INTRODUCTION
Colorectal cancer (CRC) is one of the most common malignant tumors of the digestive system[1-3]. The treatment of CRC is based on radical surgery and supplemented by chemotherapy, but nearly half of the patients are still trapped by tumor recurrence or metastasis without effective treatment. The occurrence of CRC is a process involving multi-stage mutation accumulation of multiple oncogenes, and the immune microenvironment of the tumor also plays an important role in regulation[4]. Due to its important function of disease defense and monitoring, the immune system has been occupying the hot spot of tumor treatment in recent years, which has opened up a new direction for the treatment of CRC. In the past 10 years, the clinical application of tumor immunotherapy has brought exciting results, but it also brings a series of problems with the heterogeneity within and between tumors[5]. Taking immune checkpoint block (ICB) therapy as an example, programmed death protein-1 (PD-1) blockers have shown considerable efficacy in patients with metastatic CRC with mismatch repair deficiency and high microsatellite instability, who exhibit high levels of tumor mutation load and immune infiltration[6-8]. In contrast, CRC patients with mismatch repair and microsatellite stabilization do not respond to ICB therapy, and their low tumor mutation load and immune infiltration are important reasons for treatment resistance. Therefore, the immune microenvironment largely determines the difficulty of treatment and the survival prognosis of patients with CRC[9]. In the long term, the synergistic anti-tumor effect of T cell as the main guide, supplemented by dendritic cell, macrophage and other antigen presentation has long occupied the mainstream research direction of immunotherapy. However, the role of B cells, which are multifunctional cells capable of both antigen presentation and foreign body defense, in the tumor microenvironment, especially in CRC, has not been adequately studied[10-12].
With the in-depth research in recent years, the diversified role of B cells in the tumor immune microenvironment has gradually been paid attention to. B cells are the source of a variety of cytokines and chemokines and have important roles in regulating immune responses, including guiding lymphoid tissue development, shaping and promoting effects and memory T cell responses, it is also possible to induce cellule-driven three-stage gonadal organization (TLS) independent of gonadal organization by producing Lα1β2. In addition, B cells produce interleukin (IL)-10 and IL-35 negatively to modulate the immune response[13]. B cells and plasma cells can support anti-tumor immune responses through a variety of mechanisms. Recent studies have shown that B cells infiltrated by tumor tissue and B cells and plasma cells in lymph nodes drained by tumor play an important role in anti-tumor immune response[14]. A study of CRC-infiltrated B cells showed that CD20+B cells have good prognostic value for the progression of CRC and demonstrated a significant correlation between CD20+B cells and CD8+T cells[15].
The mechanism of circulating and infiltrating B cells in the immune microenvironment of CRC has been the focus of much attention. Much of the controversy has focused on the role and mechanism of B cells in anti-tumor immune responses. On the one hand, some scholars believe that B cells have a positive impact on the immune response of CRC by regulating the activity of immune cells and secreting antibodies. On the other hand, there are also studies supporting the inhibitory role of B cells in the immune microenvironment, which may promote the immune escape of tumors by inhibiting the activity of T cells. Recent research advances have shown that single-cell sequencing technology has made remarkable achievements in revealing circulating and infiltrating B-cell heterogeneity. This technique not only enables researchers to dig deeper into the specific expression profile and function of B cell subsets but also reveals their interactions in the immune microenvironment of CRC. This provides entirely new cell-level insight into further understanding of the complex regulatory mechanisms of B cells in cancer immunity. We want to settle current debates in the field by learning more about B-cell heterogeneity at the cellular level and using multi-omics approaches in a planned way. This will help us make a unique contribution to understanding how the immune system controls CRC. This research innovatively integrates advanced sequencing technology and has profound implications for the field of cancer immunotherapy.
Based on the analysis of single-cell transcriptome and antibody library data, this study explored the role of B-cell subtypes in the immune microenvironment and tumor progression of CRC, compared the differences in the composition of tumor and paracancer invasive B-cell subsets, studied the potential molecular driving mechanism, and analyzed the changes in the interaction between B-cells and immune cells in the microenvironment. To explore the possible regulatory pathway to promote the anti-tumor effect of B cells, in order to provide a new idea for tumor immunotherapy of CRC.
MATERIALS AND METHODS
Samples were collected and suspensions prepared for single-cell sequencing
Surgical samples were collected from 23 patients (All stage I/II CRC patients) from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). After the sampling criteria were informed, tumor, adjacent normal tissue and peripheral blood samples of each patient were collected under the same environmental conditions, with a total of 9 samples. The fresh tissue samples were digested and prepared into single-cell suspension. CD45+ cells were obtained by flow sorting. Peripheral blood mononuclear cells (PBMC) were obtained from blood samples after derubing treatment. After quality control, the transcriptome and VDJ libraries were constructed with 10XGenomics technology for single cell sequencing. The transcriptome and B cell anti receptor library (BCR) data were assembled and compared with the genome.
To construct transcriptome maps of infiltrating and circulating B-cell subsets in CRC
The preliminary analysis of transcriptome data was performed using the Seurat 4.0 software package. First, a single sample is processed according to a standardized process, which consists of the following steps: Quality control (elimination of cells with less than 200 and more than 8000 expressing genes, elimination of genes expressed in less than 10 cells), standardization of sequencing data by sequencing depth, log transformation, search for highly variable genes (2000), principal component analysis, K-means unsupervised clustering. Finally, the visualization is realized by using the unified stream approach and projection. According to the classification of marker genes provided in experiments, immune cells were divided into the following groups: (1) T/NK cells, The tagged genes were CD3 delta subunit of T-cell receptor complex (CD3D), granulysin (GNLY), and granuleprotein 7, a natural killer cell (NKG7); (2) B cells and plasma cells, The tagged genes were CD79a molecule, membrane spanning 4-domains A1, PR/SET domain 1 and syndecan 1; and (3) myeloid cells, labeled as interleukin 1β, S100 calcium binding proteinA8 and cystatin C. After removing batch effect, Harmony 3.4 software package was used to integrate all samples, and after processing according to the above standardized process, all B cells including plasma cells were screened out. BCR data were matched according to the cell name, and then re-clustered and grouped according to the standardized process, and subgroups were divided according to the reference markers. Calculate the proportion of each subpopulation in each sample. At the same time, T/NK cells and myeloid cell populations were grouped according to the above process, and the results were used for cell communication analysis.
Differential gene and pathway enrichment analysis of B cell subsets in normal and tumor tissues
The differential genes of B cell subpopulations in tumor and adjacent normal tissue samples were calculated using MAST 1.20 package. |log2 (fold change) | (|log2 (FC) |) genes with a corrected P value less than 0.001 were selected. The differential factors of up-modulation and down-modulation were sorted out, and the pathway enrichment analysis of differential genes in each subpopulation was performed with clusterProfiler 3.14 package. The pathway enrichment analysis was conducted according to the corrected P-value from smallest to largest, and the top 10 pathways among up-modulation and down-modulation pathways were selected for biological analysis.
Based on TCGA-colon cancer data, the changes of B-cell subpopulations in tumor and normal samples were verified
In order to further verify the accuracy of the results, we applied the CIBERSORTx 1.0 deconvolution method to map the gene expression profiles of each subgroup of B cells into the colon cancer data (COAD) of TCGA for analysis. The characteristic gene composition scores of B-cell subsets in each sample were calculated, and their changes in tumor and normal tissues were compared. At the same time, according to the information of disease progression of patients, the differences of each subgroup in tumor progression of patients were analyzed.
