mutational landscape of the disease[24,25]. Representative genomic profiling studies report that APOBEC signatures account for approximately 25%-45% of the total mutational burden in Chinese ESCC samples, compared with 10%-20% in African ESCC cohorts[26]. Studies have shown that mutational signatures, such as those related to alcohol metabolism, correlate with genetic variants in alcohol-metabolizing genes like ALDH2 and ADH1B, especially in populations where alcohol consumption is prevalent[15-17]. Additionally, genomic analyses reveal that mutations in KMT2D and NOTCH1 occur in 20%-30% and 15%-25% of northern Chinese ESCC cases, respectively, but appear at substantially lower frequencies (< 10%) in several African datasets, highlighting geographic and ethnic variation in somatic mutation spectra[27-30]. These mutations, often influenced by regional environmental exposures, contribute to the heterogeneity of ESCC, underscoring the complexity of its pathogenesis across different populations[12,21].
Gene-environment interactions play a pivotal role in determining the susceptibility to ESCC, with environmental factors such as alcohol, tobacco, and dietary habits influencing the effects of genetic variants. Polymorphisms in ADH1B and ALDH2, which modulate alcohol metabolism, are strongly associated with increased ESCC risk in East Asian populations, particularly among those who consume alcohol[21,31]. These findings highlight the importance of gene-environment interactions in modulating disease risk and suggest that genetic predispositions may amplify the carcinogenic effects of environmental exposures such as tobacco smoking and alcohol consumption[15,32]. In African populations, although tobacco and alcohol consumption are prevalent, gene-environment interaction studies remain sparse[33,34], limiting our understanding of the full scope of these interactions. Some studies have also investigated other lifestyle factors, such as oral health and diet, in Chinese cohorts, showing that genetic risk can be modulated by environmental exposures like smoking and diet[35]. Mechanistically, ALDH2 and ADH1B risk alleles impair the clearance of acetaldehyde, leading to increased intracellular accumulation of this carcinogenic metabolite, which in turn promotes DNA adduct formation, mutagenesis, and chronic epithelial inflammation[36,37]. Tobacco-related carcinogens such as N-nitrosamines and polycyclic aromatic hydrocarbons activate oxidative-stress pathways and overwhelm detoxification systems, particularly in individuals with reduced-function alleles in detoxification genes, thereby enhancing genomic instability[38,39]. Environmental exposures can also induce epigenetic alterations, including promoter hypermethylation of tumor suppressor genes and dysregulated histone modifications, which further contribute to aberrant transcriptional programs in ESCC. In addition, chronic alcohol and tobacco exposure triggers pro-inflammatory signaling and immune microenvironment remodeling, creating a permissive milieu for malignant transformation, especially in genetically susceptible populations. These findings emphasize the need for comprehensive studies that include a broader range of environmental factors, particularly in underrepresented populations.
Transcriptomic analyses across various ethnic populations have uncovered significant differences in gene expression patterns that may contribute to ethnic disparities in ESCC. For instance, African American ESCC tumors exhibit differential expression in immune-related pathways and detoxification processes, such as the NRF2 pathway, which is critical for cellular response to oxidative stress and carcinogen exposure[24,40,41]. Epigenomic studies have also revealed consistent tumor-specific methylation alterations, which are potential biomarkers for early detection and prognosis. For example, methylation changes in genes like PAX9 and SIM2 have been identified across multiple high-incidence populations, suggesting their potential utility as diagnostic or prognostic markers[42]. However, despite these advances, the integration of multi-omics data, including transcriptomic and epigenomic profiles, remains limited, particularly in non-Asian populations. Moreover, there is a lack of longitudinal data to assess how these epigenetic changes evolve during tumor progression[40,41,43].
The identification of specific genetic mutations and mutational signatures in ESCC has significant clinical implications for diagnosis, prognosis, and treatment. For instance, mutations in genes like NOTCH1, KMT2D, and PIK3CA have been linked to poor prognosis and resistance to chemoradiotherapy in Chinese ESCC patients, highlighting the potential of these markers for guiding treatment decisions[44,45]. Moreover, the discovery of recurrent somatic mutations and copy number alterations, such as epidermal growth factor receptor mutations in African American ESCC, offers promising therapeutic targets[24]. These findings suggest that precision medicine, informed by genetic profiling, could improve treatment outcomes by selecting therapies tailored to the unique genomic landscape of each patient's tumor[46]. However, the clinical application of these findings is often hindered by disparities in access to genomic testing and targeted therapies across different populations[47].
