Published online Nov 15, 2025. doi: 10.4251/wjgo.v17.i11.112838
Revised: September 19, 2025
Accepted: October 27, 2025
Published online: November 15, 2025
Processing time: 99 Days and 6 Hours
Colorectal cancer (CRC) is the second leading cause of cancer-related death, lar
To investigate the molecular heterogeneity of primary aCRC in order to identify clinically relevant genomic alterations.
We conducted a retrospective molecular analysis of 73 consecutive patients with histologically confirmed primary aCRC (stage pT4a-b). All molecular findings were correlated with available clinicopathological data. In addition, we performed survival analyses using publicly available datasets and tools.
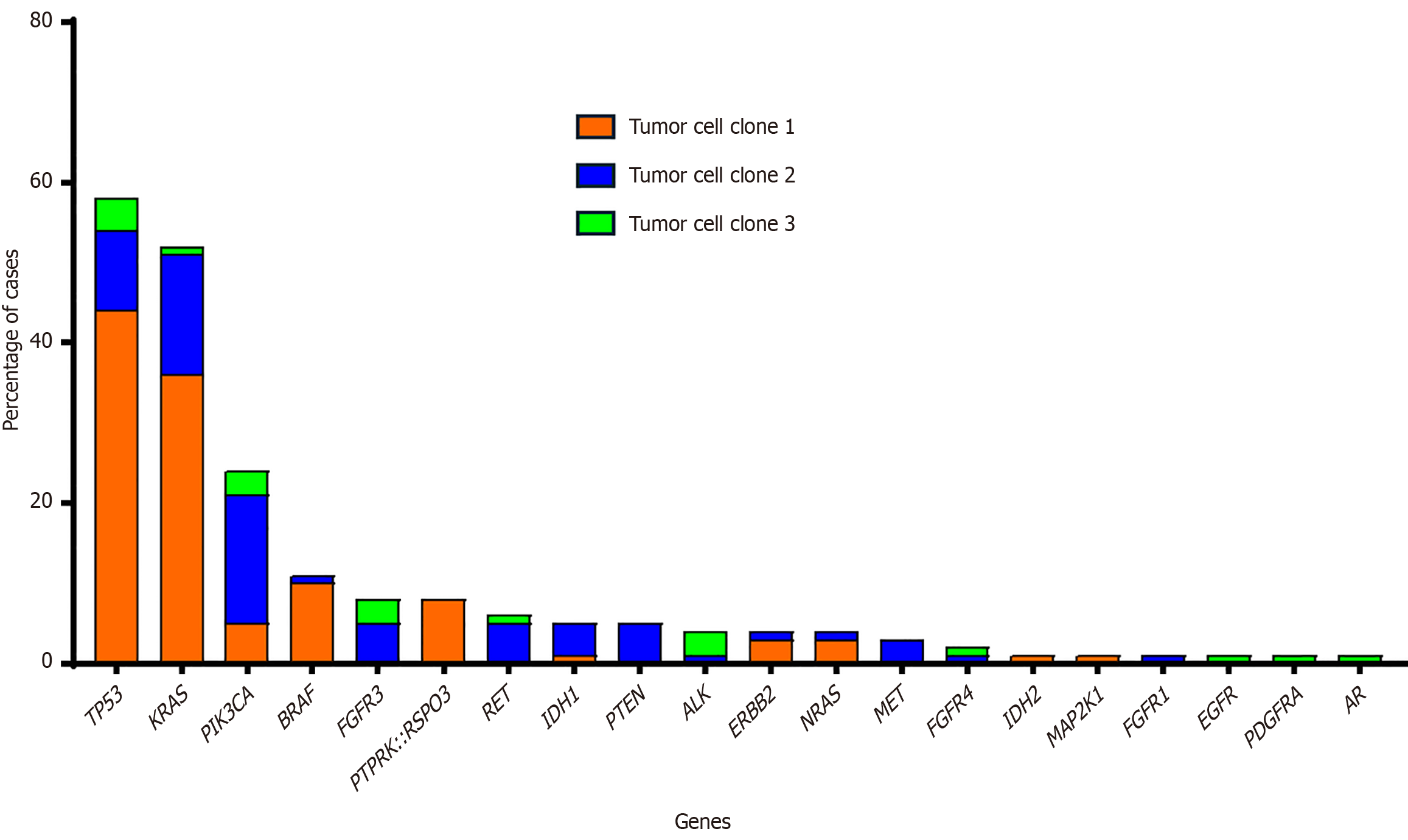
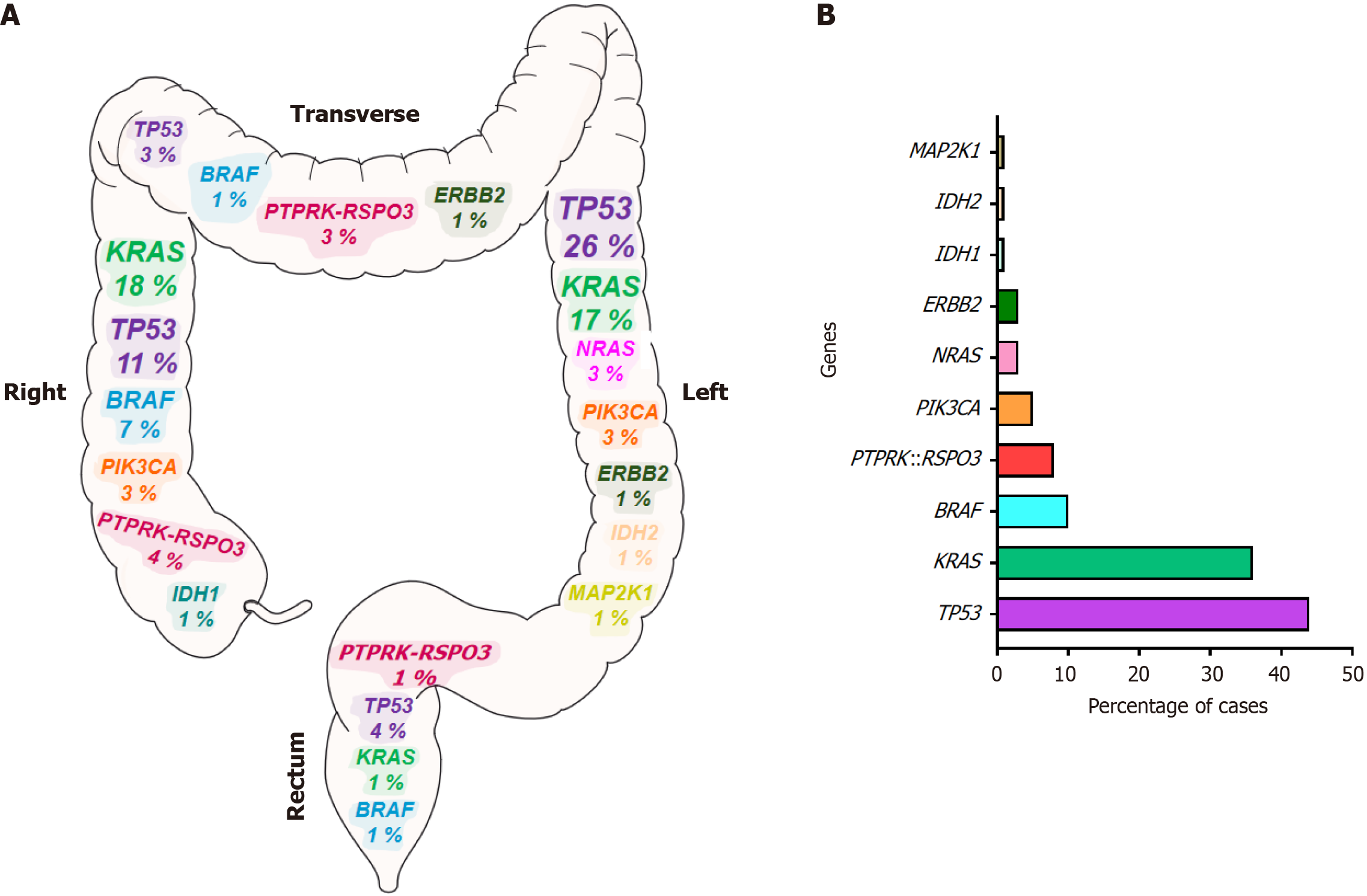
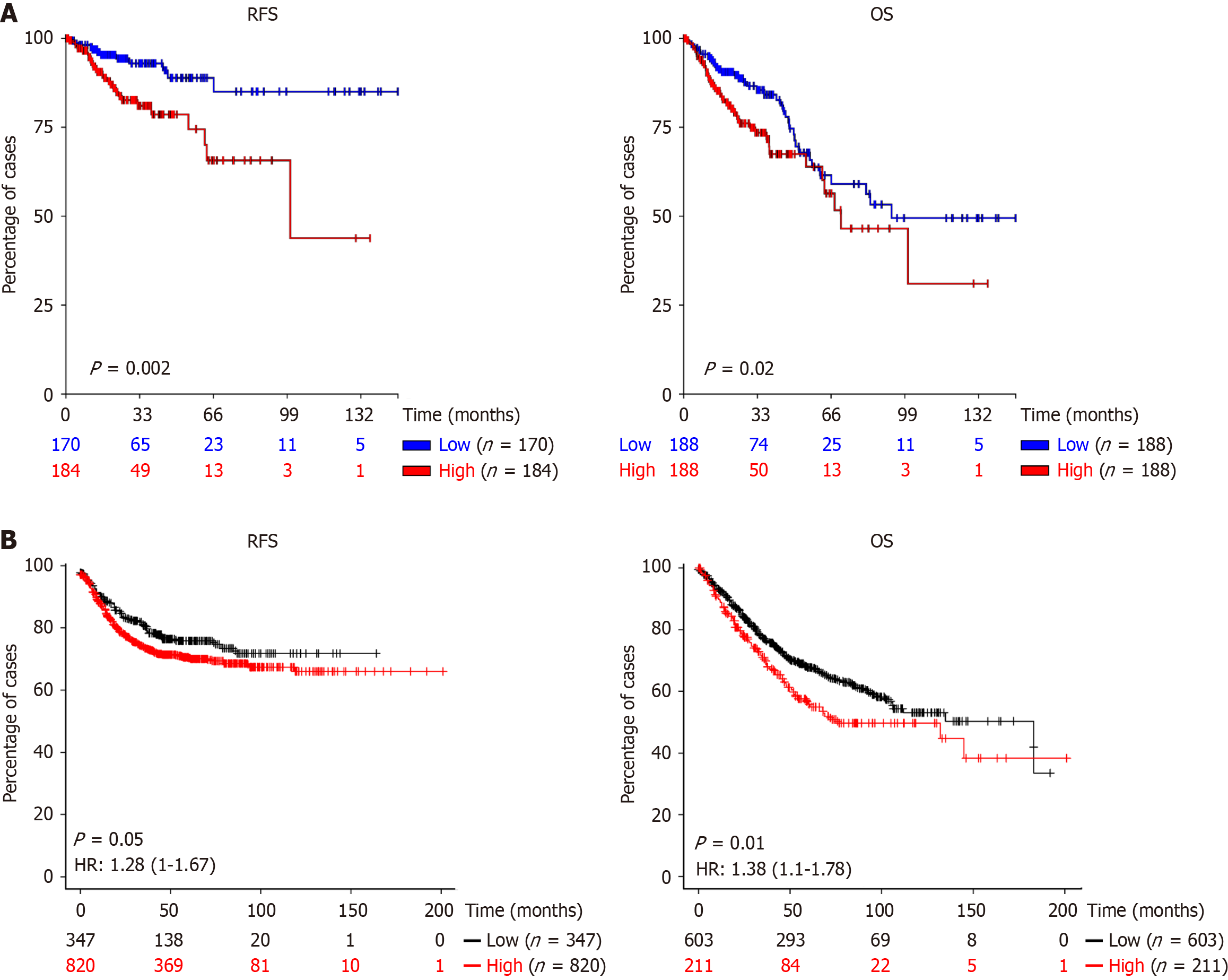
Genetic abnormalities identified in primary tumors were most frequently mutations in tumor protein p53 (58% of cases), Kirsten rat sarcoma viral oncogene homolog (52%), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (25%), B-Raf kinase (11%) and fibroblast growth factor receptor 3 (8%), as well as R-spondin 3 (RSPO3) fusions (8%). Alterations in the tumor protein p53 and neuroblastoma RAS viral oncogene homolog genes were predominantly observed in tumors from the left colon, whereas B-Raf kinase mutations and RSPO3 fusions were more frequently detected in the right or transverse colon. We also show a strong association between the presence of RSPO3 rearrangements and patients with small tumors, normal carcinoembryonic antigen levels, and microsatellite stable tumors. Furthermore, aCRC patients with protein tyrosine phosphatase receptor type k::RSPO3 fusions exhibited a higher mortality rate. Elevated RSPO3 gene expression levels were also significantly correlated with poorer OS across two large, independent CRC cohorts.
This study identifies a relatively high incidence of RSPO3 rearrangements in aCRC and a strong association with clinical features. Furthermore, we find that RSPO3 fusions are associated with poorer OS.
Core Tip: In our study, we identified key genomic alterations in advanced colorectal cancer using next-generation seque