Analysis of interaction between B cell subsets and immune cells
T/NK cells and myeloid cells were selected for re-clustering and grouping, and statistically significant molecular pairs among cell subsets were calculated using cell Phone EDB v3.0 cell communication software. Molecular pairs with corrected P value less than 0.05 were selected for analysis, focusing on germinal center B cells (GCB). To identify important regulatory molecules and analyze their roles in the immune microenvironment.
B cell VDJ group data analysis
On a patient-by-patient basis, the cloning index of each subpopulation of B cells was calculated and migration was assessed. The statistical difference between the groups was compared using the paired T-test.
Among them, m represents the number of clones whose cell number is greater than 2. n represents the number of cells contained in clone type i; N represents the total number of cells per cell subpopulation in a single sample.
m represents the number of clonotypes shared in subpopulations C1 and C2; ni-C represents the number of cells contained in clonotype i in subpopulation C; N represents the total number of cells per cell subpopulation in a single sample.
B cell antigen is subject to mutation level analysis of weight chain and light chain
The tool pRESTO v0.70 was used to analyze the mutation level of BCR, and the following quality control was performed: Sequences that were not recognized by primers or had poor scores were read, and sequences that appeared at least twice in a single sample were retained; Use the International Immune Gene Database as a reference; non-functional sequences were removed and sequences with more than 30 mutations were removed; A symmetrically weighted number of nucleotide point mutations was selected as the number of mutation sites. The average number of mutation sites of each group of B cells represented the mutation level of the cell population, and the changes in tumor and normal tissue were compared.
Statistical method
R software package states v0.3.1 was used for statistical analysis. Quantitative data included cell composition ratio, clonal expansion index and B-cell heavy chain mutation number, which were expressed in mean ± SD form Kruskal-Wallis test was used to compare the difference in the proportion of cell population composition in tumor, normal and blood samples. In the TCGA-COAD validation dataset, the difference in subpopulation composition between tumor and normal tissue was analyzed using independent sample t test. Kruskal-Wallis test was used to analyze the difference of B-cell clonal expansion index between tumor and normal tissue. In the analysis of B-cell heavy chain mutation levels, statistical differences between samples were analyzed using the independent sample t test. P < 0.05 indicated that the difference was statistically significant.
RESULTS
Transcriptome map of global immune cells and B cell subsets
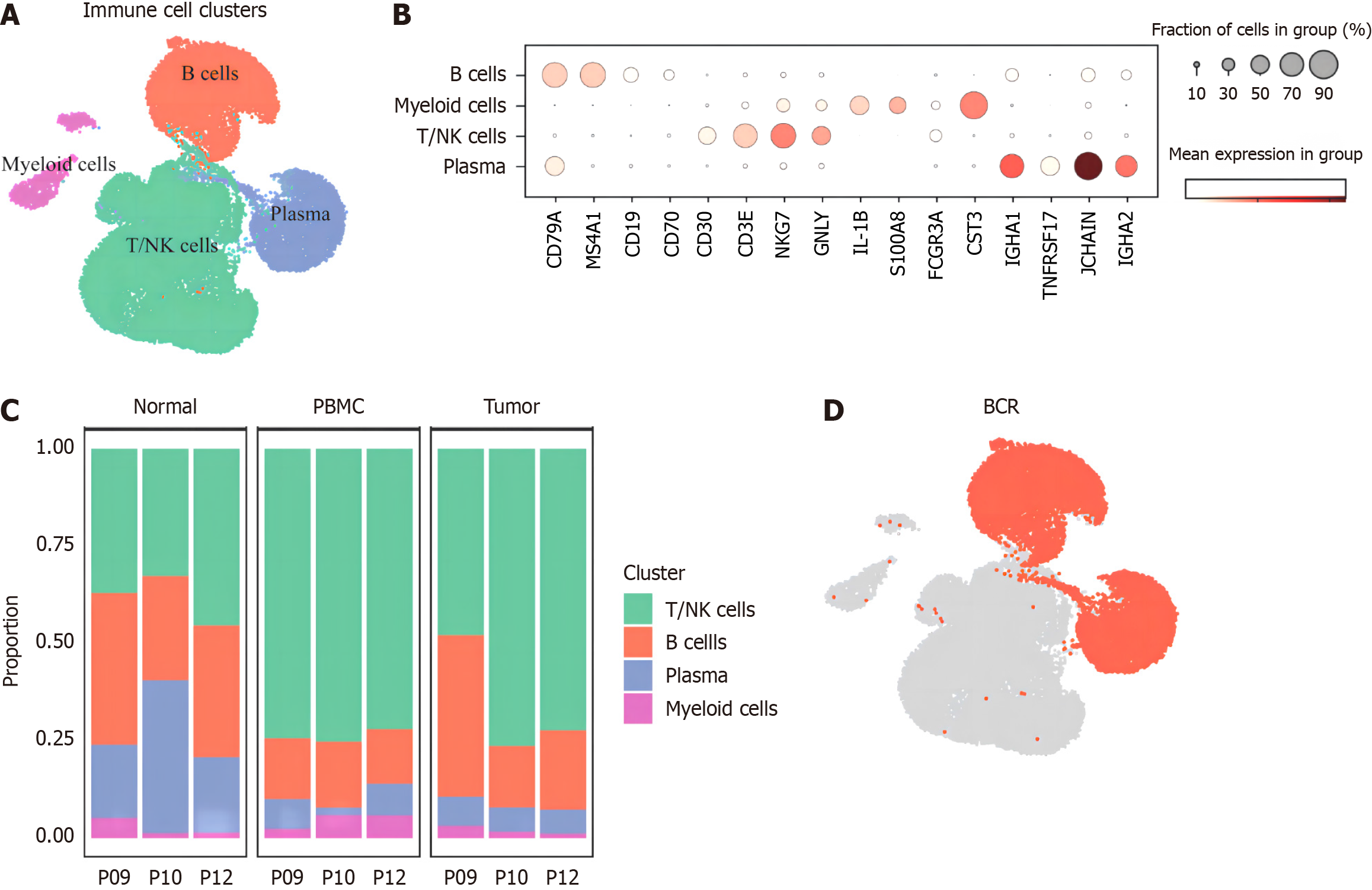
Our study integrated a total of 69 samples of CD45+ immune cells of untreated CRC patients from TCGA database (https://portal.gdc.cancer.gov/). After quality control and batch correction and integration, a total of 35619 cells were obtained. There were 18246 T/NK cells, 14244 B and plasma cells, and 3 129 myeloid cells (Figure 1A). The marker genes were shown in Figure 1B. It can be seen from the proportion maps of cell composition in the three tissues (Figure 1C) that T/NK cells generally increased in tumor samples, while B cells, especially plasma cells, generally decreased, initially reflecting a weakening trend in the role of plasma cells in CRC immune microenvironment. Next, all B cells and plasma cells were sorted out, combined with 8 804 BCR-positive cells assisted sorting (Figure 1D), for subsequent analysis.
Figure 1 Transcriptome map of main immune cells.
A: Distribution of main immune cell clusters; B: Marker genes' expression of main clusters; C: Cluster proportion among patients and samples; D: Total collected 88 04 B cells for paired B cell anti receptor library. BCR: B cell anti receptor library.
