Published online Oct 15, 2025. doi: 10.4251/wjgo.v17.i10.111971
Revised: August 6, 2025
Accepted: September 4, 2025
Published online: October 15, 2025
Processing time: 92 Days and 0 Hours
Patients harboring gene mutations like KRAS, NRAS, and BRAF demonstrate highly variable responses to chemotherapy, posing challenges for treatment optimization. Multiparametric magnetic resonance imaging (MRI), with its non-invasive capability to assess tumor characteristics in detail, has shown promise in evaluating treatment response and predicting therapeutic outcomes. This technology holds potential for guiding personalized treatment strategies tailored to individual patient profiles, enhancing the precision and effectiveness of colorectal cancer care.
To create a multiparametric MRI-based predictive model for assessing chemotherapy efficacy in colorectal cancer patients with gene mutations.
This retrospective study was conducted in a tertiary hospital, analyzing 157 colorectal cancer patients with gene mutations treated between August 2022 and December 2023. Based on chemotherapy outcomes, the patients were categorized into favorable (n = 60) and unfavorable (n = 50) response groups. Univariate and multivariate logistic regression analyses were performed to identify independent predictors of chemotherapy efficacy. A predictive nomogram was constructed using significant variables, and its performance was assessed using the area under the receiver operating characteristic curve (AUC) in both training and validation sets.
Univariate analysis identified that tumor differentiation, T2 signal intensity ratio, tumor-to-anal margin distance, and MRI-detected lymph node metastasis as significantly associated with chemotherapy response (P < 0.05). Multivariate Logistics regression confirmed these four parameters as independent predictors. The predictive model demonstrated strong discrimination, with an AUC of 0.938 (sensitivity: 86%; specificity: 92%) in the training set, and 0.942 (sensitivity: 100%; specificity: 83%) in the validation set.
We established and validated a multiparametric MRI-based model for predicting chemotherapy response in colorectal cancer patients with gene mutations. This model holds promise for guiding individualized treatment strategies.
Core Tip: This study pioneers a multiparametric magnetic resonance imaging (MRI)-based predictive model tailored for colorectal cancer patients with gene mutations (e.g., KRAS, NRAS, and BRAF) to evaluate chemotherapy efficacy. By integrating tumor differentiation, T2 signal intensity ratio, tumor-to-anal margin distance, and MRI-detected lymph node metastasis, the model achieved high accuracy (area under the receiver operating characteristic curve > 0.93) in both training and validation sets, offering a non-invasive tool for personalized treatment planning.
- Citation: Kang WY, Deng WM, Ye XQ, Zhong YH, Li XJ, Feng LL, Luo DH. Multiparametric magnetic resonance imaging-based predictive model for chemotherapy response in colorectal cancer patients with gene mutations. World J Gastrointest Oncol 2025; 17(10): 111971
- URL: https://www.wjgnet.com/1948-5204/full/v17/i10/111971.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i10.111971
Colorectal cancer is a common malignancy of the gastrointestinal tract, arising from the colon or rectum. Its incidence and mortality rates rank among the highest of digestive system cancers, second only to gastric, esophageal, and primary liver cancers. The incidence has been increasing over the years, with a trend toward older age at onset[1,2]. Early-stage colorectal cancer is often asymptomatic. As the disease progresses, patients may present with changes in bowel habits, hematochezia, diarrhea, alternating constipation and diarrhea, and localized abdominal pain. In advanced stages, systemic symptoms such as anemia and weight loss may occur[3]. For individuals with recent changes in bowel habits or rectal bleeding, prompt colonoscopic evaluation is recommended, as colonoscopy remains the most effective diagnostic tool for early detection of colorectal cancer[4].
Colorectal cancer is frequently associated with somatic gene mutations, particularly in oncogenes such as KRAS, NRAS, and BRAF. Among these, KRAS and NRAS mutations are commonly associated with resistance to chemotherapeutic agents and poor clinical outcomes. These mutations may lead to abnormal activation of intracellular signaling, promoting uncontrolled tumor cell proliferation and reducing responsiveness to chemotherapy[5]. In some cases, these genetic alterations exist independently, while in others they may interact with additional genetic or environmental factors, collectively contributing to colorectal cancer pathogenesis. Clinical studies have demonstrated that patients harboring such mutations exhibit significant variability in their response to chemotherapy[6,7]. Notably, research by Stec et al[8] showed that the presence of KRAS mutations may serve as a prognostic marker in patients receiving irinotecan- or oxaliplatin-based chemotherapy regimens.
Currently, the prognosis of colorectal cancer during chemotherapy is primarily assessed using genetic testing, laboratory biomarkers, and clinicopathological parameters. In recent years, multiparametric magnetic resonance imaging (MRI)—including high-resolution MRI, dynamic contrast-enhanced imaging, and diffusion-weighted imaging—has emerged as a non-invasive tool for evaluating tumor response and predicting treatment efficacy, thereby supporting the development of personalized therapeutic strategies[9]. However, limited research has explored the utility of multiparametric MRI-based models specifically in colorectal cancer patients with genetic mutations, and further studies are warranted to evaluate their predictive value in this context.
In this study, we aimed to investigate the relationship between gene mutations and chemotherapy efficacy in patients with colorectal cancer. Based on the findings, we developed and validated a multiparametric MRI-based predictive model to assess chemotherapy response in this patient population. This model may assist clinicians in better evaluating treatment outcomes and tailoring individualized therapeutic strategies.