- Citation: Tur R, Abad M, Filipovich E, Rivas MB, Rodriguez M, Montero JC, Sayagués JM. RSPO3 rearrangements in advanced colorectal cancer patients and their relationship with disease characteristics. World J Gastrointest Oncol 2025; 17(11): 112838
- URL: https://www.wjgnet.com/1948-5204/full/v17/i11/112838.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i11.112838
Sporadic colorectal cancer (sCRC) is the third most frequently diagnosed cancer worldwide and is the second most common cause of cancer-related deaths. Mortality is largely due to limited treatment options available for patients diagnosed at advanced stages of the disease[1]. In fact, the five-year survival rate for advanced colorectal cancer (aCRC) is less than 10%[2]. Up to 25% of sCRC cases present with stage IV disease and approximately 25%-50% who initially present with early-stage sCRC go on to develop metastases over the course of the disease[2]. This urgent clinical need has meant that the research currently being carried out by various research groups is focused on the development of safe, effective, and tolerable therapies for advanced disease. Since 2004, cetuximab and panitumumab have been introduced as monoclonal anti-epidermal growth factor receptor (EGFR) antibody therapeutic drugs to treat aCRC[3]. Mutations in any of the genes that are part of the EGFR signaling pathway [Kirsten rat sarcoma viral oncogene homolog (KRAS) and neuroblastoma RAS viral oncogene homolog (NRAS) genes] determine the response to this therapy, for which reason they are used as predictive biomarkers[4]. However, data on the prevalence of mutations in these genes, in different locations of aCRC tumors, are limited. More recently, aCRC patients with deficient mismatch repair (15%) have shown an excellent response to treatment with immune checkpoint inhibitors[5]. Additional therapeutic targets, such as human epidermal growth factor receptor 2 amplification or overexpression (3%-5% of cases), have also led to new molecularly based treatment options, including those with monoclonal antibodies (trastuzumab and pertuzumab) and tyrosine kinase inhibitors (tucatinib)[3]. Likewise, 0.5%-1% of colorectal cancer (CRC) tumors present