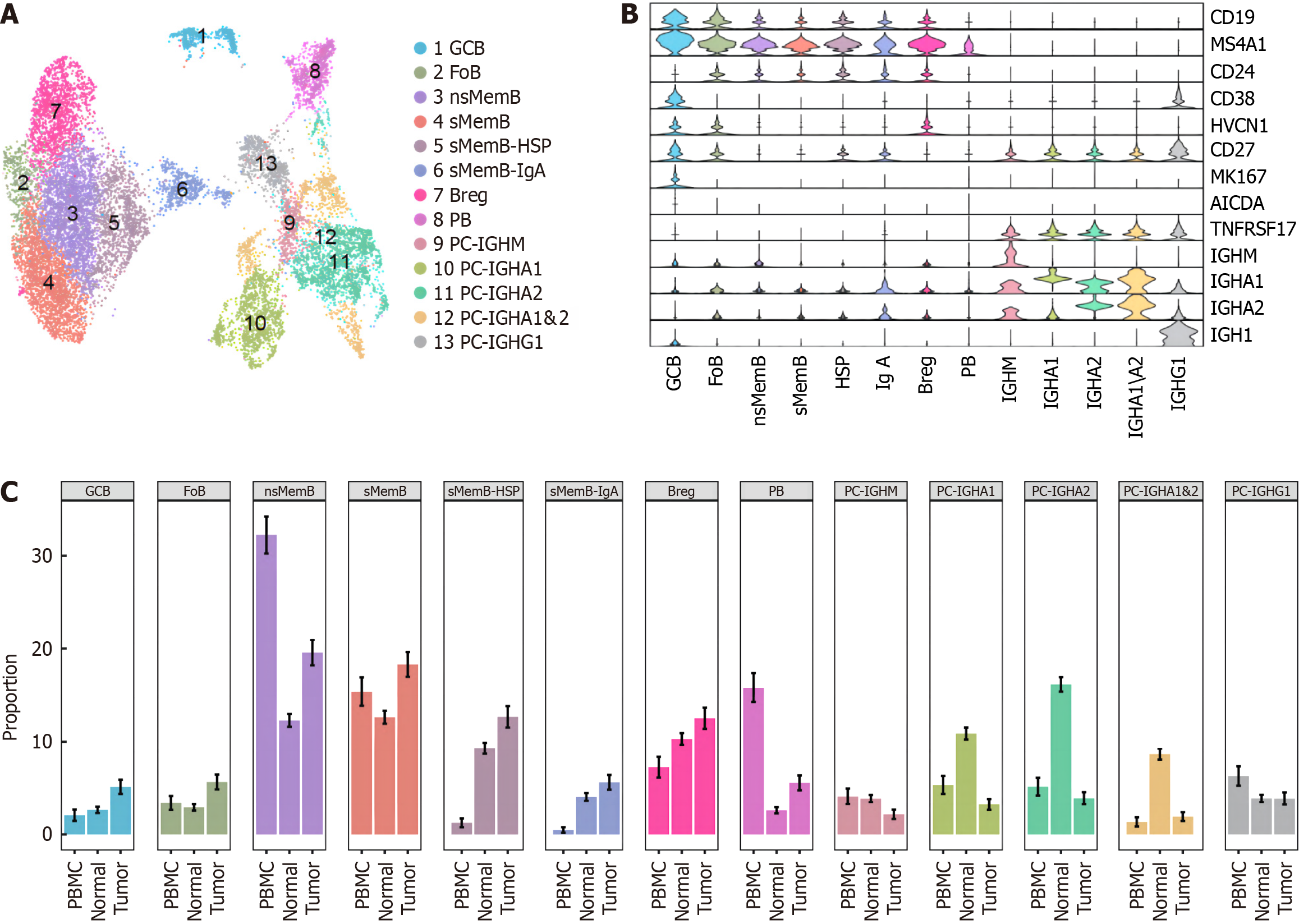
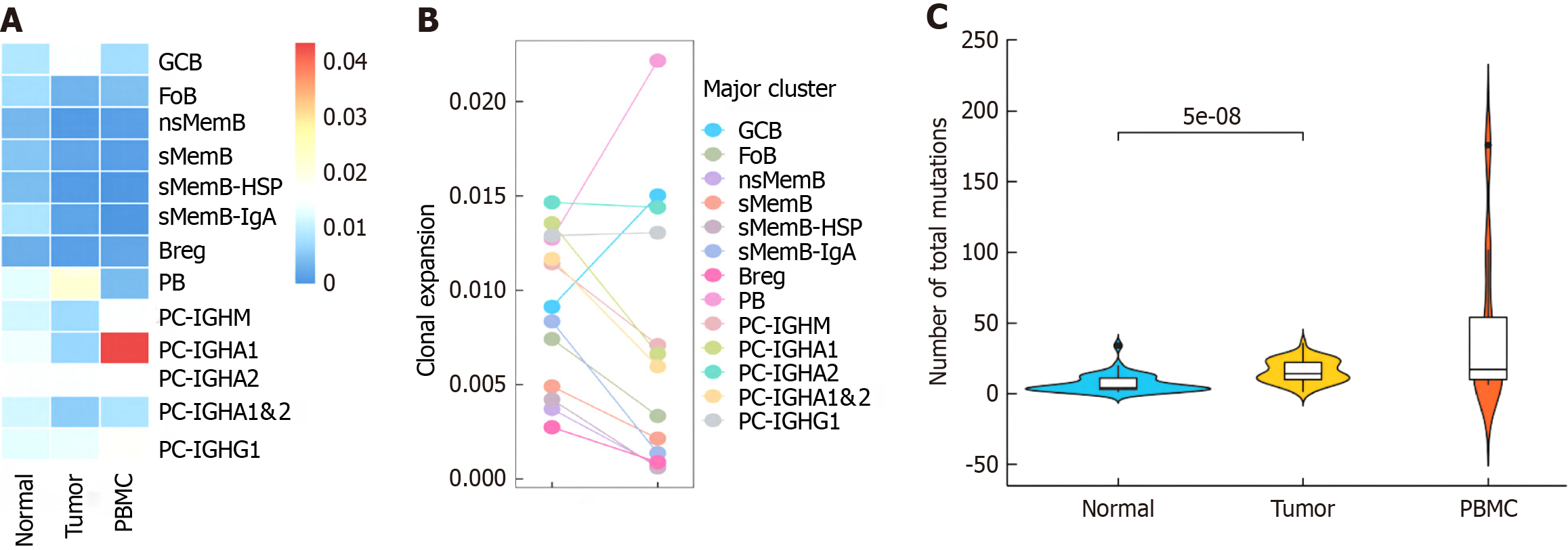
After processing 14 244 B cells according to the standardized process, our study regrouped them and got the following 13 cell subsets: GCB, follicular B lymphocyte, untransformed memory B cell (nsMemB), transformed memory B cell (sMemB), memory B with high epidarheuk egg white (SMEMB-HSP), memory B with high immunoglobulin IgA expression Cell (sMemB-IgA), modulatory B cell (Breg), plasmoblast cell (PB), and plasmocyte expressing various types of immune globular egg white subtypes. The marker genes are shown in Figure 2A and B. The subpopulations of B cells showed different infiltration patterns in the three tissue samples (Figure 2C). Compared with normal tissues, more CD20+B cells were infiltrated in tumor samples, and the levels of GCB, nsMemB, sMemB and SMEMB-HSP were significantly increased (P = 0.023, 0.002, 0.002, 0.031, respectively). The results showed that the role of CD20+B cells in the tumor microenvironment was significantly increased, while plasma cells (Especially those expressing immunoglobulin IgA types) showed a significant decline, indicating that the role of plasma cells in the microenvironment was significantly weakened. In addition, the composition of peripheral blood samples was mainly composed of memory B cells, especially untransformed B cells, indicating that memory B cells are the main force of circulating B cells to exert anti-tumor effects.