The sample size calculation of this study was performed using the overall ratio formula. According to previous literature, π is taken as 4.35%[10], the test level α is 0.05, and the allowable error δ is taken as 0.06. The calculated sample size is at least about 44 cases. Taking into account the 20% sample inefficiency, the final sample size is at least 54 cases. This study also requires a verification group. Therefore, the total sample size is at least 108 cases. Final, a total of 157 patients with colorectal cancer carrying gene mutations were retrospectively included at our hospital between August 2022 and December 2023. The inclusion criteria were as follows: (1) Patients met the clinical diagnostic criteria for colorectal cancer with confirmed RAS family and BRAF gene mutations, based on colonoscopic pathology findings[11], including mutations in KRAS, NRAS, or BRAF; and (2) No prior history of chemotherapy[12]. The exclusion criteria were: (1) Presence of contraindications to chemotherapy; and (2) Diagnosis of other malignant tumors.
Patients were randomly divided into a modeling group (n = 110) and a verification group (n = 47) according to a ratio of 7:3. Within the modeling group, patients were further classified into a favorable response group (n = 60) and an unfavorable response group (n = 50) based on their chemotherapy outcomes. The evaluation of treatment efficacy was based on the Response Evaluation Criteria in Solid Tumors: Complete response (CR) was defined as the disappearance of all target lesions, with all lymph nodes (whether target or non-target) measuring less than 10 mm in short axis; partial response (PR) was defined as a ≥ 30% reduction in the sum of the diameters of target lesions; stable disease (SD) referred to cases where changes in lesion size did not meet the criteria for either PR or progressive disease (PD); PD was defined as a ≥ 20% increase in the sum of lesion diameters or the appearance of new lesions. Patients achieving CR or PR were categorized into the favorable group, while those with SD or PD were classified into the unfavorable group. Tumor locations were categorized as left-sided colon, right-sided colon, or rectum, based on the primary site of tumor involvement. All patients received six cycles of FOLFOX-based chemotherapy before evaluation. T-staging data were collected according to pre-treatment MRI findings, and most patients were classified as T3 or T4. Regarding molecular characteristics, KRAS mutations accounted for the majority of cases, followed by NRAS and BRAF mutations.
This study was approved by the Ethics Committee of our hospital in accordance with applicable regulatory and ethical guidelines for retrospective studies (Approval No. YW2022-21-3). The requirement for informed consent was waived due to the retrospective nature of the study and the exclusive use of de-identified patient data, which posed no risk to patient privacy or clinical care.
All patients received neoadjuvant chemotherapy prior to surgical treatment. The chemotherapy protocol consisted of the following agents: Oxaliplatin was administered at a dose of 85 mg/m2via intravenous infusion over 2 hours on Day 1 of each chemotherapy cycle; calcium folinate was given at 200 mg/m² via intravenous infusion over 2 hours on days 1 and 2; fluorouracil (5-FU) was administered in two phases: An initial intravenous bolus of 400 mg/m2 on days 1 and 2, followed by a continuous intravenous infusion of 600 mg/m2 over 22 hours on the same days. In our institution, neoadjuvant chemotherapy is considered for patients with high-risk features on imaging, such as large tumor size, close proximity to the mesorectal fascia, or suspected extramural vascular invasion—even in the absence of lymph node metastasis. Therefore, some N0 patients also underwent neoadjuvant therapy based on multidisciplinary team evaluation. The FOLFOX regimen was selected as the standard neoadjuvant chemotherapy protocol due to its wide clinical use, favorable tolerance, and ease of administration in our hospital setting. CAPEOX was not used primarily because capecitabine is orally administered and requires close monitoring of gastrointestinal toxicity, which posed logistical challenges in our patient population. No targeted therapies were administered to the patients during the chemotherapy period analyzed in this study.
All MRI scans were performed to assess the primary colorectal tumor, which ensured accurate evaluation of local anatomical features. Although MRI is more commonly used in rectal cancer, our institution routinely performs pelvic-abdominal MRI across all colorectal cancer cases—including right-sided tumors—to achieve consistent imaging assessment and obtain quantitative parameters required for model development. GEHDX 3.0 TMR MRI was used. All patients were fasted for 12-14 hours and underwent bowel preparation before scanning and were informed of the precautions in the examination process. Patients were asked to cooperate with the physician during the examination. A small amount of air was injected through the anus before scanning. Routine plain scan was performed first. Imaging protocols included cross-sectional turbo spin echo T1 weighted imaging (repetition time [TR] = 984 ms, echo time [TE] = 13 ms, field of view [FOV] = 200 mm × 200 mm, slice thickness = 3 mm) and turbo spin echo T2 weighted imaging with fat suppression (TR = 4940 ms, TE = 96 ms, FOV = 200 mm × 200 mm, slice thickness = 3 mm). Transverse T1WI-FS sequences were also obtained. For contrast-enhanced imaging, gadopentetate dimeglumine was administered via intravenous injection into the antecubital vein. Dynamic contrast-enhanced scans were obtained during the arterial (25-30 seconds), venous (55-60 seconds), and delayed (180 seconds) phases. The distance from the lower edge of the tumor to the anal verge was measured on sagittal T2-weighted images using electronic calipers within the Picture Archiving and Communication System. The measurement was performed by two experienced radiologists, and the final value was determined by consensus in cases of discrepancy.
Clinical and imaging data were collected from the hospital’s electronic medical record system. Patient information included age, sex, body mass index (BMI), tumor location, presence of comorbidities (e.g., hypertension and diabetes), family history of colorectal cancer, pathological type, clinical stage, and degree of tumor differentiation.
MRI parameters included T2 signal intensity grade, the distance from the lower margin of the tumor to the anal verge, and the presence of lymph node metastasis. Tumor location was determined based on the site that accounted for the majority of tumor volume when multiple locations were involved. Comorbidities were defined as the presence of chronic conditions such as hypertension or diabetes. Pathological type was determined based on histopathological findings from biopsy specimens. T2 signal intensity was classified as follows: (1) Grade 0: No signal enhancement; (2) Grade 1: Mild and poorly defined signal enhancement; and (3) Grade 2: Marked and well-defined signal enhancement. Lymph node status was assessed using MRI and categorized into four stages: (1) N0: No lymph node metastasis; (2) N1: Metastasis to 1-3 regional lymph nodes; (3) N2: Metastasis to 4-6 regional lymph nodes; and (4) N3: Metastasis to more than 7 regional lymph nodes or to distant lymph nodes. For analysis, N0 was considered non-metastatic, while N1-N3 were grouped as metastatic.
