Published online Aug 15, 2024. doi: 10.4251/wjgo.v16.i8.3376
Revised: May 3, 2024
Accepted: May 22, 2024
Published online: August 15, 2024
Processing time: 154 Days and 16.6 Hours
Long non-coding RNAs (lncRNAs), with transcript lengths exceeding 200 nucleotides and little or no protein-coding capacity, have been found to impact colorectal cancer (CRC) through various biological processes. LncRNA expression can regulate autophagy, which plays dual roles in the initiation and progression of cancers, including CRC. Abnormal expression of lncRNAs is associated with the emergence of chemoresistance. Moreover, it has been confirmed that targeting autophagy through lncRNA regulation could be a viable approach for combating chemoresistance. Two recent studies titled “Human β-defensin-1 affects the mammalian target of rapamycin pathway and autophagy in colon cancer cells through long non-coding RNA TCONS_00014506” and “Upregulated lncRNA PRNT promotes progression and oxaliplatin resistance of colorectal cancer cells by regulating HIPK2 transcription” revealed novel insights into lncRNAs associated with autophagy and oxaliplatin resistance in CRC, respectively. In this editorial, we particularly focus on the regulatory role of lncRNAs in CRC-related auto
Core Tip: Long non-coding RNAs (lncRNAs) expression can regulate both autophagy and chemoresistance in colorectal cancer (CRC). Autophagy exerts dual effects on chemotherapy resistance through multiple mechanisms, including the regulation of autophagy-related lncRNAs. In this editorial, we focus on the role of lncRNAs in the regulation of autophagy and chemoresistance in CRC and explore the feasibility of modulating chemosensitivity by interfering with lncRNAs involved in the autophagic process.
- Citation: Yu JM, Sun CQ, Xu HH, Jiang YL, Jiang XY, Ni SQ, Zhao TY, Liu LX. Navigating the labyrinth of long non-coding RNAs in colorectal cancer: From chemoresistance to autophagy. World J Gastrointest Oncol 2024; 16(8): 3376-3381
- URL: https://www.wjgnet.com/1948-5204/full/v16/i8/3376.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i8.3376
Colorectal cancer (CRC) is the third most common malignancy and the second leading cause of cancer-related death worldwide[1]. It is paramount to extensively and intensively investigate the pathogenesis of CRC to facilitate the development of effective therapeutic strategies. Two recent studies by Li et al[2] and Zhao et al[3] have provided fresh perspectives on the molecular intricacies of CRC and presented novel therapeutic targets. Li et al[2] focused on the long non-coding RNA (lncRNA) PRNT/ZNF184/HIPK2 axis and its role in oxaliplatin resistance, while Zhao et al[3] investigated the influence of human β-defensin-1 on autophagy through the lncRNA TCONS_00014506.
LncRNAs are implicated in the onset and progression of various human malignancies, including CRC[4]. These molecules can interact with RNA-binding proteins and act as competing endogenous RNAs (ceRNAs) for microRNAs (miRNAs), which are involved in epigenetic modification of DNA and RNA, thereby affecting the expression of relevant genes. These lncRNA-mediated changes can induce or inhibit autophagy in tumour cells and mediate drug resistance[4-6].
Autophagy is a cellular process in which components such as proteins and organelles are transported to lysosomes for degradation[7]. Previous studies have shown that autophagy plays dual roles in tumorigenesis[8]. On the one hand, the expression of lncRNAs has been shown to regulate autophagy in CRC, and on the other hand, lncRNAs and autophagy have cross-regulatory and dual effects on tumours: lncRNAs can increase or decrease autophagy, and changes in autophagy can further promote or inhibit tumour growth[6].
Chemotherapy is essential for patients with locally advanced or metastatic CRC[9]. Oxaliplatin-based chemotherapy has been used worldwide as a first-line treatment for CRC[10]. However, the prognosis of CRC patients is not as satisfactory as expected. Endogenous and acquired oxaliplatin resistance is now considered a major obstacle limiting treatment success[11]. Studies have shown that lncRNAs are key factors in oxaliplatin resistance, and their dysregulation induces the activation of several signalling pathways, which ultimately leads to chemoresistance[12].