structural rearrangements in the neurotrophic receptor tyrosine kinase genes and generally exhibit a high tumor mutational load and high degree of microsatellite instability (MSI), which is associated with an adverse prognosis[6]; however, there are two Food and Drug Administration (FDA)-approved neurotrophic receptor tyrosine kinase inhibitor molecules, larotrectinib and entrectinib, that have demonstrated excellent results for patients whose disease has progressed under standard therapy or for those without treatment options[7]. Similarly, selpercatinib received FDA tumor-agnostic approval for refractory advanced solid tumors with a rearranged during transfection gene fusion, based on the results from the LIBRETTO-001 trial[8], reported in < 1% of patients with sCRC[9]. Finally, the FDA approved the combination of encorafenib with cetuximab for patients who received prior treatment for aCRC with B-Raf kinase (BRAF) V600E mutations (8%-10% of cases)[10]. This clearly indicates that, as our understanding of the genomic landscape of tumors deepens, the therapeutic paradigm for aCRC expands. In fact, in the present study we found that of the 73 patients with aCRC analyzed by next-generation sequencing (NGS) techniques, 6 (8% of cases) showed R-spondin 3 (RSPO3) fusions. Several studies have reported that RSPO3 fusions are mutually exclusive with adenomatous polyposis coli mutations and also contribute to the activation of Wnt signaling and the development of sCRC in this subgroup of patients[11-14]. Interestingly, blocks of secretion of Wnt ligand and RSPO3 neutralizing antibodies have shown an antitumoral effect in in vitro and in vivo models of CRC expressing the RSPO3 fusion[15-19]. However, despite abundant evidence that hyperactivation of the Wnt pathway drives CRC and that therapies targeting this pathway have been effective in preclinical models of CRC, targeted Wnt therapies have not progressed in clinical settings[20]. The arrival of effective therapies in the clinical setting that target the Wnt signaling pathway implies that, as part of the daily routine, it will be necessary to evaluate whether components of this pathway are altered in CRC tumors.