To calculate the T2 signal intensity ratio, a region of interest (ROI) was manually drawn within the most hyperintense area of the tumor on axial T2-weighted images. A second ROI of similar size was placed in the adjacent gluteus maximus muscle on the same slice level, avoiding vessels and artifacts. The mean signal intensity values from both ROIs were measured, and the ratio of tumor signal intensity to muscle signal intensity was computed. Two radiologists independently performed all measurements, and the averaged values were used for analysis.
Statistical analyses were performed using SPSS version 27.0 (IBM Corp., Armonk, NY, United States). The Shapiro-Wilk test was applied to assess the normality of continuous variables, and all data were confirmed to follow a normal distribution. Normally distributed continuous variables (e.g., age, BMI, T2 signal intensity grading, and tumor-to-anal margin distance) are expressed as the mean ± SD, and comparisons between groups were conducted using independent-samples t-tests. Categorical variables (e.g., sex, tumor location, comorbidities, family history of colorectal cancer, pathological type, clinical stage, tumor differentiation, and MRI-detected lymph node metastasis) are presented as counts and percentages. The χ² test or Fisher’s exact test was used for group comparisons, as appropriate. Collinearity diagnostics were performed using tolerance and variance inflation factor values to assess potential multicollinearity before conducting multivariate logistic regression analysis. Univariate and multivariate Logistical regression models were used to identify factors associated with chemotherapy response. A receiver operating characteristic (ROC) curve was generated to evaluate the predictive performance of the multiparametric MRI-based nomogram model in estimating chemotherapy efficacy among patients with colorectal cancer harboring KRAS, NRAS, or BRAF mutations.
The average age of the 157 patients was 56.29 ± 2.97 years, including 68 males and 42 females. There was no significant difference between the favorable and unfavorable response groups in terms of age, sex, BMI, tumor location, comorbidities, family history, pathological type, or clinical stage (P > 0.05). However, significant differences were observed in tumor differentiation, T2 signal intensity ratio, tumor-to-anal margin distance, and MRI-detected lymph node metastasis (P < 0.05), as detailed in Table 1.
| Baseline data | Favorable group (n = 60) | Unfavorable group (n = 50) | t/χ2 | P value | |
| Age (years) | 56.34 ± 3.11 | 56.27 ± 2.73 | 0.124 | 0.901 | |
| Sex | Male | 36 | 32 | 0.185 | 0.912 |
| Female | 24 | 18 | |||
| BMI (kg/m2) | 21.13 ± 1.34 | 21.27 ± 1.41 | 0.533 | 0.595 | |
| Tumor location | Left side | 24 | 21 | 0.064 | 0.996 |
| Right side | 18 | 15 | |||
| Rectum | 18 | 14 | |||
| Comorbid underlying diseases | Yes | 15 | 10 | 0.388 | 0.824 |
| N/A | 45 | 40 | |||
| Family history of colorectal cancer | Yes | 3 | 2 | 0.063 | 0.969 |
| N/A | 57 | 48 | |||
| Type of pathology | Adenocarcinoma | 34 | 29 | 0.020 | 0.888 |
| Miscellaneous | 26 | 21 | |||
| Clinical stage | Stage II | 37 | 24 | 2.077 | 0.354 |
| Stage III | 23 | 26 | |||
| Degree of tumor differentiation | Poorly differentiated | 23 | 10 | 8.952 | 0.030 |
| Moderately differentiated | 25 | 11 | |||
| Well-differentiated | 12 | 19 | |||
| T2 signal strength grade | 2.62 ± 0.51 | 3.43 ± 0.64 | 7.387 | < 0.001 | |
| Distance from the lower edge of the tumor to the anus (anal margin; cm) | 6.86 ± 1.31 | 5.11 ± 1.08 | 7.546 | < 0.001 | |
| MRI lymph node metastasis | Yes | 11 | 32 | 23.891 | < 0.001 |
| N/A | 49 | 18 |
Variables that were statistically significant in the univariate analysis—including tumor differentiation, T2 signal intensity ratio, tumor-to-anal margin distance, and MRI-detected lymph node metastasis—were entered into the multivariate logistic regression model. Chemotherapy response was defined as the dependent variable (unfavorable response = 1; favorable response = 0). The analysis identified all four variables as independent predictors of chemotherapy efficacy in patients with colorectal cancer harboring KRAS, NRAS, or BRAF mutations (P < 0.05), as presented in Table 2.