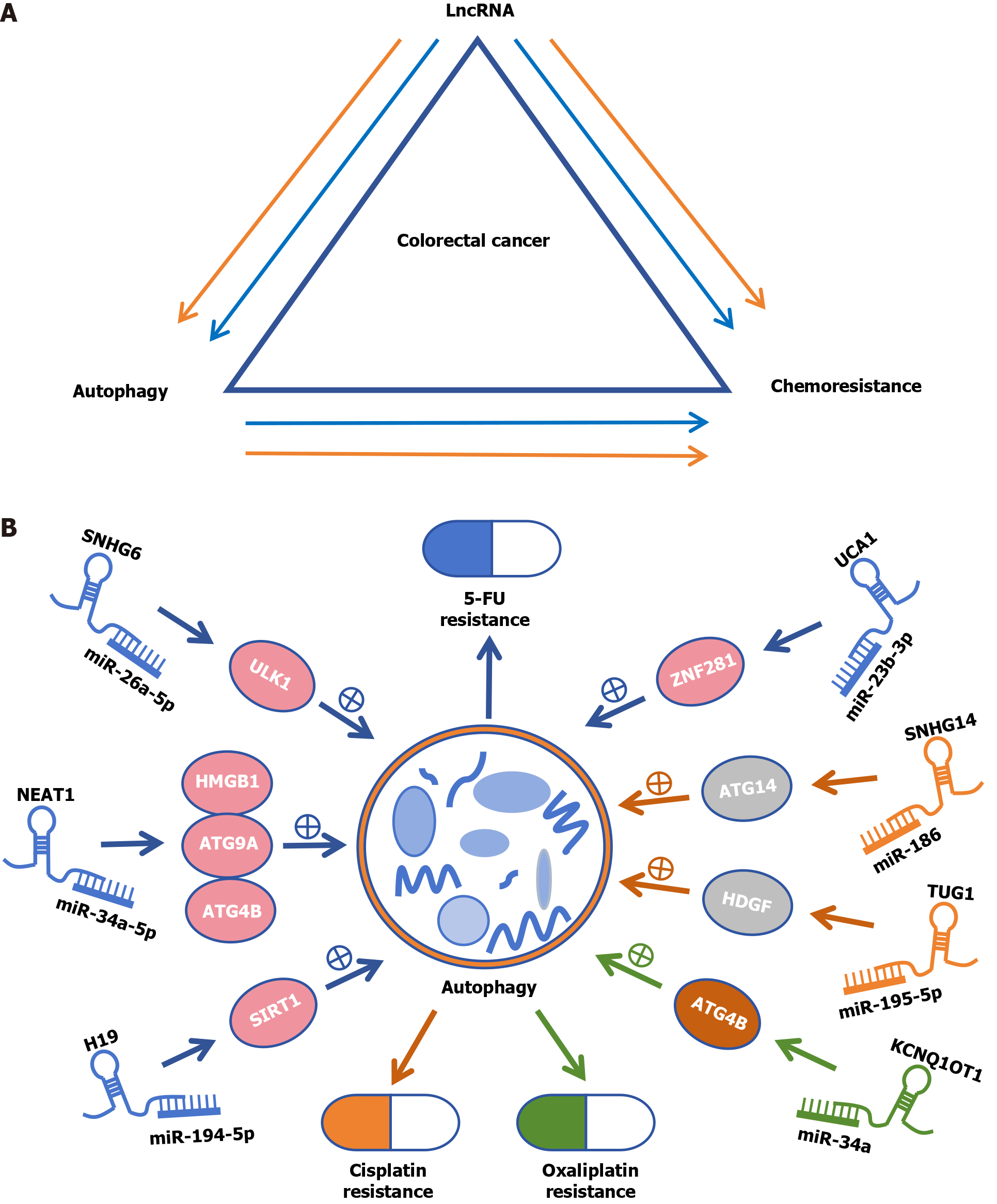
Autophagy also plays dual roles in the development of chemotherapeutic resistance, either by promoting or inhibiting resistance through many complex mechanisms, including the regulation of autophagy-related lncRNAs (ARlncRNAs)[6] (Figure 1A). In this editorial, we focus on the role of lncRNAs in the regulation of autophagy and chemoresistance in CRC and explore the feasibility of modulating chemosensitivity by interfering with lncRNAs involved in the autophagic process.