Figure 2 Atlas of B cell subclusters in colorectal cancer.
A: Distribution of 13 B cell subclusters; B: Marker genes' expression of subgroups; C: Proportion of B cell subclusters among peripheral blood mononuclear cells, normal and tumor samples.
Based on TCGA-COAD data, the differences of B-cell subpopulations in tumor and normal tissues were verified
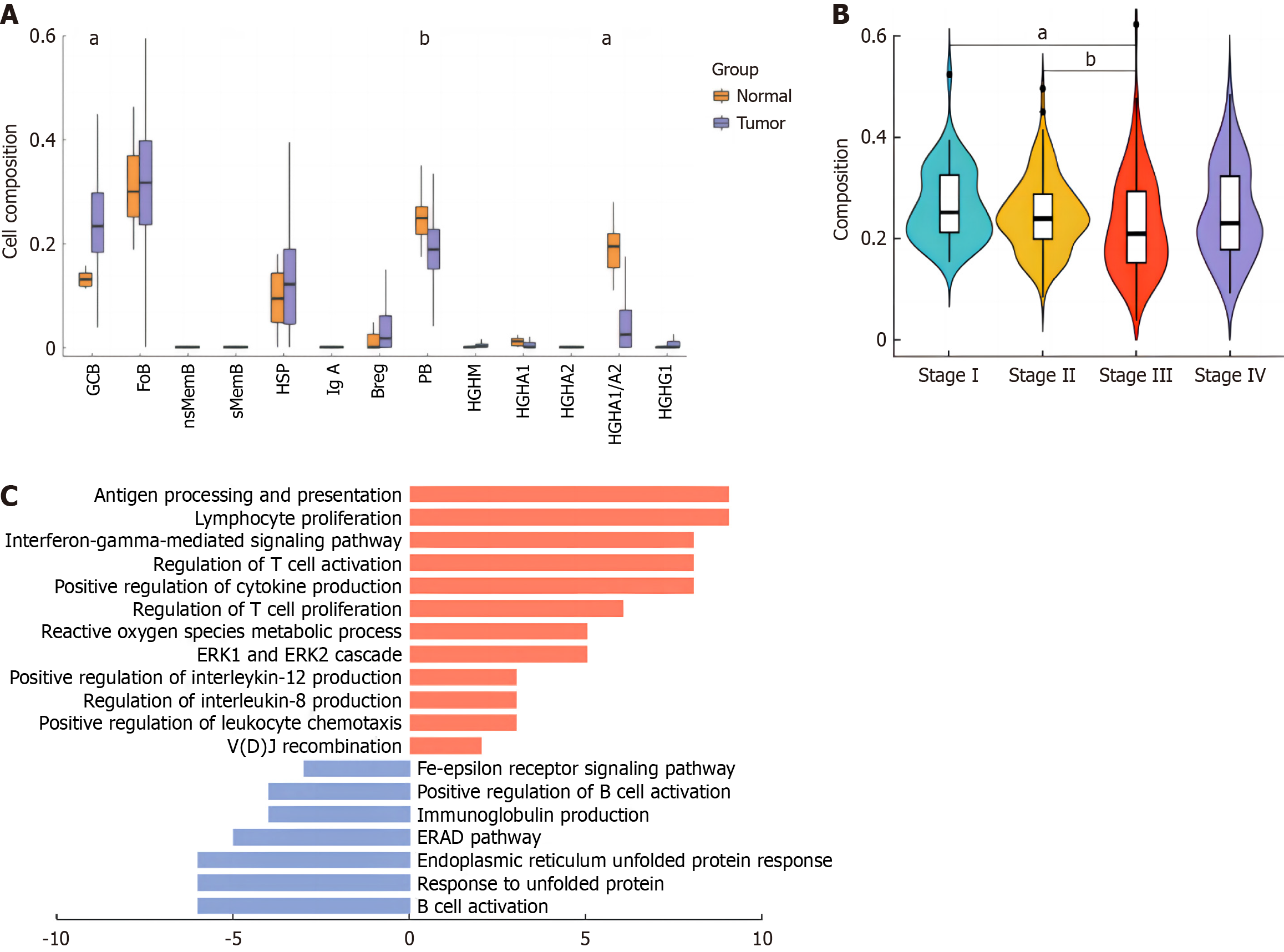
Our study mapped the single-cell expression profile of B cell subsets to the COAD data of TCGA to calculate the composition score of B cell subsets. As shown in Figure 3A, GCB increased significantly in tumors, and memory B cells after transformation increased significantly, which was consistent with the results of single-cell analysis. At the same time, the expression of IGHA1/2 and IGHG1 plasma cells decreased significantly, which further confirmed that the role of plasma cells in the microenvironment was limited. Background studies have reported that IGHG1 plasma cells are the main secreting cells of tumor-specific antibodies in the microenvironment, and can exercise anti-tumor effects through the dependent cytoplasmic mediated cytotoxicity (ADCC) of antibodies from NK cells and the induction of phagocytosis of macrophages. It is inferred that the antitumor effect of plasma cells is inhibited. GCB is the source of generation of memory B cells and plasma cells in the immune microenvironment. We considered whether the development status of GCB in the tumor immune microenvironment was changed, and then focused on the changes of GCB in the microenvironment.
Figure 3 Composition of B cell subsets in tumor and normal tissues by The Cancer Genome Atlas colon cancer data profiling.
A: Composition scores of B cell subsets in tumor vs normal tissues of The Cancer Genome Atlas-colon cancer data; B: Germinal center B cell (GCB) composition scores among progressing stages; C: Pathway enrichment analysis of GCB cluster. aP < 0.01; bP < 0.05.
Considering that our samples were all from early CRC patients, we combined the disease course information of TCGA patients to analyze the changes of GCB. As shown in Figure 3B, with the deterioration of the disease, GCB scores gradually decreased during stages I to III, and the differences were statistically significant (P values were 0.003 and 0.024, respectively), suggesting that the function of GCB was gradually inhibited during the development of tumors. In stage IV, although GCB characteristics showed an upward trend, there was no statistical significance. In order to further explore the changes of GCB driven by tumor microenvironment, we performed pathway enrichment analysis of GCB differential genes in tumor samples. Results as shown in Figure 3C, the antigen-presenting function of GCB in tumors was significantly increased, favourably supporting the activation and proliferation of T cells. At the same time, GCB itself was carrying out an effective VDJ rearrangement to improve the affinity of specific antigens, which had a positive anti-tumor effect.