| Risk factor | Β value | SE value | Ward value | OR value | 95%CI | P value | Tolerance | VIF |
| Degree of tumor differentiation | 0.785 | 0.348 | 5.092 | 2.193 | 1.109-4.338 | < 0.001 | 0.954 | 1.021 |
| T2 signal strength grading | 0.126 | 0.064 | 3.861 | 1.134 | 1.000-1.286 | < 0.001 | 0.971 | 1.012 |
| Distance between lower edge of tumor and anus | 0.748 | 0.264 | 8.020 | 2.112 | 1.259-3.543 | < 0.001 | 0.967 | 1.031 |
| MRI-detected lymph node metastasis | 0.449 | 0.211 | 4.532 | 1.567 | 1.036-2.370 | < 0.001 | 0.975 | 1.027 |
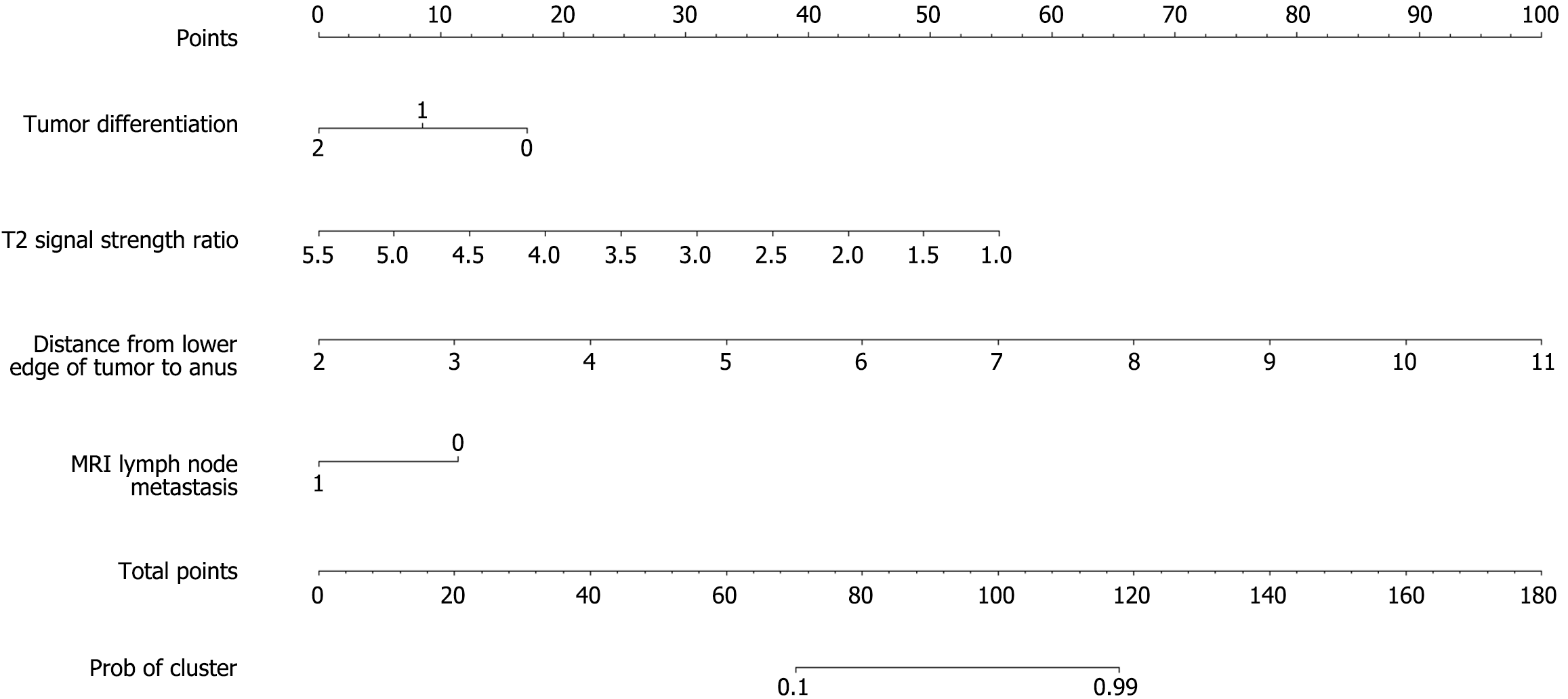
Based on the results of the multivariate logistic regression analysis, a nomogram was constructed to visualize the predictive model (Figure 1). Each independent predictor was assigned a score on a calibrated scale according to its regression coefficient. The individual scores were summed to generate a total score, which corresponded to the estimated probability of an unfavorable chemotherapy response. The total score ranged from 0 to 100, with the predicted probability of poor response ranging from 0.1 to 0.9. A higher total score indicated an increased likelihood of chemotherapy inefficacy.