The effects of lncRNAs on autophagy and the influence of lncRNA-regulated autophagy on CRC pathogenesis are complicated[13]. Inhibition of autophagy by linc-POU3F3 in LOVO and SW480 CRC cells may enhance the malignant phenotype of CRC[14]. Similarly, the inhibition of LINC00858 induces colon cancer cell apoptosis, senescence, and autophagy. LINC00858 functions as a tumour-promoting lncRNA in colon cancer through the downregulation of WNK2[15]. Moreover, the lncRNA CPS1-IT might suppress metastasis and EMT by inhibiting hypoxia-induced autophagy through the inactivation of HIF-1α in CRC[16]. In addition, the lncRNAs SLCO4A1-AS1[17], UCA1[18], and MALAT1[19] promote autophagy through the miRNA-related axis, thus enhancing the proliferation of CRC cells. The dual regulatory role of ARlncRNAs, determined by the dual nature of autophagy, can serve as an effective therapeutic target for CRC[20].
Human β-defensin 1 (hBD-1) is a multifaceted antimicrobial peptide that acts as a tumour suppressor[21]. Zhao et al[3] verified that hBD-1 may induce autophagy in colon cancer SW620 cells by inhibiting the phosphorylation of mTOR through the lncRNA TCONS_00014506 at the cellular level. However, this study focused on a single cell line, and the impact of hBD-1 was not assessed; this is a limitation that could be addressed in future research. Moreover, an investigation of the expression and function of TCONS_00014506 in patient-derived tumour samples could provide clinically relevant support for the in vitro findings. Bioinformatics analysis of autophagy-related differentially expressed lncRNAs was performed to offer novel insights on strategies that could complement existing treatments to potentially overcome resistance mechanisms and improve patient outcomes.
In recent years, emerging evidence has shown that lncRNAs play irreplaceable roles in drug resistance[4,6,22]. However, we have limited knowledge of the lncRNAs that are closely related to oxaliplatin resistance in CRC. A novel lncRNA, Linc00152 antagonizes oxaliplatin sensitivity by acting as a competing ceRNA to modulate the expression of miR-193a-3p and subsequently modulate the expression of erb-b2 receptor tyrosine kinase 4[23]. The lncRNAs CACS15[24], KCNQ1OT1[25], MEG3[26], CRNDE[27], LINC00525[28], and MALAT1[29,30] promote oxaliplatin resistance by sponging specific miRNAs. In addition, the lncRNA CCAL facilitates resistance to oxaliplatin in CRC cells by increasing the level of β-catenin expression[31]. The lncRNA LUCAT1 affects oxaliplatin sensitivity through the p53 signalling pathway. Knockdown of LUCAT1 renders CRC cells hypersensitive to oxaliplatin treatment[32].
A recent study by Li et al[2] revealed that the lncRNA PRNT is upregulated in oxaliplatin-resistant CRC cells and modulates the expression of HIPK2 by sponging ZNF184, marring bioinformatics analyses with robust in vitro and in vivo experiments. However, there is a pressing need to validate the clinical relevance of the PRNT/ZNF184/HIPK2 axis in a larger cohort of CRC patients. Considering the role of PRNT in chemoresistance, a systematic characterization of lncRNAs may redefine our approach to cancer treatment. Furthermore, examining the interplay between lncRNAs and other signalling pathways could uncover additional layers of regulation and points of intervention.
ARlncRNAs can mediate both sides of autophagy and thus can positively or negatively affect drug resistance[20]. Autophagy-mediated drug resistance is a complex phenomenon involving multiple factors, including the recirculation of cytoplasmic components, gene repair mechanisms, alterations in drug concentration and metabolism, changes in the expression or activity of key proteins, and modifications in apoptotic and survival signalling pathways[6].