Despite the wealth of information emerging from genomic studies on ESCC, several challenges remain. One significant limitation is the underrepresentation of certain ethnic groups, particularly African populations, in genomic research. This gap in diversity hinders the development of comprehensive, population-wide risk models and therapeutic strategies[34]. Additionally, many studies suffer from small sample sizes, which limit the statistical power of the findings and their generalizability to larger populations[22,44]. Methodological variability across studies, including differences in sequencing platforms, data processing pipelines, and analytical approaches, introduces further challenges in comparing results across studies[34]. The lack of longitudinal data also restricts the ability to track the progression of genetic mutations and their impact on clinical outcomes[26,48].
To address these challenges, future research must prioritize large-scale, multi-ethnic studies that incorporate diverse populations, particularly African and mixed-ancestry cohorts, to provide a more comprehensive understanding of ESCC pathogenesis. Additionally, integrating multi-omics approaches, including genomic, transcriptomic, and epigenomic data, will be crucial for uncovering the full complexity of ESCC and its ethnic variations. More robust models of gene-environment interactions are also needed, particularly those that include a broader range of environmental exposures, to improve risk prediction and prevention strategies. Furthermore, functional validation of identified genetic variants and biomarkers is essential for translating genomic findings into clinical practice. Finally, the development of population-specific genetic risk scores, incorporating both genetic and environmental factors, could significantly enhance precision medicine strategies for ESCC.
In conclusion, the genetic landscape of ESCC is profoundly shaped by ethnic variations, with distinct mutational signatures and susceptibility loci identified across different populations. While advances in genomic, transcriptomic, and epigenomic profiling have shed light on the complex interplay between genetic predisposition and environmental exposures, significant gaps remain, particularly in underrepresented populations like Africans. These disparities underscore the need for more inclusive, multi-ethnic studies that integrate genetic and environmental factors to better understand ESCC pathogenesis. Future research should focus on functional validation of genetic variants, incorporation of multi-omics approaches, and the development of precision medicine strategies tailored to diverse ethnic groups. Addressing these gaps will be crucial for improving risk stratification, early detection, and personalized therapies for ESCC, ultimately contributing to better clinical outcomes across all populations.
| 1. | Chen WC, Brandenburg JT, Choudhury A, Hayat M, Sengupta D, Swiel Y, Babb de Villiers C, Ferndale L, Aldous C, Soo CC, Lee S, Curtis C, Newton R, Waterboer T, Sitas F, Bradshaw D, Abnet CC, Ramsay M, Parker MI, Singh E, Lewis CM, Mathew CG. Genome-wide association study of esophageal squamous cell cancer identifies shared and distinct risk variants in African and Chinese populations. Am J Hum Genet. 2023;110:1690-1703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 2. | Van Loon K, Mmbaga EJ, Mushi BP, Selekwa M, Mwanga A, Akoko LO, Mwaiselage J, Mosha I, Ng DL, Wu W, Silverstein J, Mulima G, Kaimila B, Gopal S, Snell JM, Benz SC, Vaske C, Sanborn Z, Sedgewick AJ, Radenbaugh A, Newton Y, Collisson EA. A Genomic Analysis of Esophageal Squamous Cell Carcinoma in Eastern Africa. Cancer Epidemiol Biomarkers Prev. 2023;32:1411-1420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 3. | Karagoz K, Lehman HL, Stairs DB, Sinha R, Arga KY. Proteomic and Metabolic Signatures of Esophageal Squamous Cell Carcinoma. Curr Cancer Drug Targets. 2016;. [PubMed] |