Analysis of communication molecular pairs between GCB and T cells and myeloid cells
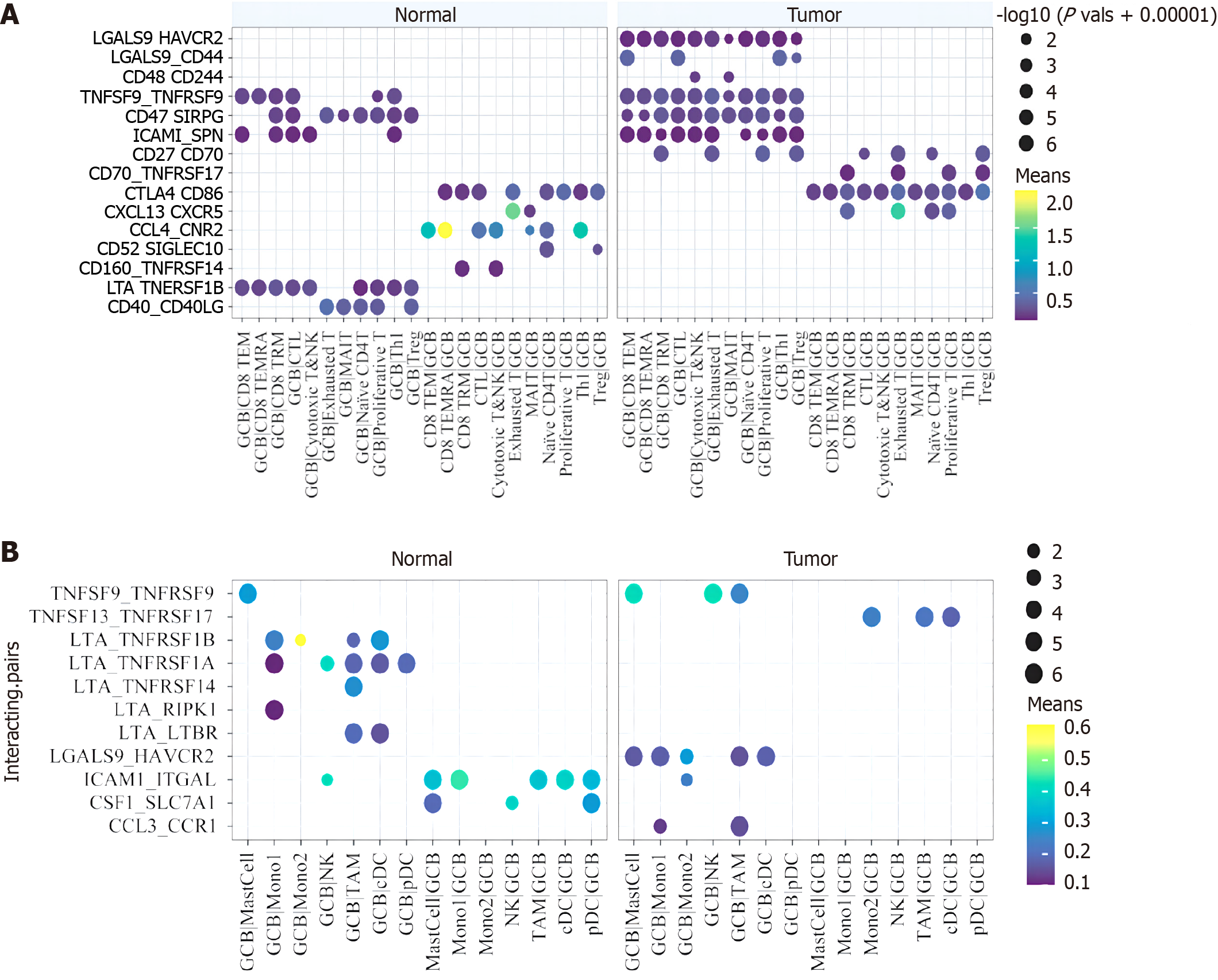
In the above study, we found that GCB may play a positive role in the immune microenvironment of CRC. In order to explore the influence factors of external environment, we conducted cell communication analysis. Figure 4A shows the communication molecular pairs with statistical significance in the T cell-GCB relationship network. It can be seen that in tumor samples, molecule pairs that inhibit T cell function are significantly decreased. The tumor effect is positive. In addition, elevated signals in tumors include molecular pairs that promote T and B cell activation and development, such as CD27-CD70 and CXCL13-CXCR5.
Figure 4 Interecting pairs between B cell subsets with T cells and myeloid cells.
A: Significant protein-protein interaction pairs between B cell subsets and T cell subsets; B: Significant protein-protein interaction pairs between B cell subsets and myeloids subsets.
However, there are also some weak inhibitory signals that are enhanced in tumors, such as LGALS9-CD44, which has been reported to promote and stabilize the development of regulatory T cells (Tregs). This suggests that the function of GCB in the germatogenic center is multifaceted, but mainly promotes and activates the antitumor effect of T cells. Perhaps taking measures against GCB to specifically activate and enhance the signal of T cell function and inhibit its association with Tregs or depleted T cells can effectively promote the anti-tumor effect of the immune microenvironment as a whole. In myeloid cells, however, the situation in which B cell subsets communicate with them changes less in tumors and normal tissues. Of concern is the up-regulation of tumor necrosis factor (TNF) receptor family signaling in tumors, suggesting that GCB cells have a weak role in promoting macrophage function in the microenvironment (Figure 4B).
Analysis of antigen receptor libraries of each subgroup of B cells
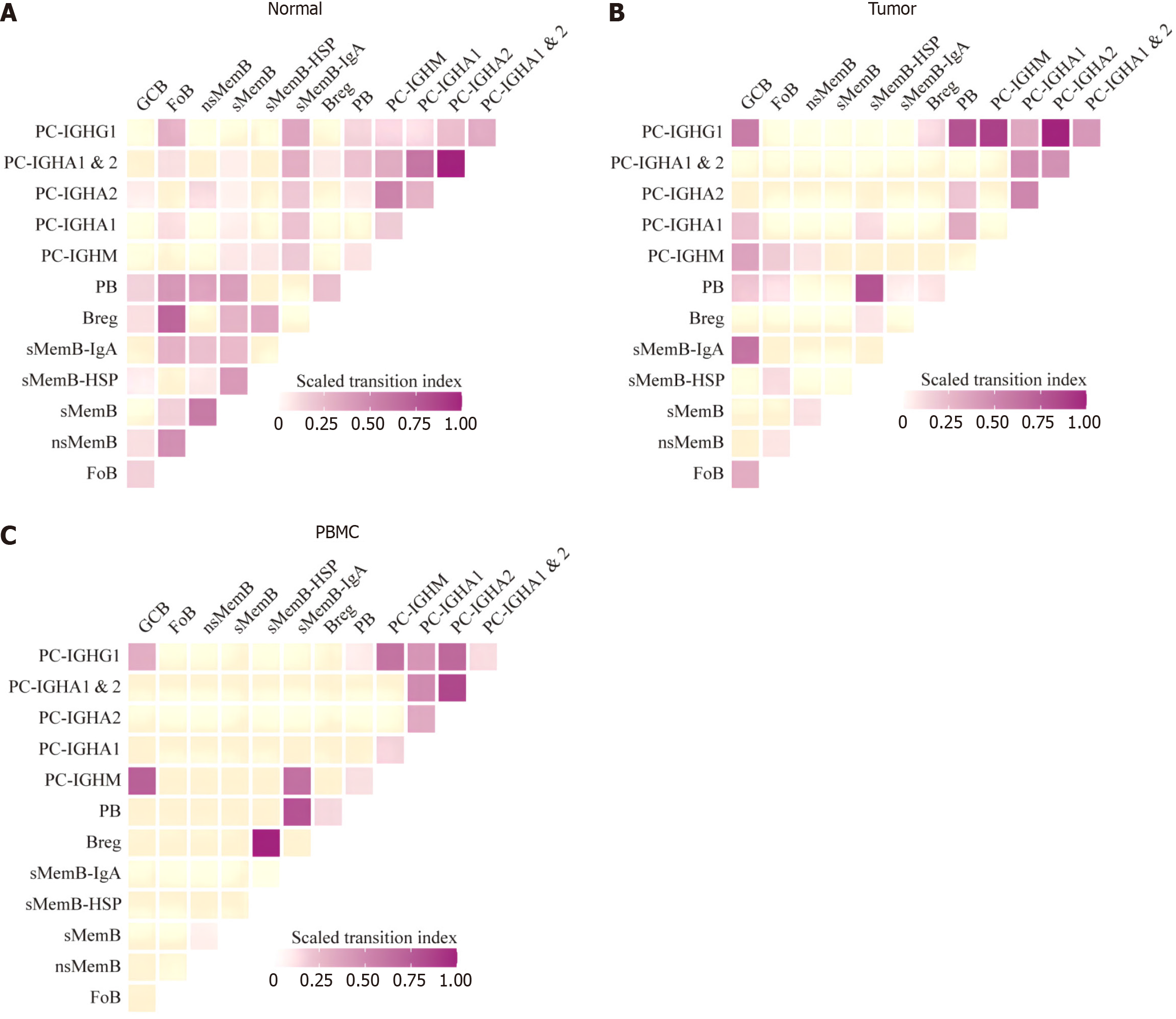
Figure 5A shows the clonal expansion of each subpopulation of B cells in three tissue samples. In general, the clonal index of B cells in normal and tumor tissues was lower than that of PBMC, while the clonal index of most B cell subgroups in tumor samples was lower than that of normal tissues, but the two subgroups of GCB and PB were both increased in tumors, and the difference was statistically significant (P = 0.001). It is suggested that