LncRNA H19 promotes SIRT1-dependent autophagy via miR-194-5p, thereby inducing 5-Fluorouracil (5-FU) resistance in CRC[33]. The knockdown of NEAT1 attenuates autophagy by targeting miR-34a to increase 5-FU sensitivity[34]. SNHG6 may promote chemoresistance through ULK1-induced autophagy by sponging miR-26a-5p in CRC cells[35]. In vivo and in vitro, the lncRNA UCA1 promotes autophagy through the miR-23b-3p/ZNF281 axis, which mediates resistance to 5-FU in CRC cells[36]. Similarly, the lncRNA SNHG14 stimulates cellular autophagy through interaction with the miR-186/ATG14 axis to promote cisplatin resistance in CRC[37]. The lncRNA TUG1 targets miR-195-5p and blocks its expression. MiR-195-5p promotes the HDGF/DDX/β-catenin axis, thereby triggering autophagy, which promotes resistance to cisplatin[38] (Figure 1B).
Both recent studies are noteworthy, as they revealed the roles of lncRNAs in CRC progression and treatment resistance. A more comprehensive approach is necessary to elucidate the network of lncRNA interactions in CRC since they focused on individual lncRNA. ARlncRNAs act by regulating specific downstream miRNAs[20]. However, individual lncRNA can regulate more than one miRNA, so broader exploration of the numerous targets or signalling pathways downstream of ARlncRNAs is essential. Furthermore, the interaction between autophagy and chemoresistance remains an underexplored topic, and studies in this area may reveal innovative CRC therapies.
Although modulating chemosensitivity by interfering with lncRNAs involved in the autophagy process is a promising new approach for cancer treatment, multicentre validations based on sufficient samples are still necessary. With our increasing knowledge of lncRNAs in CRC, there is promise that some lncRNAs might be applied in biomarker-directed precision medicine approaches to improve survival outcomes in CRC patients.
| 1. | Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 10895] [Article Influence: 3631.7] [Reference Citation Analysis (2)] |
| 2. | Li SN, Yang S, Wang HQ, Hui TL, Cheng M, Zhang X, Li BK, Wang GY. Upregulated lncRNA PRNT promotes progression and oxaliplatin resistance of colorectal cancer cells by regulating HIPK2 transcription. World J Gastrointest Oncol. 2024;16:1564-1577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 3. | Zhao YX, Cui Y, Li XH, Yang WH, An SX, Cui JX, Zhang MY, Lu JK, Zhang X, Wang XM, Bao LL, Zhao PW. Human β-defensin-1 affects the mammalian target of rapamycin pathway and autophagy in colon cancer cells through long non-coding RNA TCONS_00014506. World J Gastrointest Oncol. 2024;16:1465-1478. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 4. | Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77:3965-3981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2088] [Cited by in RCA: 2229] [Article Influence: 247.7] [Reference Citation Analysis (0)] |
| 5. | Chan JJ, Tay Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 528] [Cited by in RCA: 850] [Article Influence: 106.3] [Reference Citation Analysis (0)] |
| 6. | Chang H, Zou Z. Targeting autophagy to overcome drug resistance: further developments. J Hematol Oncol. 2020;13:159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 188] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 7. | Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol Cancer. 2020;19:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 304] [Cited by in RCA: 1202] [Article Influence: 200.3] [Reference Citation Analysis (0)] |
| 8. | Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1283] [Cited by in RCA: 1365] [Article Influence: 65.0] [Reference Citation Analysis (0)] |
| 9. | Biller LH, Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA. 2021;325:669-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 1733] [Article Influence: 346.6] [Reference Citation Analysis (1)] |