| 4. | Shi Y, Ge X, Ju M, Zhang Y, Di X, Liang L. Circulating Tumor Cells in Esophageal Squamous Cell Carcinoma - Mini Review. Cancer Manag Res. 2021;13:8355-8365. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 5. | Xie F, Qiu J, Sun C, Feng L, Jun Y, Luo C, Guo X, Zhang B, Zhou Y, Wang Y, Zhang L, Wang Q. Development of a Specific Aptamer-Modified Nano-System to Treat Esophageal Squamous Cell Carcinoma. Adv Sci (Weinh). 2024;11:e2309084. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 6. | Tehranifar P, Neugut AI, Phelan JC, Link BG, Liao Y, Desai M, Terry MB. Medical advances and racial/ethnic disparities in cancer survival. Cancer Epidemiol Biomarkers Prev. 2009;18:2701-2708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 91] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 7. | Coker AL, Desimone CP, Eggleston KS, White AL, Williams M. Ethnic disparities in cervical cancer survival among Texas women. J Womens Health (Larchmt). 2009;18:1577-1583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 8. | Bandera EV, Lee VS, Rodriguez-Rodriguez L, Powell CB, Kushi LH. Racial/Ethnic Disparities in Ovarian Cancer Treatment and Survival. Clin Cancer Res. 2016;22:5909-5914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 94] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 9. | Erkizan C, Wadleigh R. Abstract 7356: 3 UTR variations in esophageal squamous cell carcinoma in African Americans. Cancer Res. 2024;84:7356. [DOI] [Full Text] |
| 10. | Chen S, Zhou K, Yang L, Ding G, Li H. Racial Differences in Esophageal Squamous Cell Carcinoma: Incidence and Molecular Features. Biomed Res Int. 2017;2017:1204082. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 11. | Deng J, Chen H, Zhou D, Zhang J, Chen Y, Liu Q, Ai D, Zhu H, Chu L, Ren W, Zhang X, Xia Y, Sun M, Zhang H, Li J, Peng X, Li L, Han L, Lin H, Cai X, Xiang J, Chen S, Sun Y, Zhang Y, Zhang J, Chen H, Zhang S, Zhao Y, Liu Y, Liang H, Zhao K. Comparative genomic analysis of esophageal squamous cell carcinoma between Asian and Caucasian patient populations. Nat Commun. 2017;8:1533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 86] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 12. | Guo J, Huang J, Zhou Y, Zhou Y, Yu L, Li H, Hou L, Zhu L, Ge D, Zeng Y, Guleng B, Li Q. Germline and somatic variations influence the somatic mutational signatures of esophageal squamous cell carcinomas in a Chinese population. BMC Genomics. 2018;19:538. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 13. | Deiana G, Sun R, Huang J, Napolioni V, Ciccocioppo R. Contribution of infectious diseases to the selection of ADH1B and ALDH2 gene variants in Asian populations. Alcohol Clin Exp Res (Hoboken). 2024;48:855-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 14. | Ahn KS, Abdiev S, Rahimov B, Malikov Y, Bahramov S, Okada R, Naito M, Hamajima N. Alcohol dehydrogenase 1B and Aldehyde dehydrogenase 2 Polymorphisms in Uzbekistan. Asian Pac J Cancer Prev. 2009;10:17-20. [PubMed] |
| 15. | Tanaka F, Yamamoto K, Suzuki S, Inoue H, Tsurumaru M, Kajiyama Y, Kato H, Igaki H, Furuta K, Fujita H, Tanaka T, Tanaka Y, Kawashima Y, Natsugoe S, Setoyama T, Tokudome S, Mimori K, Haraguchi N, Ishii H, Mori M. Strong interaction between the effects of alcohol consumption and smoking on oesophageal squamous cell carcinoma among individuals with ADH1B and/or ALDH2 risk alleles. Gut. 2010;59:1457-1464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 112] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 16. | Zhang GH, Mai RQ, Huang B. Meta-analysis of ADH1B and ALDH2 polymorphisms and esophageal cancer risk in China. World J Gastroenterol. 2010;16:6020-6025. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 17. | Zhang G, Mai R, Huang B. ADH1B Arg47His polymorphism is associated with esophageal cancer risk in high-incidence Asian population: evidence from a meta-analysis. PLoS One. 2010;5:e13679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 18. | Hirohashi K, Ohashi S, Amanuma Y, Nakai Y, Ida T, Baba K, Mitani Y, Mizumoto A, Yamamoto Y, Kikuchi O, Matsubara J, Yamada A, Miyamoto S, Seno H, Matsuda T, Muto M. Protective effects of Alda-1, an ALDH2 activator, on alcohol-derived DNA damage in the esophagus of human ALDH2*2 (Glu504Lys) knock-in mice. Carcinogenesis. 2020;41:194-202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 19. | Ding JH, Li SP, Cao HX, Wu JZ, Gao CM, Liu YT, Zhou JN, Chang J, Yao GH. Alcohol dehydrogenase-2 and aldehyde dehydrogenase-2 genotypes, alcohol drinking and the risk for esophageal cancer in a Chinese population. J Hum Genet. 2010;55:97-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 20. | Seitz HK, Stickel F. Acetaldehyde as an underestimated risk factor for cancer development: role of genetics in ethanol metabolism. Genes Nutr. 2010;5:121-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 204] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 21. | Wu C, Kraft P, Zhai K, Chang J, Wang Z, Li Y, Hu Z, He Z, Jia W, Abnet CC, Liang L, Hu N, Miao X, Zhou Y, Liu Z, Zhan Q, Liu Y, Qiao Y, Zhou Y, Jin G, Guo C, Lu C, Yang H, Fu J, Yu D, Freedman ND, Ding T, Tan W, Goldstein AM, Wu T, Shen H, Ke Y, Zeng Y, Chanock SJ, Taylor PR, Lin D. Genome-wide association analyses of esophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene-environment interactions. Nat Genet. 2012;44:1090-1097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 221] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 22. | Erkizan HV, Sukhadia S, Natarajan TG, Marino G, Notario V, Lichy JH, Wadleigh RG. Exome sequencing identifies novel somatic variants in African American esophageal squamous cell carcinoma. Sci Rep. 2021;11:14814. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 23. | Chen WC, Bye H, Matejcic M, Amar A, Govender D, Khew YW, Beynon V, Kerr R, Singh E, Prescott NJ, Lewis CM, Babb de Villiers C, Parker MI, Mathew CG. Association of genetic variants in CHEK2 with oesophageal squamous cell carcinoma in the South African Black population. Carcinogenesis. 2019;40:513-520. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 24. | Erkizan HV, Johnson K, Ghimbovschi S, Karkera D, Trachiotis G, Adib H, Hoffman EP, Wadleigh RG. African-American esophageal squamous cell carcinoma expression profile reveals dysregulation of stress response and detox networks. BMC Cancer. 2017;17:426. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 25. | Du P, Huang P, Huang X, Li X, Feng Z, Li F, Liang S, Song Y, Stenvang J, Brünner N, Yang H, Ou Y, Gao Q, Li L. Comprehensive genomic analysis of Oesophageal Squamous Cell Carcinoma reveals clinical relevance. Sci Rep. 2017;7:15324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 26. | Moody S, Senkin S, Islam SMA, Wang J, Nasrollahzadeh D, Cortez Cardoso Penha R, Fitzgerald S, Bergstrom EN, Atkins J, He Y, Khandekar A, Smith-Byrne K, Carreira C, Gaborieau V, Latimer C, Thomas E, Abnizova I, Bucciarelli PE, Jones D, Teague JW, Abedi-Ardekani B, Serra S, Scoazec JY, Saffar H, Azmoudeh-Ardalan F, Sotoudeh M, Nikmanesh A, Poustchi H, Niavarani A, Gharavi S, Eden M, Richman P, Campos LS, Fitzgerald RC, Ribeiro LF, Soares-Lima SC, Dzamalala C, Mmbaga BT, Shibata T, Menya D, Goldstein AM, Hu N, Malekzadeh R, Fazel A, McCormack V, McKay J, Perdomo S, Scelo G, Chanudet E, Humphreys L, Alexandrov LB, Brennan P, Stratton MR. Mutational signatures in esophageal squamous cell carcinoma from eight countries with varying incidence. Nat Genet. 2021;53:1553-1563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 117] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 27. | Zhang R, Li C, Wan Z, Qin J, Li Y, Wang Z, Zheng Q, Kang X, Chen X, Li Y, He J, Li Y. Comparative genomic analysis of esophageal squamous cell carcinoma among different geographic regions. Front Oncol. 