In the present study, we focused on the search for new biomarkers using NGS techniques in patients with aCRC to find new targets for the treatment of patients with advanced disease. For this purpose, we analyzed the genetic abnormalities (mutations, copy number and fusions) of 50 genes in 73 patients with aCRC, with a median follow-up of 62 months. We find that RSPO3 fusions occur at a high frequency in advanced disease and are associated with poorer overall survival (OS). Through in silico analyses, we also show that elevated levels of RSPO3 transcripts have an adverse influence on recurrence-free survival (RFS) and OS in two independent series, making them important markers for future therapeutic decision-making.
Seventy-three consecutive aCRC patients who had undergone surgical resection of primary tumor tissues between January 2013 and December 2018 in the Surgical Department of Hospital Nuestra Señora de Sonsoles (Avila, Spain) were included in the study. Informed consent was given by all patients before they entered the study, in accordance with the Declaration of Helsinki. The study was approved by the local ethics committee of the Hospital Nuestra Señora de Sonsoles. Formalin-fixed, paraffin-embedded (FFPE) tissue samples were obtained from surgical tumor resection in all patients (n = 73). The tumors were diagnosed and classified according to the American Joint Committee on Cancer criteria[21] by a pathologist from the Pathology Department of the Hospital Nuestra Señora de Sonsoles. Primary tumors were localized in the rectum (n = 6), or in the transverse (n = 5), right (caecum, ascending; n = 27) or left (descending and sigmoid; n = 35) colon. The median size of the primary tumors was 6.1 ± 2.2 cm. Most patients exhibited tumors on the surface of the visceral peritoneum (T4a; 75% of cases; Table 1).
| Variable | PTPRK::RSPO3 fusion, yes | PTPRK::RSPO3 fusion, no | P value | Total (n = 73) |
| Age (years)1 | 71 (62-90) | 67 (36-94) | NS | 73 (36-94) |
| Gender | ||||
| Female | 3 (50) | 32 (48) | NS | 35 (52) |
| Male | 3 (50) | 35 (52) | 38 (48) | |
| Site of primary tumor | ||||
| Right colon | 3 (50) | 24 (36) | 0.01 | 27 (37) |
| Transverse colon | 2 (33) | 3 (4) | 5 (7) | |
| Left colon | 0 (0) | 35 (52) | 35 (48) | |
| Rectum | 1 (17) | 5 (8) | 6 (8) | |
| Histopathological grade | ||||
| Low | 5 (83) | 59 (88) | NS | 64 (86) |
| High | 1 (17) | 8 (12) | 9 (14) | |
| Lymph node involvement | ||||
| pN0 | 2 (33) | 28 (42) | 29 (40) | |
| pN1 | 3 (50) | 22 (32) | NS | 25 (35) |
| pN2 | 1 (17) | 17 (26) | 18 (25) | |
| Metastasis status | ||||
| M0 | 3 (50) | 47 (70) | NS | 50 (68) |
| M1 | 3 (50) | 20 (30) | 23 (32) | |
| Size (cm) | ||||
| < 5 | 5 (83) | 39 (58) | 0.08 | 44 (60) |
| ≥ 5 | 1 (17) | 28 (42) | NS | 29 (40) |
| Microsatellite instability | ||||
| Yes | 0 (0) | 10 (15) | 0.008 | 10 (14) |
| No | 6 (100) | 57 (85) | 63 (86) | |
| CEA serum levels (ng/mL)1 | 5.7 (0.7-23) | 25 (0.6-825) | 0.009 | 23 (0.6-825) |
| Number of deaths | 5 (83) | 41 (61) | 0.04 | 46 (63) |
| OS (months)1 | 42 (3-53) | 52 (2-133) | 0.002 | 62 (2-133) |
NGS was performed in the University Hospital of Salamanca (Anatomical Pathology Service, Salamanca, Spain) as follows. From each FFPE sample, 1 to 6 sections (5 μm thick) were obtained, deparaffinized, and subjected to tissue lysis. Automated nucleic acid extraction was then carried out using the Genexus™ Purification System (Thermo Fisher, MA, United States). Finally, 20 μL of DNA and RNA (10 ng, quantified with qubit) were used for sequencing on the Ion Torrent™ Genexus™ Integrated Sequencer. This sequencer is a fully automated NGS system that integrates library preparation, template preparation, sequencing, and data analysis. The panel used was the Oncomine Precision Assay, which allows simultaneous detection of hotspot mutations (substitutions, insertions, and deletions), copy number variations, and gene fusions across 50 cancer-related genes using the Ion Torrent GX5 Chip (Supplementary Table 1). Data were analyzed automatically using Oncomine™ Reporter software (Thermo Fisher, MA, United States). The results were filtered using the Variant Matrix Summary filter (version 5.16), which provides a summary of hotspot mutations including the amino acid changes and allelic frequencies of each variant. Furthermore, it indicates the number of readings of detected fusions and provides numerical values for copy number variations. Only results from samples that met the quality parameters for FFPE samples recommended by the manufacturer were considered.