| 10. | Yang AD, Fan F, Camp ER, van Buren G, Liu W, Somcio R, Gray MJ, Cheng H, Hoff PM, Ellis LM. Chronic oxaliplatin resistance induces epithelial-to-mesenchymal transition in colorectal cancer cell lines. Clin Cancer Res. 2006;12:4147-4153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 438] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 11. | Huang H, Zhu L, Reid BR, Drobny GP, Hopkins PB. Solution structure of a cisplatin-induced DNA interstrand cross-link. Science. 1995;270:1842-1845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 238] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 12. | Qi FF, Yang Y, Zhang H, Chen H. Long non-coding RNAs: Key regulators in oxaliplatin resistance of colorectal cancer. Biomed Pharmacother. 2020;128:110329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 13. | Cheng L, Han T, Zhang Z, Yi P, Zhang C, Zhang S, Peng W. Identification and Validation of Six Autophagy-related Long Non-coding RNAs as Prognostic Signature in Colorectal Cancer. Int J Med Sci. 2021;18:88-98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 14. | Shan TD, Xu JH, Yu T, Li JY, Zhao LN, Ouyang H, Luo S, Lu XJ, Huang CZ, Lan QS, Zhong W, Chen QK. Knockdown of linc-POU3F3 suppresses the proliferation, apoptosis, and migration resistance of colorectal cancer. Oncotarget. 2016;7:961-975. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 15. | Wu J, Meng X, Gao R, Jia Y, Chai J, Zhou Y, Wang J, Xue X, Dang T. Long non-coding RNA LINC00858 inhibits colon cancer cell apoptosis, autophagy, and senescence by activating WNK2 promoter methylation. Exp Cell Res. 2020;396:112214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 16. | Zhang W, Yuan W, Song J, Wang S, Gu X. LncRNA CPS1-IT1 suppresses EMT and metastasis of colorectal cancer by inhibiting hypoxia-induced autophagy through inactivation of HIF-1α. Biochimie. 2018;144:21-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 90] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 17. | Wang Z, Jin J. LncRNA SLCO4A1-AS1 promotes colorectal cancer cell proliferation by enhancing autophagy via miR-508-3p/PARD3 axis. Aging (Albany NY). 2019;11:4876-4889. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 18. | Song F, Li L, Liang D, Zhuo Y, Wang X, Dai H. Knockdown of long noncoding RNA urothelial carcinoma associated 1 inhibits colorectal cancer cell proliferation and promotes apoptosis via modulating autophagy. J Cell Physiol. 2019;234:7420-7434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 19. | Si Y, Yang Z, Ge Q, Yu L, Yao M, Sun X, Ren Z, Ding C. Long non-coding RNA Malat1 activated autophagy, hence promoting cell proliferation and inhibiting apoptosis by sponging miR-101 in colorectal cancer. Cell Mol Biol Lett. 2019;24:50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 85] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 20. | Zhang Y, Tang J, Wang C, Zhang Q, Zeng A, Song L. Autophagy-related lncRNAs in tumor progression and drug resistance: A double-edged sword. Genes Dis. 2024;11:367-381. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 27] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 21. | Álvarez ÁH, Martínez Velázquez M, Prado Montes de Oca E. Human β-defensin 1 update: Potential clinical applications of the restless warrior. Int J Biochem Cell Biol. 2018;104:133-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 22. | Chen B, Dragomir MP, Yang C, Li Q, Horst D, Calin GA. Targeting non-coding RNAs to overcome cancer therapy resistance. Signal Transduct Target Ther. 2022;7:121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 306] [Article Influence: 76.5] [Reference Citation Analysis (0)] |
| 23. | Yue B, Cai D, Liu C, Fang C, Yan D. Linc00152 Functions as a Competing Endogenous RNA to Confer Oxaliplatin Resistance and Holds Prognostic Values in Colon Cancer. Mol Ther. 2016;24:2064-2077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 158] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 24. | Gao R, Fang C, Xu J, Tan H, Li P, Ma L. LncRNA CACS15 contributes to oxaliplatin resistance in colorectal cancer by positively regulating ABCC1 through sponging miR-145. Arch Biochem Biophys. 2019;663:183-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 79] [Article Influence: 11.3] [Reference Citation Analysis (1)] |