they may actively proliferate after being stimulated by antigens in the immune microenvironment (Figure 5B and C). This is positive for the anti-tumor effect of B cells. At the same time, the mutation frequency of GCB B cell receptor chain in tumor was also higher than that in normal tissue, and the difference was statistically significant (P = 0.001), suggesting GCB mutation levels are increasing, which in turn strongly enhances affinity with tumor antigens. The migration index between subpopulations is shown in Figure 6, which shows the analysis results consistent with the transcriptome. In normal tissues, the conversion index between CD20+ subgroups was low, and GCB mainly shared more clonotypes with plasma blasts. However, in tumors, GCB shares more clonotypes with memory B cells expressing IgA immunoglobulins, which once again proves that GCB is more oriented to memory B cells in tumors.
Figure 5 Clonal expansion index of B cell subsets and mutation level of germinal center B cell.
A: Zscored clonal expansion index among three samples; B: Comparsion of clonal expansion between normal and tumor tissues; C: Heavy chain mutation level of germinal center B cell among samples.
Figure 6 Transition index of B cell subsets.
A: Normal tissue; B: Tunor tissue; C: Peripheral blood mononuclear cell sample.
T-cell receptor clonotype analysis of CRC tumor tissue and normal mucosa
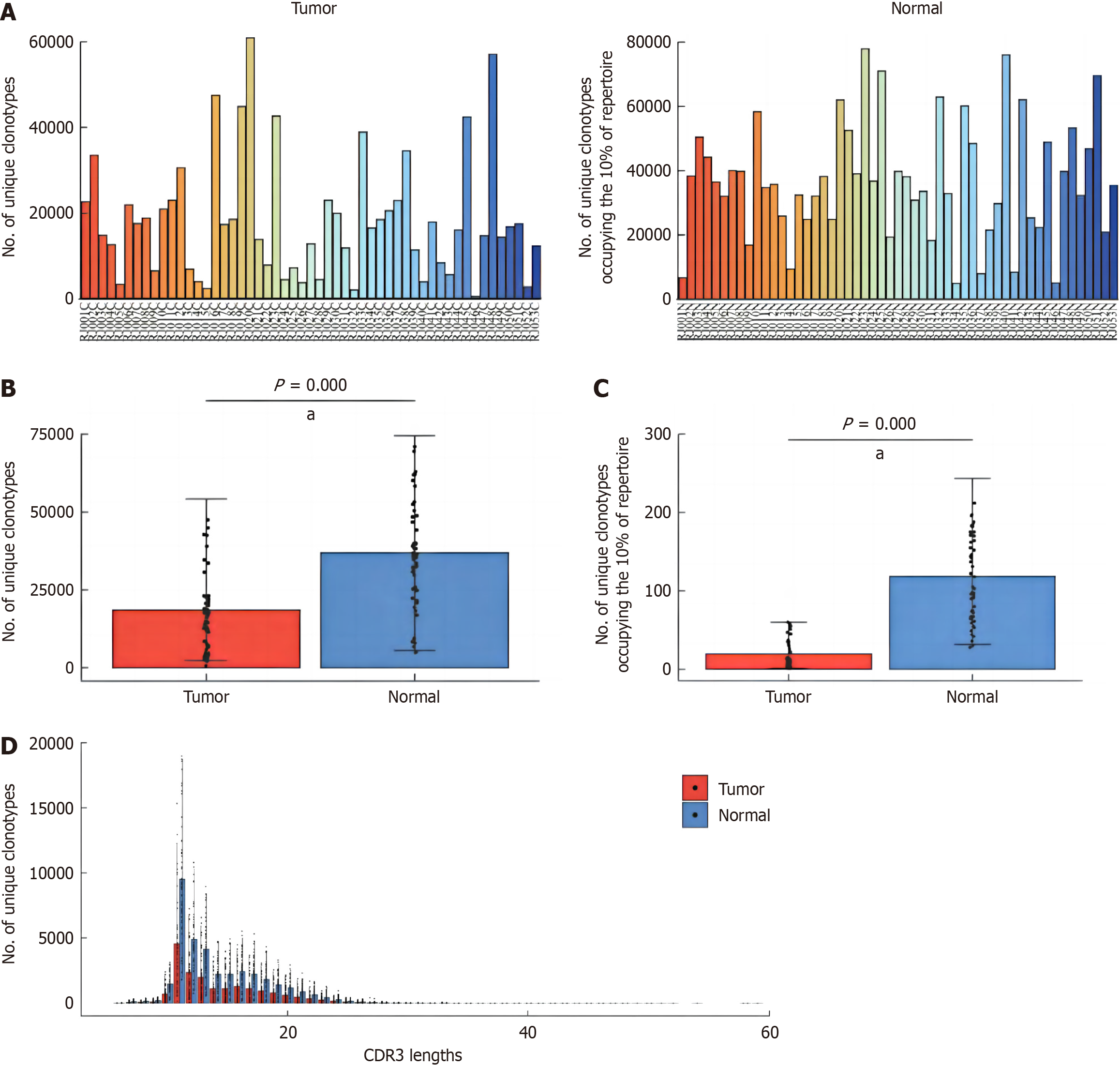
The number of T-cell receptor (TCR) clones in the samples was analyzed. Generally speaking, the number of clones in the tumor tissues was lower than that in the normal mucosa tissues around cancer (Figure 7A). The results of statistical analysis showed that the number of TCR clones in tumor tissue was significantly lower than that in pericancerous normal mucosal tissue, and the difference was statistically significant (P = 0.001, Figure 7B), and the number of clones in tumor tissue, which accounted for 10% of the immune pool, was also lower than that in pericancerous normal mucosal tissue (P = 0.001, Figure 7C). In addition, the number of clonotypes in different length distributions of TCRβ chain CDR3 amino acid sequences in tumor tissues and pericarcinoma normal mucosal tissues was analyzed, and the results showed that the number of clonotypes in tumor tissues was lower than that in pericarcinoma normal mucosal tissues on different length of CDR3 sequences (Figure 7D). These results suggested that the diversity of TCR group was higher in normal tissues around cancer.