Kaplan-Meier analysis was performed using the UCSC Xena Functional Genomics Explorer online tools at https://xena.ucsc.edu/ and the KMplot online tool (https://kmplot.com/analysis/)[22], accessed on July 15, 2025. These analyses were carried out to assess the prognostic value of RSPO3 gene expression in CRC. For the UCSC Xena analysis, the selected dataset was The Cancer Genome Atlas colon and rectal cancer. Patients were stratified into high- and low-expression groups based on median dichotomization. RFS and OS were used as endpoints for survival assessment. Statistical significance of the differences between survival curves was assessed using one-sided log-rank tests.
All the values of clinical/biological, morphological, genetic, and evolutionary variables were collected in a database. The mean, SD and range of all continuous variables were calculated in SPSS v.21 (IBM Corp., Armonk, NY, United States); dichotomous variables were reported as n (%). To evaluate the statistical significance of group differences, Mann-Whitney U test was used for non-normally distributed continuous variables. For dichotomous variables, the χ2 test was used. Statistical significance was concluded for values of P < 0.05.
Overall, 73 patients diagnosed with aCRC (38 males and 35 females; median age of 73 years, ranging from 36 to 94 years) at the Hospital Nuestra Señora de Sonsoles (Avila; Spain) were studied. Histologically, all cases were adenocarcinomas, with 63 (86%) classified as low-grade and 10 (14%) as high-grade. In all cases, the histopathological grade was inde
All patients underwent complete surgical tumor resection. aCRC cases were more frequently located in the left colon (48% of cases) and showed a higher incidence of lymph node metastases (60%) as well as elevated serum carcinoembryonic antigen (CEA) levels (median of 23.2 ng/mL, ranging from 0.6 ng/mL to 825 ng/mL). As shown in Table 1, most patients (63%) had died by the end of the study. The median follow-up time was 62 months (range 2-133 months).
To identify new biomarkers in patients with aCRC, we performed NGS on 73 primary tumors from these patients. The genetic abnormalities found in primary tumors were most frequently mutations of tumor protein p53 (TP53) (42/73 cases; 58%), KRAS (38/73 cases; 52%), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) (18/73 cases; 25%), BRAF (8/73 cases; 11%) and fibroblast growth factor receptor 3 (6/73 cases; 8%), followed by RSPO3 fusions (6/73; 8%) (Figure 1). Furthermore, NGS techniques revealed the presence of two or more tumor cell clones in 37 (51%) and 11 (15%) tumors, respectively; in each tumor, each of the clones represented ≥ 3% of cells in the sample (Supple
Next, we investigated the association between different alterations detected and the primary tumor’s location. A significant correlation was observed between the genetic alterations detected in the ancestral clone and at the site of the primary tumor (Figure 2). Thus, while alterations at the level of the TP53 and NRAS genes were more frequently observed in the left colon, BRAF mutations and RSPO3 fusions were detected more commonly in the right or transverse colon (P = 0.03). By contrast, no significant differences were found between the location of the primary tumor and other genetic anomalies observed in the ancestral clone of the patient’s tumor sample.
Of all the alterations detected, we focused on the RSPO3 fusion because it may represent a therapeutic target in this pathology, and its association with disease parameters remains unknown in patients with aCRC. PTPRK::RSPO3 fusions were more often associated with the presence of small tumors, without reaching statistical significance (< 5 cm; P = 0.08), localized in the right colon (P = 0.01), and without MSI (P = 0.008; Table 1). From a prognostic point of view, aCRC patients with PTPRK::RSPO3 fusions also showed a higher frequency of deaths (P = 0.04), which was associated with significantly shorter OS (median of 42 months vs 52 months, respectively; P < 0.002). In contrast, no significant differences were found between aCRC cases with and without the PTPRK::RSPO3 fusions with respect to patient age, gender, lymph node involvement, or metastatic status (Table 1).
It has been reported that RSPO3 fusion results in increased RNA transcript levels[11,23,24], so we decided to evaluate whether elevated RSPO3 expression could influence OS and RFS in two independent cohorts of consecutive CRC patients. For this purpose, we analyzed publicly available datasets from UCSC Xena (https://xena.ucsc.edu/) (354 and 376 CRC patients for RFS and OS, respectively) and KMplot (https://kmplot.com/analysis/index.php?p=service) (1167 and 814 CRC patients for RFS and OS, respectively). High RSPO3 expression was significantly associated with worse patient outcomes in terms of RFS and OS in both cohorts (P ≤ 0.05; Figure 3), suggesting a potential prognostic role for RSPO3 in CRC.
The search for novel therapies in oncology is a medical need, particularly for the treatment of advanced disease. In this context, aCRC exhibits elevated mortality mainly due to the limited efficacy of currently available antitumor agents. The identification of novel genetic alterations in tumor samples that are amenable to targeted therapies is critical for improving the clinical outcomes of cancer patients. NGS has revolutionized molecular tumor testing in oncology by offering the simultaneous assessment of many gene regions using FFPE clinical samples. Currently, NGS is performed in pathology services to determine molecular alterations that can be actioned with drugs, mainly in non-small cell lung cancer. As the impact of NGS on aCRC has not been thoroughly studied or clearly defined, we initiated a study of NGS to identify new potentially actionable molecular alterations in a subgroup of 73 aCRC patients with limited therapeutic options.