2022;12:999424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 28. | Naseri A, Salehi-Pourmehr H, Majidazar R, Seraji P, Rezazadeh-Gavgani E, Zehtabi M, Kiani-Kezbin H, Salehnia F, Hassannezhad S, Hajikamanj A, Raeisi M. Systematic Review and Meta-analysis of the Most Common Genetic Mutations in Esophageal Squamous Cell Carcinoma. J Gastrointest Cancer. 2022;53:1040-1049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 29. | Zhang N, Shi J, Shi X, Chen W, Liu J. Mutational Characterization and Potential Prognostic Biomarkers of Chinese Patients with Esophageal Squamous Cell Carcinoma. Onco Targets Ther. 2020;13:12797-12809. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 30. | Salem ME, Puccini A, Xiu J, Raghavan D, Lenz HJ, Korn WM, Shields AF, Philip PA, Marshall JL, Goldberg RM. Comparative Molecular Analyses of Esophageal Squamous Cell Carcinoma, Esophageal Adenocarcinoma, and Gastric Adenocarcinoma. Oncologist. 2018;23:1319-1327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 148] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 31. | Chen YJ, Chen C, Wu DC, Lee CH, Wu CI, Lee JM, Goan YG, Huang SP, Lin CC, Li TC, Chou YP, Wu MT. Interactive effects of lifetime alcohol consumption and alcohol and aldehyde dehydrogenase polymorphisms on esophageal cancer risks. Int J Cancer. 2006;119:2827-2831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 73] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 32. | Lee CH, Lee JM, Wu DC, Goan YG, Chou SH, Wu IC, Kao EL, Chan TF, Huang MC, Chen PS, Lee CY, Huang CT, Huang HL, Hu CY, Hung YH, Wu MT. Carcinogenetic impact of ADH1B and ALDH2 genes on squamous cell carcinoma risk of the esophagus with regard to the consumption of alcohol, tobacco and betel quid. Int J Cancer. 2008;122:1347-1356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 92] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 33. | Matejcic M, Mathew CG, Parker MI. The Relationship Between Environmental Exposure and Genetic Architecture of the 2q33 Locus With Esophageal Cancer in South Africa. Front Genet. 2019;10:406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 34. | Simba H, Kuivaniemi H, Lutje V, Tromp G, Sewram V. Systematic Review of Genetic Factors in the Etiology of Esophageal Squamous Cell Carcinoma in African Populations. Front Genet. 2019;10:642. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 35. | Zhao R, Li X, Yang X, Zhang T, Lu M, Ye W, Jin L, Suo C, Chen X. Association of Esophageal Squamous Cell Carcinoma With the Interaction Between Poor Oral Health and Single Nucleotide Polymorphisms in Regulating Cell Cycles and Angiogenesis: A Case-Control Study in High-Incidence Chinese. Cancer Control. 2022;29:10732748221075811. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 36. | Yukawa Y, Muto M, Hori K, Nagayoshi H, Yokoyama A, Chiba T, Matsuda T. Combination of ADH1B*2/ALDH2*2 polymorphisms alters acetaldehyde-derived DNA damage in the blood of Japanese alcoholics. Cancer Sci. 2012;103:1651-1655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 37. | Yu HS, Oyama T, Isse T, Kitagawa K, Pham TT, Tanaka M, Kawamoto T. Formation of acetaldehyde-derived DNA adducts due to alcohol exposure. Chem Biol Interact. 2010;188:367-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 119] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 38. | Korenjak M, Temiz NA, Keita S, Chavanel B, Renard C, Sirand C, Cahais V, Mayel T, Vevang KR, Jacobs FC, Guo J, Smith WE, Oram MK, Tăbăran FA, Ahlat O, Cornax I, O'Sullivan MG, Das S, Nandi SP, Cheng Y, Alexandrov LB, Balbo S, Hecht SS, Senkin S, Virard F, Peterson LA, Zavadil J. Human cancer genomes harbor the mutational signature of tobacco-specific nitrosamines NNN and NNK. bioRxiv. 