Figure 7 Comparison of the number of T-cell receptor clonotypes between tumor and adjacent normal tissues.
A: Estimation of the number of T-cell receptor (TCR) clonotypes in tumor and adjacent normal tissues; B: Comparison of TCR clonotypes between tumor and adjacent normal tissues; C: Comparison of the number of TCR clonotypes occupying 10% of repertoire between tumor and adjacent normal tissues; D: Comparison of TCR clonotypes with different CDR3 sequence lengths between tumor and adjacent normal tissues. aP < 0.01.
Clonotype analysis of different amplification levels
TCR clones were ranked from highest to lowest amplification level, and the proportion of clones ranked in the top 10, 11-100, 101-1000, 1001-3000, 3001-10000, 10001-30000, 30 001-50 000 were analyzed. The proportion of TCR clones with the top 10 amplification levels in tumor tissues was significantly higher than that in normal pericancerous mucosa tissues, and the difference was statistically significant (P = 0.001, Figure 8), suggesting that the degree of TCR amplification in tumor tissues was higher than that in normal pericancerous mucosa tissues.
Figure 8 Comparison of proportion of T-cell receptor clonotypes with different amplification levels between tumor and adjacent normal tissues.
aP < 0.01.
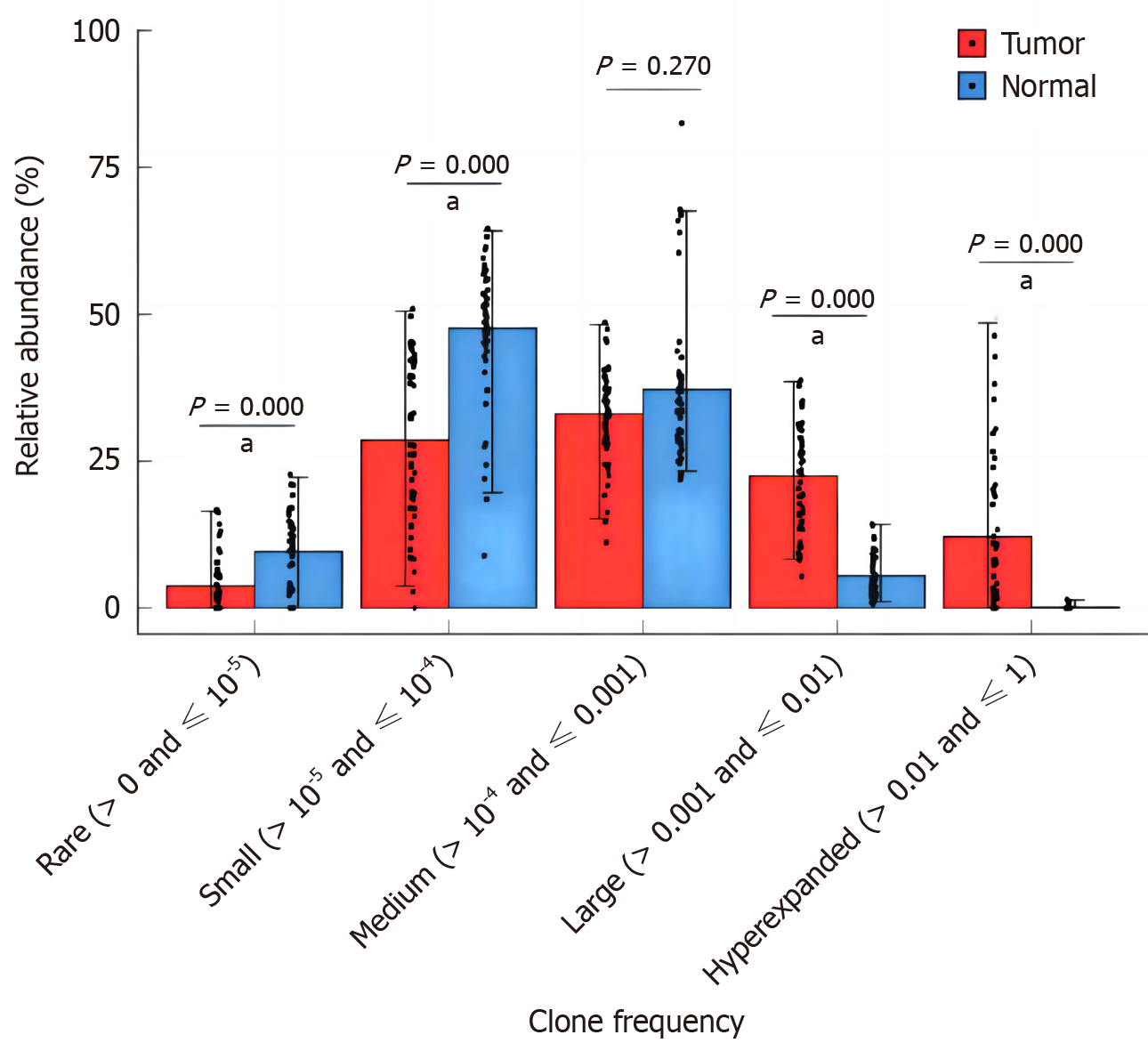
Analysis of relative abundance of clonotypes with different frequencies
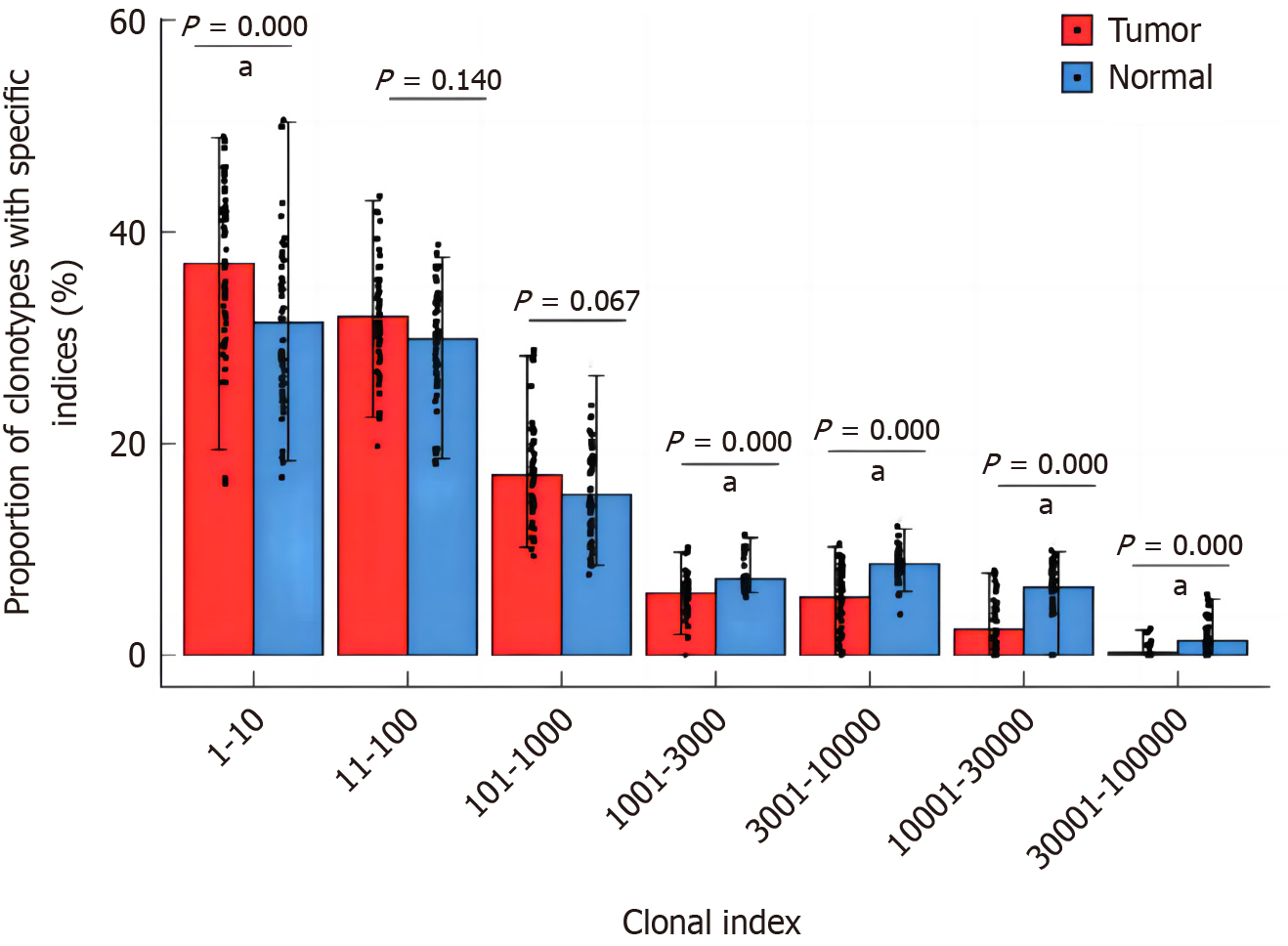
The relative abundance of different frequency TCR clones in the whole clonal space of tumor tissue and normal mucosa were further analyzed. The frequencies of clones include rare frequency (> 0 and ≤ 10−5), small frequency (> 10−5 and ≤ 10−4), medium frequency (> 10−4 and ≤ 0.001), high frequency (> 0.001 and ≤ 0.01), and ultra-high frequency (> 0.01 and ≤ 1). In both tumor tissue and pericancerous normal mucosal tissue, TCR clones were mainly of low frequency (> 10−5 and ≤ 10−4) and medium frequency (> 10−4 and ≤ 0.001). The relative abundance of high frequency (> 0.001 ≤ 0.01) and ultra high frequency (> 0.01 and ≤ 1) clonotypes in tumor tissues was higher than that of pericancerous normal mucosa tissues, and the relative abundance of rare frequency (> 0 ≤ 10−5) and low frequency (> 10−5 ≤ 10−4) clonotypes was lower than that of pericancerous normal mucosa tissues (P = 0.001, Figure 9).
Figure 9 Comparison of relative abundances in different T-cell receptor clonotypes with specific frequencies between tumor and adjacent normal tissues.
aP < 0.01.
DISCUSSION
In this study, the infiltrating and circulating B cells of CRC were analyzed in detail based on the single cell transcriptome and immune bank data through bioinformatics analysis[16-18]. It was found that CD20+B cells increased significantly in tumor tissues, with the most significant effect of GCB. The positive effects of GCB were explained by means of pathway analysis, communication analysis and VDJ data analysis, and it was found that GCB had positive anti-tumor effects in the microenvironment[19]. On the one hand, it played a strong supporting role in the anti-tumor effect of T cells[20]. On the other hand, GCB in tumor tissues was also carrying out a high level of BCR heavy chain mutation. To adapt to the high-affinity evolution of tumor antigens[21-23]. On the contrary, the role of plasma cells in tumor was greatly reduced, mainly plasma cells expressing IgA, and the evolution of GCB into plasma cells gradually decreased in tumor tissues, instead, it gradually developed into memory B cells[24]. In conclusion, B cells also play an important role in the tumor immune microenvironment of CRC, which is not a simple bystander cell, but an active participant in fundamentally coordinating the immune response[25-28].