Analysis of these patients revealed that the most frequently observed genetic alterations in primary tumors were TP53, KRAS, PIK3CA and BRAF, consistent with previous NGS studies[25-27]. Interestingly, we identified the PTPRK::RSPO3 rearrangement in 8% of our cases (6/73). This fusion, together with the PTPRK::RSPO2 fusion, was initially identified in CRC by Seshagiri et al[11] using RNA-seq; who reported a comparable frequency of RSPO3 rearrangements (5 out of 68 cases). In contrast, lower incidences of 1.22%, 1.33%, 1.38% and 2.35% for the RSPO3 rearrangements in CRC were reported in four other studies by Seeber et al[28], Shinmura et al[24], Nanishi et al[14] and Hashimoto et al[12], res
It is well known that activating mutations in the KRAS (35% of cases), BRAF (8%) and NRAS (1%-3%) genes are associated with poor response to anti-EGFR therapies, which rules out anti-EGFR-directed therapy as an option for these patients. In this context, Seshagiri et al[11], Hashimoto et al[12] and Nanishi et al[14] reported that samples showing RSPO rearrangements also presented somatic mutations on KRAS, BRAF, or NRAS genes in 7/7, 27/29 and 19/24 cases, respectively. In our series, RSPO3 rearrangements coexisted with KRAS mutations in 4 out of 6 RSPO3 fusion-positive cases, supporting the frequent coexistence of these alterations. Likewise, and in line with previous studies showing that patients with RSPO3 rearrangements do not display MSI, our results confirmed this association. These findings are therapeutically relevant, as this subgroup of RSPO3-positive patients is currently unlikely to benefit from immunotherapy or anti-EGFR agents. Nevertheless, RSPO3 fusions have been proposed as a potential therapeutic target in CRC[11]. Preclinical studies in CRC models harboring RSPO3 fusions have demonstrated that both inhibition of Wnt ligand secretion and the use of RSPO3-neutralizing antibodies exert significant antitumor activity[15-19,29], supporting their clinical translation. Accordingly, RSPO3 rearrangements are now being evaluated as therapeutic targets in early-phase trials. For instance, Wnt pathway inhibitors, including Porcupine inhibitors, have enrolled biomarker-defined cohorts with RSPO2/3 fusions and reported preliminary signals of clinical activity. In addition, the anti-RSPO3 antibody rosmantuzumab (OMP-131R10) was evaluated in a phase I trial in CRC and other solid tumors[20]. Although definitive evidence of clinical efficacy is still lacking, these studies collectively support the feasibility of conducting future trials that prospectively recruit RSPO3-rearranged CRC patients to assess the therapeutic potential of Wnt pathway inhibition.
The pathogenesis of CRC depends on the location of the tumor, whereby molecular differences may exist between the right and left colon and rectum. Furthermore, it is well known that, in general terms, tumors in the left colon have a better prognosis than those in the right colon. This may be attributed to the molecular differences between them. In fact, in the present study, TP53 and NRAS were preferentially localized on the left side of the colon, whereas BRAF and MSI were more frequently detected on the right side. These findings are consistent with the results obtained by other authors who also analyzed these alterations in CRC using NGS[25-27,30,31]. Additionally, in the present study, RSPO3 fusion was preferentially detected in tumors located in the right and transverse colon, which is consistent with the findings reported by Hashimoto et al[12]. In contrast, Nanishi et al[14] and Seeber et al[28] did not observe significant differences in the tumor localization of RSPO rearrangements. These discrepancies across studies are unlikely to be explained by sample size or tumor stage, but may instead reflect methodological differences. Studies that did not report such associations also described a lower incidence of RSPO3 rearrangements, which could have limited their ability to detect this pattern. Future multicenter studies will be necessary to resolve these inconsistencies.
In this study, we also identified an association between RSPO3 rearrangements and small tumors (< 5 cm), low histopathological grade, and normal CEA levels. These clinical parameters may aid in identifying patients with aCRC harboring RSPO3 rearrangements. However, despite their potential clinical relevance, previous studies have reported limited clinical data, and thus, the clinicopathological characteristics of patients with RSPO3 rearrangements remain poorly defined.
From a prognostic point of view, RSPO3 rearrangements were significantly associated with shorter OS. Tumors harboring RSPO3 rearrangements are known to exhibit abnormally elevated RSPO3 protein expression. In fact, our in silico analysis of consecutive cases of sporadic CRC (pT1-pT4) showed that increased RSPO3 gene expression is associated with poorer disease-free survival and OS. However, to date, no studies have specifically evaluated the prognostic relevance of RSPO3 rearrangements in patients with aCRC. To our knowledge, this is the first study to demonstrate an association between RSPO3 rearrangements and clinicopathological characteristics in patients with advanced-stage disease.
We acknowledge that our study has certain limitations. First, the relatively small sample size (n = 73) may have contributed to the comparatively high incidence of RSPO3 rearrangements observed, as well as the absence of RSPO2 rearrangements in our cohort. Second, the NGS panel used may not detect novel RSPO2 or RSPO3 fusions involving uncharacterized partners or breakpoints, which could potentially affect the overall detection rate of these rearrangements. Third, our study is a single-center, retrospective analysis of pT4a-b CRC patients, which may limit the generalizability of our findings to other stages, patient populations, or healthcare settings. Nevertheless, several of our observations align with previous reports on RSPO3 fusions, supporting their relevance in this specific subgroup. Future multicenter studies with larger and more diverse cohorts, including patients across all stages and ethnic backgrounds, will be essential to better define the clinical features and prognostic value of RSPO3 fusions in CRC, as well as to explore whether this subgroup could benefit from targeted therapeutic strategies.