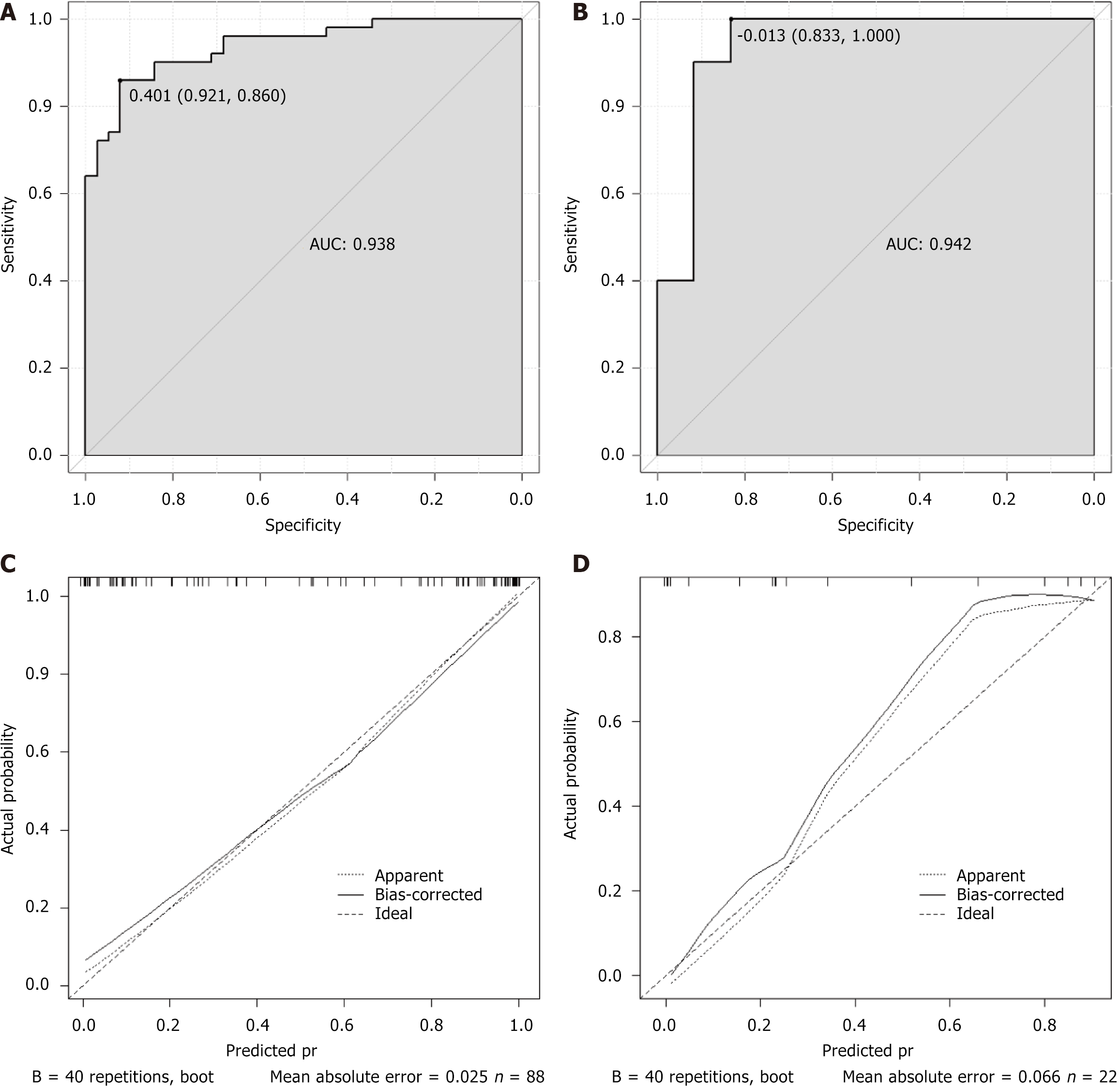
ROC curve analysis was performed to evaluate the predictive performance of the multiparametric MRI-based nomogram. In the training cohort, the model achieved an area under the ROC curve (AUC) of 0.938, with a sensitivity of 86% and specificity of 92% (Figure 2A). In the validation cohort, the AUC was 0.942, with a sensitivity of 100% and specificity of 83% (Figure 2B). Calibration curves for both the training and validation sets demonstrated slopes close to 1, showing strong agreement between predicted and observed probabilities (Figure 2C and D).
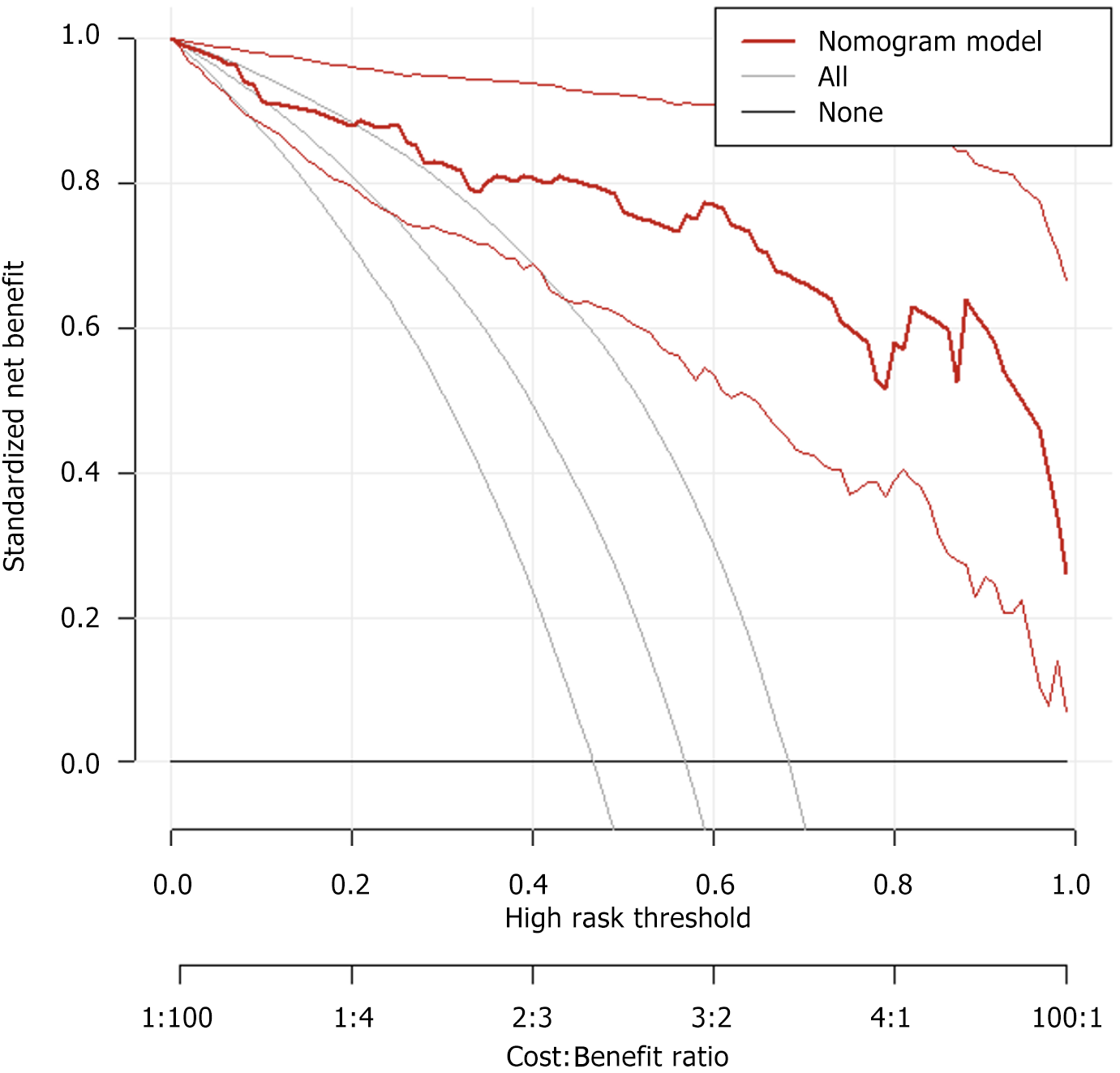
Decision curve analysis (DCA) curve was performed to assess the clinical utility of the nomogram in predicting chemotherapy efficacy. The DCA curve demonstrated a clear net benefit across a wide range of threshold probabilities, indicating that the model has favorable clinical applicability and can provide meaningful support for clinical decision-making (Figure 3).