2024;2024.06.28.600253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 39. | Ding YS, Zhang L, Jain RB, Jain N, Wang RY, Ashley DL, Watson CH. Levels of tobacco-specific nitrosamines and polycyclic aromatic hydrocarbons in mainstream smoke from different tobacco varieties. Cancer Epidemiol Biomarkers Prev. 2008;17:3366-3371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 75] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 40. | Sharip A, Rakhimova S, Molkenov A, Seisenova A, Kozhamkulov U, Akhmetollayev I, Zhukov Y, Omarov M, Tuleutaev M, Akilzhanova A, Kairov U. Transcriptome profiling and examination of individuals diagnosed with esophageal squamous cell carcinoma originating from Kazakhstan. Eurasian J Appl Biotechnol. 2024;3S:65. [DOI] [Full Text] |
| 41. | Sharip A, Rakhimova S, Molkenov A, Ashenova A, Kozhamkulov U, Akhmetollayev I, Zinovyev A, Zhukov Y, Omarov M, Tuleutaev M, Rakhmetova V, Terwilliger JD, Lee JH, Zhumadilov Z, Akilzhanova A, Kairov U. Transcriptome profiling and analysis of patients with esophageal squamous cell carcinoma from Kazakhstan. Front Genet. 2024;15:1249751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 42. | Talukdar FR, Soares Lima SC, Khoueiry R, Laskar RS, Cuenin C, Sorroche BP, Boisson A, Abedi-ardekani B, Carreira C, Menya D, Dzamalala CP, Assefa M, Aseffa A, Miranda-gonçalves V, Jerónimo C, Henrique RM, Shakeri R, Malekzadeh R, Gasmelseed N, Ellaithi M, Gangane N, Middleton DR, Le Calvez-Kelm F, Ghantous A, Roux ML, Schüz J, Mccormack V, Parker MI, Pinto LFR, Herceg Z. Data from Genome-Wide DNA Methylation Profiling of Esophageal Squamous Cell Carcinoma from Global High-Incidence Regions Identifies Crucial Genes and Potential Cancer Markers. Mar 31, 2023. [cited 8 December 2025]. Available from: https://aacr.figshare.com/collections/Data_from_Genome-Wide_DNA_Methylation_Profiling_of_Esophageal_Squamous_Cell_Carcinoma_from_Global_High-Incidence_Regions_Identifies_Crucial_Genes_and_Potential_Cancer_Markers/6512658. |
| 43. | Talukdar FR, Soares Lima SC, Khoueiry R, Laskar RS, Cuenin C, Sorroche BP, Boisson AC, Abedi-Ardekani B, Carreira C, Menya D, Dzamalala CP, Assefa M, Aseffa A, Miranda-Gonçalves V, Jerónimo C, Henrique RM, Shakeri R, Malekzadeh R, Gasmelseed N, Ellaithi M, Gangane N, Middleton DRS, Le Calvez-Kelm F, Ghantous A, Roux ML, Schüz J, McCormack V, Parker MI, Pinto LFR, Herceg Z. Genome-Wide DNA Methylation Profiling of Esophageal Squamous Cell Carcinoma from Global High-Incidence Regions Identifies Crucial Genes and Potential Cancer Markers. Cancer Res. 2021;81:2612-2624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 44. | Liu M, An H, Zhang Y, Sun W, Cheng S, Wang R, Wang X, Feng L. Molecular analysis of Chinese oesophageal squamous cell carcinoma identifies novel subtypes associated with distinct clinical outcomes. EBioMedicine. 2020;57:102831. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 45. | Zhou Y, Chu L, Li S, Chu X, Ni J, Jiang S, Pang Y, Zheng D, Lu Y, Lan F, Cai X, Yang X, Zhu Z. Prognostic value of genomic mutation signature associated with immune microenvironment in southern Chinese patients with esophageal squamous cell carcinoma. Cancer Immunol Immunother. 2024;73:141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 46. | Brown J, Stepien AJ, Willem P. Landscape of copy number aberrations in esophageal squamous cell carcinoma from a high endemic region of South Africa. BMC Cancer. 2020;20:281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 47. | Gao L, Guo RY, Lu H, Shi ZH, Liu JF. Clinical and Genomic Analysis of Patients with Short Survival after Surgery for Esophageal Squamous Cell Carcinoma. Dig Dis. 2023;41:353-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 48. | Song Y, Wang Y, Xu L, Ma J, Chen E, Zang R, Jia W, Tao X, Hu L. A genetic variant in CHRNB3-CHRNA6 increases risk of esophageal squamous cell carcinoma in Chinese populations. Carcinogenesis. 2015;36:538-542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/