In summary, we identified a relatively high incidence of RSPO3 rearrangements (8%) in aCRC and found a strong as
| 1. | Morgan E, Arnold M, Gini A, Lorenzoni V, Cabasag CJ, Laversanne M, Vignat J, Ferlay J, Murphy N, Bray F. Global burden of colorectal cancer in 2020 and 2040: incidence and mortality estimates from GLOBOCAN. Gut. 2023;72:338-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 511] [Cited by in RCA: 1311] [Article Influence: 437.0] [Reference Citation Analysis (13)] |
| 2. | Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 10886] [Article Influence: 3628.7] [Reference Citation Analysis (2)] |
| 3. | Underwood PW, Ruff SM, Pawlik TM. Update on Targeted Therapy and Immunotherapy for Metastatic Colorectal Cancer. Cells. 2024;13:245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 63] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 4. | Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A. Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov. 2014;4:1269-1280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 415] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 5. | Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz HJ, Morse MA, Desai J, Hill A, Axelson M, Moss RA, Goldberg MV, Cao ZA, Ledeine JM, Maglinte GA, Kopetz S, André T. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18:1182-1191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1775] [Cited by in RCA: 2183] [Article Influence: 242.6] [Reference Citation Analysis (9)] |
| 6. | Ratti M, Grizzi G, Passalacqua R, Lampis A, Cereatti F, Grassia R, Hahne JC. NTRK fusions in colorectal cancer: clinical meaning and future perspective. Expert Opin Ther Targets. 2021;25:677-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 7. | Wang H, Li ZW, Ou Q, Wu X, Nagasaka M, Shao Y, Ou SI, Yang Y. NTRK fusion positive colorectal cancer is a unique subset of CRC with high TMB and microsatellite instability. Cancer Med. 2022;11:2541-2549. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 46] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 8. | Subbiah V, Wolf J, Konda B, Kang H, Spira A, Weiss J, Takeda M, Ohe Y, Khan S, Ohashi K, Soldatenkova V, Szymczak S, Sullivan L, Wright J, Drilon A. Tumour-agnostic efficacy and safety of selpercatinib in patients with RET fusion-positive solid tumours other than lung or thyroid tumours (LIBRETTO-001): a phase 1/2, open-label, basket trial. Lancet Oncol. 2022;23:1261-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 288] [Article Influence: 72.0] [Reference Citation Analysis (0)] |
| 9. | Santos C, Sanz-Pamplona R, Salazar R. RET-fusions: a novel paradigm in colorectal cancer. Ann Oncol. 2018;29:1340-1343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 10. | Corcoran RB, André T, Atreya CE, Schellens JHM, Yoshino T, Bendell JC, Hollebecque A, McRee AJ, Siena S, Middleton G, Muro K, Gordon MS, Tabernero J, Yaeger R, O'Dwyer PJ, Humblet Y, De Vos F, Jung AS, Brase JC, Jaeger S, Bettinger S, Mookerjee B, Rangwala F, Van Cutsem E. Combined BRAF, EGFR, and MEK Inhibition in Patients with BRAF(V600E)-Mutant Colorectal Cancer. Cancer Discov. 2018;8:428-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 403] [Cited by in RCA: 477] [Article Influence: 59.6] [Reference Citation Analysis (7)] |
| 11. | Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, Chaudhuri S, Guan Y, Janakiraman V, Jaiswal BS, Guillory J, Ha C, Dijkgraaf GJ, Stinson J, Gnad F, Huntley MA, Degenhardt JD, Haverty PM, Bourgon R, Wang W, Koeppen H, Gentleman R, Starr TK, Zhang Z, Largaespada DA, Wu TD, de Sauvage FJ. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 724] [Cited by in RCA: 799] [Article Influence: 57.1] [Reference Citation Analysis (0)] |
| 12. | Hashimoto T, Takayanagi D, Yonemaru J, Naka T, Nagashima K, Yatabe Y, Shida D, Hamamoto R, Kleeman SO, Leedham SJ, Maughan T, Takashima A, Shiraishi K, Sekine S. Clinicopathological and molecular characteristics of RSPO fusion-positive colorectal cancer. Br J Cancer. 2022;127:1043-1050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 13. | de Lau W, Barker N, Low TY, Koo BK, Li VS, Teunissen H, Kujala P, Haegebarth A, Peters PJ, van de Wetering M, Stange DE, van Es JE, Guardavaccaro D, Schasfoort RB, Mohri Y, Nishimori K, Mohammed S, Heck AJ, Clevers H. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 909] [Cited by in RCA: 1042] [Article Influence: 69.5] [Reference Citation Analysis (0)] |