Colorectal cancer is a common malignant tumor with an increasing incidence year by year, which seriously threatens human health. Chemotherapy is one of the important means for the treatment of colorectal cancer; however, therapeutic responses vary considerably among individuals[13]. To better predict the chemotherapy efficacy in patients with colorectal cancer carrying gene mutations, a multi-parameter MRI-based model was established in this study.
This study investigated the factors influencing chemotherapy efficacy in patients with colorectal cancer harboring KRAS, NRAS, or BRAF gene mutations. The results showed no statistically significant differences between the favorable and unfavorable response groups in terms of age, sex, BMI, tumor location, comorbidities, family history, pathological type, or clinical stage (P > 0.05). However, significant differences were observed in tumor differentiation, T2 signal intensity ratio, tumor-to-anal margin distance, and MRI-detected lymph node metastasis (P < 0.05). These findings suggest that tumor differentiation may be a key factor influencing chemotherapy response[14]. Poorly differentiated tumors are typically associated with more aggressive biological behavior, including rapid proliferation and increased metastatic potential. As a result, patients with low tumor differentiation often exhibit reduced sensitivity to chemotherapeutic agents, leading to suboptimal treatment outcomes[15,16]. In contrast, well-differentiated tumors tend to exhibit slower growth and limited metastatic potential. Consequently, patients with higher tumor differentiation are generally more responsive to chemotherapy, resulting in more favorable treatment outcomes. T2 signal intensity ratio is an important parameter reflecting the biological characteristics of tumors[17]. Patients with a high T2 signal intensity ratio tend to have a higher water content in cancer tissues, greater cell density, and stronger ability to uptake and metabolize chemotherapeutic agents, thereby resulting in better chemotherapy outcomes. The distance between the lower edge of the tumor and the anus is also an factor affecting the efficacy of chemotherapy[18,19]. Patients with tumors located closer to the anal verge are more susceptible to distant metastasis via lymphatic pathways, which may contribute to reduced chemotherapy efficacy[20]. In addition, patients with MRI-detected lymph node metastases—indicating the spread of cancer cells through the lymphatic pathway—typically exhibit poor responses to chemotherapy[21]. The study further incorporated the significant variables identified in the univariate analysis into a multivariate logistic regression model. The results demonstrated that tumor differentiation, T2 signal intensity ratio, tumor-to-anal margin distance, and MRI-detected lymph node metastasis were independent predictors of chemotherapy efficacy in patients with colorectal cancer harboring gene mutations. These findings suggest that each of these parameters independently contributes to predicting treatment response[22,23].
Based on the results of multivariate logistic regression analysis, independent predictors were incorporated into R software to construct a nomogram for the MRI-based prediction model. This model offers an intuitive and clinically accessible tool for estimating the likelihood of chemotherapy response in colorectal cancer patients with gene mutations. DCA showed that the model provided a clear net clinical benefit across a wide range of threshold probabilities, indicating its strong practical utility in clinical decision-making. To further evaluate the model’s performance, ROC curve analysis was conducted. In the training cohort, the model demonstrated excellent discriminative ability with an AUC of 0.938. At the optimal cutoff value of 0.78, sensitivity and specificity were 86.0% and 92.1%, respectively, showing high accuracy in identifying patients likely to respond to chemotherapy. In the validation cohort, the model achieved an even higher AUC of 0.942. With the optimal threshold set at 0.83, sensitivity reached 100.0%, successfully identifying all patients with favorable chemotherapy outcomes, while specificity was 83.3%. Despite the slight decrease in specificity, the increase in sensitivity is clinically meaningful. Calibration curves in both the training and validation sets showed slopes close to 1, demonstrating strong agreement between predicted probabilities and actual outcomes. These results support the model’s robustness, accuracy, and potential clinical applicability in guiding individualized treatment strategies.
This study successfully established a model to predict the efficacy of chemotherapy in patients with colorectal cancer carrying genetic mutations, which has important innovative and clinical application value. Previous research results by Lim et al[24] analyzed the influencing factors of efficacy, but did not establish a model. This study established a comprehensive prediction model based on the influencing factors to further in-depth research. Compared to existing predictive models that rely solely on genetic markers or histological features, which often require invasive biopsy and may not reflect the spatial heterogeneity of the tumor, our model offers several distinct advantages. First, it utilizes only noninvasive, routinely acquired MRI parameters, making it more accessible and cost-effective in clinical settings. Second, by integrating multiple imaging biomarkers—such as T2 signal intensity ratio, tumor-to-anal margin distance, and nodal status—our model captures both anatomical and functional tumor characteristics. This multiparametric approach improves predictive accuracy and enables personalized chemotherapy decision-making without the need for additional imaging sequences or contrast agents. These strengths highlight the model’s clinical feasibility and potential for broad implementation. From an innovative perspective, there are relatively few studies to predict the effectiveness of chemotherapy in patients with colorectal cancer carrying genetic mutations. This study focused on this specific group and comprehensively used univariate and multivariate Logistic regression analysis to screen out independent factors that affect the effectiveness of chemotherapy, including tumor differentiation degree, T2 signal intensity ratio, distance between the lower edge of the tumor and the anus, and MRI lymph node metastasis., and a predictive model was constructed based on these factors. This accurate prediction model for patients with specific gene mutations provides new ideas and methods for the formulation of personalized treatment plans. It is different from traditional generalized prediction models and can better meet the needs of clinical precision medicine. In terms of clinical application value, this model performs well. Doctors can use this model to evaluate patients before chemotherapy and understand in advance the possibility of patients' response to different chemotherapy regimens, so as to select the most suitable treatment regimen for patients, improve treatment effectiveness, and reduce unnecessary treatment side effects and financial burden.