| 25. | Li Y, Li C, Li D, Yang L, Jin J, Zhang B. lncRNA KCNQ1OT1 enhances the chemoresistance of oxaliplatin in colon cancer by targeting the miR-34a/ATG4B pathway. Onco Targets Ther. 2019;12:2649-2660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 98] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 26. | Wang H, Li H, Zhang L, Yang D. Overexpression of MEG3 sensitizes colorectal cancer cells to oxaliplatin through regulation of miR-141/PDCD4 axis. Biomed Pharmacother. 2018;106:1607-1615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 27. | Gao H, Song X, Kang T, Yan B, Feng L, Gao L, Ai L, Liu X, Yu J, Li H. Long noncoding RNA CRNDE functions as a competing endogenous RNA to promote metastasis and oxaliplatin resistance by sponging miR-136 in colorectal cancer. Onco Targets Ther. 2017;10:205-216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 79] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 28. | Wang S, Li J, Yang X. Long Non-Coding RNA LINC00525 Promotes the Stemness and Chemoresistance of Colorectal Cancer by Targeting miR-507/ELK3 Axis. Int J Stem Cells. 2019;12:347-359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 29. | Li P, Zhang X, Wang H, Wang L, Liu T, Du L, Yang Y, Wang C. MALAT1 Is Associated with Poor Response to Oxaliplatin-Based Chemotherapy in Colorectal Cancer Patients and Promotes Chemoresistance through EZH2. Mol Cancer Ther. 2017;16:739-751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 217] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 30. | Xie JJ, Li WH, Li X, Ye W, Shao CF. LncRNA MALAT1 promotes colorectal cancer development by sponging miR-363-3p to regulate EZH2 expression. J Biol Regul Homeost Agents. 2019;33:331-343. [PubMed] |
| 31. | Deng X, Ruan H, Zhang X, Xu X, Zhu Y, Peng H, Kong F, Guan M. Long noncoding RNA CCAL transferred from fibroblasts by exosomes promotes chemoresistance of colorectal cancer cells. Int J Cancer. 2020;146:1700-1716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 172] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 32. | Zhou Q, Hou Z, Zuo S, Zhou X, Feng Y, Sun Y, Yuan X. LUCAT1 promotes colorectal cancer tumorigenesis by targeting the ribosomal protein L40-MDM2-p53 pathway through binding with UBA52. Cancer Sci. 2019;110:1194-1207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 87] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 33. | Wang M, Han D, Yuan Z, Hu H, Zhao Z, Yang R, Jin Y, Zou C, Chen Y, Wang G, Gao X, Wang X. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018;9:1149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 161] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 34. | Liu F, Ai FY, Zhang DC, Tian L, Yang ZY, Liu SJ. LncRNA NEAT1 knockdown attenuates autophagy to elevate 5-FU sensitivity in colorectal cancer via targeting miR-34a. Cancer Med. 2020;9:1079-1091. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 35. | Wang X, Lan Z, He J, Lai Q, Yao X, Li Q, Liu Y, Lai H, Gu C, Yan Q, Fang Y, Zhang Y, Li A, Liu S. LncRNA SNHG6 promotes chemoresistance through ULK1-induced autophagy by sponging miR-26a-5p in colorectal cancer cells. Cancer Cell Int. 2019;19:234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 36. | Xian Z, Hu B, Wang T, Zeng J, Cai J, Zou Q, Zhu P. lncRNA UCA1 Contributes to 5-Fluorouracil Resistance of Colorectal Cancer Cells Through miR-23b-3p/ZNF281 Axis. Onco Targets Ther. 2020;13:7571-7583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 37. | Han Y, Zhou S, Wang X, Mao E, Huang L. SNHG14 stimulates cell autophagy to facilitate cisplatin resistance of colorectal cancer by regulating miR-186/ATG14 axis. Biomed Pharmacother. 2020;121:109580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 38. | Xia C, Li Q, Cheng X, Wu T, Gao P, Gu Y. Insulin-like growth factor 2 mRNA-binding protein 2-stabilized long non-coding RNA Taurine up-regulated gene 1 (TUG1) promotes cisplatin-resistance of colorectal cancer via modulating autophagy. Bioengineered. 2022;13:2450-2469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/