CD20+B importance was determined from studies of different infiltration patterns of B cell subpopulations in three tissues, as shown by significant increases in GCB, nsMemB, sMemB, and SMEMB-HSP. Plasma cells expressing both IGHM and IGHA immunoglobulins decreased significantly[29]. This prompted us to think about the reasons for the transformation of B cells in the tumor microenvironment, and we chose GCB, the common source of both B cells and plasma cells, to explore. By mapping the transcription profiles of single-cell data to TCGA-COAD data, we found that GCB also increased significantly in tumor tissues, and the proportion of GCB gradually decreased with tumor progression, which we speculated might be due to the inhibition of GCB function in tumor development[30]. Through the pathway enrichment analysis, we found that the antigen-presenting function of GCB in tumors was significantly increased, favourably supporting the activation and proliferation of T cells, and at the same time, it was also carrying out effective VDJ rearrangement to improve the affinity of specific antigens, with positive anti-tumor effects. In the down-regulated pathway, we also found that the pathway related to the secretion of antibodies was significantly down-regulated, suggesting that one of the reasons for the decrease of plasma cells may be caused by the decreased trend of GCB transformation[31-33]. In order to further explore the influence of microenvironment GCB cells on their own function, we found through the analysis of cell interaction molecular pairs that the molecular pairs that inhibit T cell function decreased significantly, while the molecular pairs that promote the activation and development of T cells and B cells were also significantly up-regulated. This study demonstrated the active support of GCB cells for the anti-tumor effects of T cells in the CRC immune microenvironment. In the analysis of communication between GCB and myeloid cells, the enhancement of TNF signal suggests the potential role of GCB cells in supporting the anti-tumor effect of macrophages[34]. In summary, we suggest that GCB cells exert positive anti-tumor effects through indirect positive support in the early tumor microenvironment. Finally, we discussed the above analysis results by using the data of VDJ group database. From the two aspects of clone index and migration index, we found that GCB was significantly cloned and amplified in tumor tissues, while a high level of heavy chain mutation occurred, which reflected the trend of GCB amplification and enhancement of tumor antigen affinity[35]. From the analysis of inter-subpopulation migration, we found that GCB was more oriented towards memory B cells in tumor tissues. However, what role memory B cells play in the tumor immune microenvironment of CRC still needs to be further explored and analyzed in future work.
This study observed a range of effects affecting the immune microenvironment, including the increase or decrease of B cells and functional changes in different subpopulations[36]. The heterogeneity of the tumor microenvironment, the interaction of immune cells, and the variations in treatment approaches may all have an impact on the occurrence of these effects. In particular, we note that certain B cell subpopulations may play an important regulatory role in anti-tumor immunity, while others may exhibit inhibitory tendencies. This study further discusses the possible causes behind these effects, which relate to the diversity and heterogeneity of B cells in the immune response as well as their interaction networks in the CRC microenvironment[37-39]. Differences in biology, epigenetic variations, and the complexity of immune cell communication may all explain why we observe particular effects. For example, certain subpopulations of B cells may participate in immune responses by regulating T cell activity and secreting specific antibodies, while other subpopulations may play an important role in regulating inflammatory responses[40-43].