The sample size in this study was determined based on the availability of eligible patients within the specified study period at our institution, focusing on a genetically defined subgroup of colorectal cancer with KRAS/NRAS/BRAF mutations. Although the total number of cases (n = 157) may appear modest, it is generally acceptable for logistic regression modeling when considering the number of predictive variables included. In this analysis, four independent predictors were identified and incorporated into the model, ensuring an adequate events-per-variable ratio, which helps mitigate the risk of overfitting. Additionally, an internal validation cohort comprising 30% of the sample was used to test the model’s performance, and the results remained stable and consistent. Therefore, although this study was conducted at a single center, the sample size was deemed appropriate for the exploratory nature of the analysis and the focus on a genetically defined patient subgroup. Moving forward, we plan to collaborate with other medical centers to perform external validation using larger and more diverse cohorts. Such multicenter studies will be critical to further assess the robustness, generalizability, and clinical utility of the proposed predictive model across different clinical settings. Additionally, while this study included patients with KRAS, NRAS, and BRAF mutations, the sample sizes for NRAS and BRAF subgroups were limited, precluding meaningful stratified analysis by mutation type. We recognize that different gene mutations may influence chemotherapy response through distinct biological mechanisms. Future multicenter studies with larger and more balanced representation of these mutation types are warranted to develop refined, mutation-specific predictive models.
Although the statistical methodology (e.g., logistic regression and nomogram construction) used in this study is well-established, the innovation of our work lies in the specific patient population and the integration of multiparametric MRI-derived features for predictive modeling. Most previous prediction models in colorectal cancer did not focus on patients harboring KRAS, NRAS, or BRAF mutations, who often exhibit distinct biological behaviors and chemotherapy responses. Our study addresses this gap by identifying imaging biomarkers tailored for this genetically defined subgroup. Furthermore, the incorporation of quantitative MRI parameters—such as T2 signal intensity ratio and tumor-to-anal margin distance—offers a reproducible, non-invasive approach for pre-treatment stratification, which holds significant clinical value in precision oncology. This study has certain limitations. For instance, different types of gene mutations may exert varying influences on chemotherapy efficacy and patient prognosis. However, this study did not perform a detailed stratification based on specific mutation types. Future research should aim to investigate the differential impact of individual gene mutations—such as KRAS, NRAS, and BRAF—on chemotherapy response in colorectal cancer, which may further enhance the precision of predictive modeling[25,26]. In addition, while the tumor-to-anal verge distance was identified as a significant predictor, its distribution in the current cohort showed a trend toward favorable outcomes among patients with more proximal lesions. This may reflect intrinsic biological differences or anatomical factors related to drug sensitivity, and suggests that stratified modeling or cut-off optimization could further refine its predictive value. Moreover, the predictive model was internally validated using a split-sample approach, which demonstrated strong performance. Nonetheless, external validation in larger, multi-center cohorts will be essential to fully assess its generalizability and facilitate clinical application.
In conclusion, the multiparametric MRI-based prediction model demonstrates substantial value in forecasting chemotherapy response in patients with colorectal cancer harboring gene mutations. By providing more accurate and individualized risk assessments, the model can assist clinicians in optimizing treatment strategies. With ongoing advancements in medical imaging and data science, this predictive approach holds promise for further refinement and broader application in the management of colorectal cancer.
| 1. | Zhang Y, Saha K, Nandani R, Yuan J, Dey M, Gu Z. Isothiocyanates attenuate heparin-induced proliferation of colon cancer cells in vitro. Food Sci Nutr. 2024;12:7842-7853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 2. | Arnold M, Abnet CC, Neale RE, Vignat J, Giovannucci EL, McGlynn KA, Bray F. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology. 2020;159:335-349.e15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 857] [Cited by in RCA: 1439] [Article Influence: 239.8] [Reference Citation Analysis (16)] |
| 3. | Griffin-Sobel JP. Symptom management of advanced colorectal cancer. Surg Oncol Clin N Am. 2006;15:213-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 4. | Millien VO, Mansour NM. Bowel Preparation for Colonoscopy in 2020: A Look at the Past, Present, and Future. Curr Gastroenterol Rep. 2020;22:28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 82] [Article Influence: 13.7] [Reference Citation Analysis (2)] |