| 14. | Nanishi K, Hino H, Hatakeyama K, Shiomi A, Kagawa H, Manabe S, Yamaoka Y, Nagashima T, Ohshima K, Urakami K, Akiyama Y, Yamaguchi K. Incidence and clinical significance of 491 known fusion genes in a large cohort of Japanese patients with colorectal cancer. Int J Clin Oncol. 2023;28:785-793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 15. | Storm EE, Durinck S, de Sousa e Melo F, Tremayne J, Kljavin N, Tan C, Ye X, Chiu C, Pham T, Hongo JA, Bainbridge T, Firestein R, Blackwood E, Metcalfe C, Stawiski EW, Yauch RL, Wu Y, de Sauvage FJ. Targeting PTPRK-RSPO3 colon tumours promotes differentiation and loss of stem-cell function. Nature. 2016;529:97-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 203] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 16. | Fischer MM, Yeung VP, Cattaruzza F, Hussein R, Yen WC, Murriel C, Evans JW, O'Young G, Brunner AL, Wang M, Cain J, Cancilla B, Kapoun A, Hoey T. RSPO3 antagonism inhibits growth and tumorigenicity in colorectal tumors harboring common Wnt pathway mutations. Sci Rep. 2017;7:15270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 17. | Madan B, Ke Z, Harmston N, Ho SY, Frois AO, Alam J, Jeyaraj DA, Pendharkar V, Ghosh K, Virshup IH, Manoharan V, Ong EH, Sangthongpitag K, Hill J, Petretto E, Keller TH, Lee MA, Matter A, Virshup DM. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene. 2016;35:2197-2207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 204] [Cited by in RCA: 275] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 18. | Picco G, Petti C, Centonze A, Torchiaro E, Crisafulli G, Novara L, Acquaviva A, Bardelli A, Medico E. Loss of AXIN1 drives acquired resistance to WNT pathway blockade in colorectal cancer cells carrying RSPO3 fusions. EMBO Mol Med. 2017;9:293-303. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 55] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 19. | Phillips C, Bhamra I, Eagle C, Flanagan E, Armer R, Jones CD, Bingham M, Calcraft P, Edmenson Cook A, Thompson B, Woodcock SA. The Wnt Pathway Inhibitor RXC004 Blocks Tumor Growth and Reverses Immune Evasion in Wnt Ligand-dependent Cancer Models. Cancer Res Commun. 2022;2:914-928. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 26] [Reference Citation Analysis (0)] |
| 20. | Park WJ, Kim MJ. A New Wave of Targeting 'Undruggable' Wnt Signaling for Cancer Therapy: Challenges and Opportunities. Cells. 2023;12:1110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 34] [Reference Citation Analysis (0)] |
| 21. | Weiser MR. AJCC 8th Edition: Colorectal Cancer. Ann Surg Oncol. 2018;25:1454-1455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 715] [Article Influence: 89.4] [Reference Citation Analysis (0)] |
| 22. | Győrffy B. Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innovation (Camb). 2024;5:100625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 257] [Reference Citation Analysis (0)] |
| 23. | Conboy CB, Vélez-Reyes GL, Rathe SK, Abrahante JE, Temiz NA, Burns MB, Harris RS, Starr TK, Largaespada DA. R-Spondins 2 and 3 Are Overexpressed in a Subset of Human Colon and Breast Cancers. DNA Cell Biol. 2021;40:70-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 24. | Shinmura K, Kahyo T, Kato H, Igarashi H, Matsuura S, Nakamura S, Kurachi K, Nakamura T, Ogawa H, Funai K, Tanahashi M, Niwa H, Sugimura H. RSPO fusion transcripts in colorectal cancer in Japanese population. Mol Biol Rep. 2014;41:5375-5384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 25. | Jan YH, Tan KT, Chen SJ, Yip TTC, Lu CT, Lam AK. Comprehensive assessment of actionable genomic alterations in primary colorectal carcinoma using targeted next-generation sequencing. Br J Cancer. 2022;127:1304-1311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 26. | Bayle A, Basile D, Garinet S, Rance B, Laurent-Puig P, Blons H, Taieb J, Perkins G. Next-Generation Sequencing Targeted Panel in Routine Care for Metastatic Colon Cancers. Cancers (Basel). 2021;13:5750. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 27. | Ciepiela I, Szczepaniak M, Ciepiela P, Hińcza-Nowak K, Kopczyński J, Macek P, Kubicka K, Chrapek M, Tyka M, Góźdź S, Kowalik A. Tumor location matters, next generation sequencing mutation profiling of left-sided, rectal, and right-sided colorectal tumors in 552 patients. Sci Rep. 2024;14:4619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 28. | Seeber A, Battaglin F, Zimmer K, Kocher F, Baca Y, Xiu J, Spizzo G, Novotny-Diermayr V, Rieder D, Puccini A, Swensen J, Ellis M, Goldberg RM, Grothey A, Shields AF, Marshall JL, Weinberg BA, Sackstein PE, Lim KH, Tan GS, Nabhan C, Korn WM, Amann A, Trajanoski Z, Berger MD, Lou E, Wolf D, Lenz HJ. Comprehensive Analysis of R-Spondin Fusions and RNF43 Mutations Implicate Novel Therapeutic Options in Colorectal Cancer. Clin Cancer Res. 2022;28:1863-1870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 29. | Chartier C, Raval J, Axelrod F, Bond C, Cain J, Dee-Hoskins C, Ma S, Fischer MM, Shah J, Wei J, Ji M, Lam A, Stroud M, Yen WC, Yeung P, Cancilla B, O'Young G, Wang M, Kapoun AM, Lewicki J, Hoey T, Gurney A. Therapeutic Targeting of Tumor-Derived R-Spondin Attenuates β-Catenin Signaling and Tumorigenesis in Multiple Cancer Types. Cancer Res. 2016;76:713-723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 96] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 30. | Ward R, Meagher A, Tomlinson I, O'Connor T, Norrie M, Wu R, Hawkins N. Microsatellite instability and the clinicopathological features of sporadic colorectal cancer. Gut. 2001;48:821-829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 292] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 31. | Battaglin F, Naseem M, Lenz HJ, Salem ME. Microsatellite instability in colorectal cancer: overview of its clinical significance and novel perspectives. Clin Adv Hematol Oncol. 2018;16:735-745. [PubMed] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/