| 5. | Bovio F, Epistolio S, Mozzi A, Monti E, Fusi P, Forcella M, Frattini M. Role of NEU3 Overexpression in the Prediction of Efficacy of EGFR-Targeted Therapies in Colon Cancer Cell Lines. Int J Mol Sci. 2020;21:8805. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 6. | Dotolo S, Marabotti A, Rachiglio AM, Esposito Abate R, Benedetto M, Ciardiello F, De Luca A, Normanno N, Facchiano A, Tagliaferri R. A multiple network-based bioinformatics pipeline for the study of molecular mechanisms in oncological diseases for personalized medicine. Brief Bioinform. 2021;22:bbab180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 7. | Grbčić P, Fučkar Čupić D, Gamberi T, Kraljević Pavelić S, Sedić M. Proteomic Profiling of BRAFV600E Mutant Colon Cancer Cells Reveals the Involvement of Nucleophosmin/c-Myc Axis in Modulating the Response and Resistance to BRAF Inhibition by Vemurafenib. Int J Mol Sci. 2021;22:6174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 8. | Stec R, Bodnar L, Charkiewicz R, Korniluk J, Rokita M, Smoter M, Ciechowicz M, Chyczewski L, Nikliński J, Kozłowski W, Szczylik C. K-Ras gene mutation status as a prognostic and predictive factor in patients with colorectal cancer undergoing irinotecan- or oxaliplatin-based chemotherapy. Cancer Biol Ther. 2012;13:1235-1243. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 9. | Guo L, Wang Y, Yang W, Wang C, Guo T, Yang J, Shao Z, Cai G, Cai S, Zhang L, Hu X, Xu Y. Molecular Profiling Provides Clinical Insights Into Targeted and Immunotherapies as Well as Colorectal Cancer Prognosis. Gastroenterology. 2023;165:414-428.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 56] [Reference Citation Analysis (0)] |
| 10. | Manabe T, Takii Y, Oyanagi H, Nogami H, Maruyama S. Prognosis for Metastatic Colorectal Cancer Patients Achieving Complete Response After Systemic Chemotherapy. J Gastrointest Cancer. 2023;54:501-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 11. | Expert Group on Early Diagnosis and Treatment of Cancer; Chinese Society of Oncology; Chinese Medical Association. [Expert consensus on the early diagnosis and treatment of colorectal cancer in China (2023 edition)]. Zhonghua Yi Xue Za Zhi. 2023;103:3896-3908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 12. | Serup-Branda L, Laurberg JM, Bjerregaard NC, Laurberg S. [Danish National Guidelines for diagnostic strategy of patients with colorectal cancer symptoms]. Ugeskr Laeger. 2008;170:4058. [PubMed] |
| 13. | Biller LH, Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA. 2021;325:669-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 1736] [Article Influence: 347.2] [Reference Citation Analysis (1)] |
| 14. | Li C, Sun YD, Yu GY, Cui JR, Lou Z, Zhang H, Huang Y, Bai CG, Deng LL, Liu P, Zheng K, Wang YH, Wang QQ, Li QR, Wu QQ, Liu Q, Shyr Y, Li YX, Chen LN, Wu JR, Zhang W, Zeng R. Integrated Omics of Metastatic Colorectal Cancer. Cancer Cell. 2020;38:734-747.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 204] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 15. | London M, Gallo E. Critical role of EphA3 in cancer and current state of EphA3 drug therapeutics. Mol Biol Rep. 2020;47:5523-5533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 16. | Mammana M, Bergamo F, Procaccio L, Schiavon M, Loupakis F, Lonardi S, Manai C, Schirripa M, Fassan M, Dei Tos AP, Calabrese F, Rea F, Zagonel V. Outcome of patients with colorectal cancer undergoing lung metastases resection: a single-institution retrospective analysis. Tumori. 2021;107:46-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 17. | Nie H, Yu Y, Wang F, Huang X, Wang H, Wang J, Tao M, Ning Y, Zhou J, Zhao Q, Xu F, Fang J. Comprehensive analysis of the relationship between ubiquitin-specific protease 21 (USP21) and prognosis, tumor microenvironment infiltration, and therapy response in colorectal cancer. Cancer Immunol Immunother. 2024;73:156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 18. | Osumi H, Muroi A, Sakahara M, Kawachi H, Okamoto T, Natsume Y, Yamanaka H, Takano H, Kusama D, Shinozaki E, Ooki A, Yamaguchi K, Ueno M, Takeuchi K, Noda T, Nagayama S, Koshikawa N, Yao R. Evaluation of the RAS signaling network in response to MEK inhibition using organoids derived from a familial adenomatous polyposis patient. Sci Rep. 2020;10:17455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 19. | Pinto C, Biffoni M, Popoli P, Marchetti A, Marchetti P, Martini N, Normanno N. Molecular tests and target therapies in oncology: recommendations from the Italian workshop. Future Oncol. 2021;17:3529-3539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 20. | Tian X, Wang Q, Cai W. A Novel Mutation in MYH Gene Associated with Aggressive Colorectal Cancer in a Child: A Case Report and Review of Literature. Onco Targets Ther. 2020;13:8557-8565. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 21. | Volkov NM, Yanus GA, Ivantsov AO, Moiseenko FV, Matorina OG, Bizin IV, Moiseyenko VM, Imyanitov EN. Efficacy of immune checkpoint blockade in MUTYH-associated hereditary colorectal cancer. Invest New Drugs. 2020;38:894-898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 22. | Xiao J, Wang X, Liu Y, Liu X, Yi J, Hu J. Lactate Metabolism-Associated lncRNA Pairs: A Prognostic Signature to Reveal the Immunological Landscape and Mediate Therapeutic Response in Patients With Colon Adenocarcinoma. Front Immunol. 2022;13:881359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 23. | Xu X, Liu M, Peng K, Yu Y, Liu T. Asparaginyl endopeptidase contributes to cetuximab resistance via MEK/ERK signaling in RAS wide-type metastatic colorectal cancer. Clin Transl Oncol. 2023;25:776-785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 24. | Lim SH, Cho HJ, Kim KM, Lim HY, Kang WK, Lee J, Park YS, Kim HC, Kim ST. Comprehensive molecular analysis to predict the efficacy of chemotherapy containing bevacizumab in patients with metastatic colorectal cancer. Oncol Res. 2023;31:855-866. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 25. | Yang M, Davis TB, Pflieger L, Nebozhyn MV, Loboda A, Wang H, Schell MJ, Thota R, Pledger WJ, Yeatman TJ. An integrative gene expression signature analysis identifies CMS4 KRAS-mutated colorectal cancers sensitive to combined MEK and SRC targeted therapy. BMC Cancer. 2022;22:256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 26. | Zhong C, Xie T, Chen L, Zhong X, Li X, Cai X, Chen K, Lan S. Immune depletion of the methylated phenotype of colon cancer is closely related to resistance to immune checkpoint inhibitors. Front Immunol. 2022;13:983636. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/