Published online Jul 15, 2023. doi: 10.4251/wjgo.v15.i7.1149
Peer-review started: February 11, 2023
First decision: March 28, 2023
Revised: April 11, 2023
Accepted: May 8, 2023
Article in press: May 8, 2023
Published online: July 15, 2023
Processing time: 150 Days and 17.6 Hours
Genomic instability and inflammation are considered to be two enabling characteristics that support cancer development and progression. G-quadruplex structure is a key element that contributes to genomic instability and inflammation. G-quadruplexes were once regarded as simply an obstacle that can block the transcription of oncogenes. A ligand targeting G-quadruplexes was found to have anticancer activity, making G-quadruplexes potential anticancer targets. However, further investigation has revealed that G-quadruplexes are widely distributed throughout the human genome and have many functions, such as regulating DNA replication, DNA repair, transcription, translation, epigenetics, and inflammatory response. G-quadruplexes play double regulatory roles in transcription and translation. In this review, we focus on G-quadruplexes as novel targets for the treatment of gastrointestinal cancers. We summarize the application basis of G-quadruplexes in gastrointestinal cancers, including their distribution sites, structural characteristics, and physiological functions. We describe the current status of applications for the treatment of esophageal cancer, pancreatic cancer, hepatocellular carcinoma, gastric cancer, colorectal cancer, and gastrointestinal stromal tumors, as well as the associated challenges. Finally, we review the prospective clinical applications of G-quadruplex targets, providing references for targeted treatment strategies in gastrointestinal cancers.
Core Tip: G-quadruplexes are widely distributed in the human genome and have many functions. G-quadruplexes play double regulatory roles in transcription and translation. We focus on G-quadruplexes as novel therapeutic targets for gastrointestinal cancers. We summarize the application basis of G-quadruplexes in gastrointestinal cancers, including their distribution sites, structural characteristics, and physiological functions. We describe the current status of applications for the treatment of esophageal cancer, pancreatic cancer, hepatocellular carcinoma, gastric cancer, colorectal cancer, and gastrointestinal stromal tumors, as well as the associated challenges. We review prospective clinical applications of G-quadruplex targets, providing references for targeted treatment in gastrointestinal cancers.
- Citation: Han ZQ, Wen LN. Application of G-quadruplex targets in gastrointestinal cancers: Advancements, challenges and prospects. World J Gastrointest Oncol 2023; 15(7): 1149-1173
- URL: https://www.wjgnet.com/1948-5204/full/v15/i7/1149.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i7.1149
Gastrointestinal cancers seriously affect the quality of life of patients and are among the cancers with the highest incidence and mortality worldwide. There currently remains a lack of effective therapeutic methods for these cancers, despite the development of many anticancer strategies. This is mainly because the etiology and molecular mechanisms associated with the occurrence and development of many cancer types are unclear, despite tumor immunotherapy and molecular targeted therapies having achieved promising results. In the multistep development of various human cancers, 14 characteristics were summarized as the latest hallmarks of cancer: Sustaining proliferative signaling, evading growth suppression signals, avoiding immune destruction, enabling replicative immortality, tumor-promoting inflammation, activating invasion and metastasis, inducing or accessing the vasculature, genomic instability and mutation, resisting cell death, deregulating cellular metabolism, unlocking phenotypic plasticity, nonmutational epigenetic reprogramming, polymorphic microbiomes, and senescent cells[1]. Genomic instability and inflammation have been considered as the two enabling characteristics that allow cancer to acquire these hallmarks[2]. Importantly, inflammation itself can induce genomic instability[3]. The role of inflammation in the transformation of gastrointestinal cancers, such as gastric, intestinal and liver cancers, should not be ignored. Hence, in the future, anticancer strategies targeting genomic instability may have potentially broad applications for the treatment of cancer. The molecular and cellular elements affecting genomic instability may become effective anticancer targets.
In 1962, the unusual four-stranded helix structures of guanine-rich DNA sequences with a high tendency to self-assemble into planar guanine quartets (G-quartets) were first reported and named as G-quadruplexes[4]. Afterwards, DNA G-quadruplexes were found in telomeres[5], oncogene promoters[6,7], microsatellite fragments[8], and additional regions. In 1992, tetraplex formation of nucleotide sequences in the 3’ terminus of the 5s RNA were found in Escherichia coli in the presence of K+ solution[9]. Subsequently, > 3000 RNA G-quadruplex component elements in the mRNA 3’ and 5’ untranslated regions (UTRs)[10,11] and exons[12,13], as well as in other noncoding RNAs[14], were discovered in the human genome. As a nucleic acid secondary structure, a G-quadruplex differs from the typical A-, B-, C- or Z- of duplex DNA, conventional RNA, and was the supplement to nucleic acid structure type. The crystal or solution structures of various DNA or RNA G-quadruplexes have been increasingly resolved, with their physiological functions gradually clarified, especially their roles in various forms of cancers, such as breast cancer, osteosarcoma, and cervical carcinoma[3,15-19]. G-quadruplexes can regulate DNA replication[20,21], repair[22], methylation[23], and gene transcription and translation[24], and correlate with genomic instability[3,25]. In this review, we summarize the literature on G-quadruplexes and their ligands from 1962 to 2023 and describe G-quadruplex characteristics, including the existing sites, structural details, and physiological functions, and their potential applications in gastrointestinal cancer therapy. In addition, we summarize the challenges and prospects of targeting G-quadruplexes in digestive tumors to potentially prevent and treat gastrointestinal cancers.
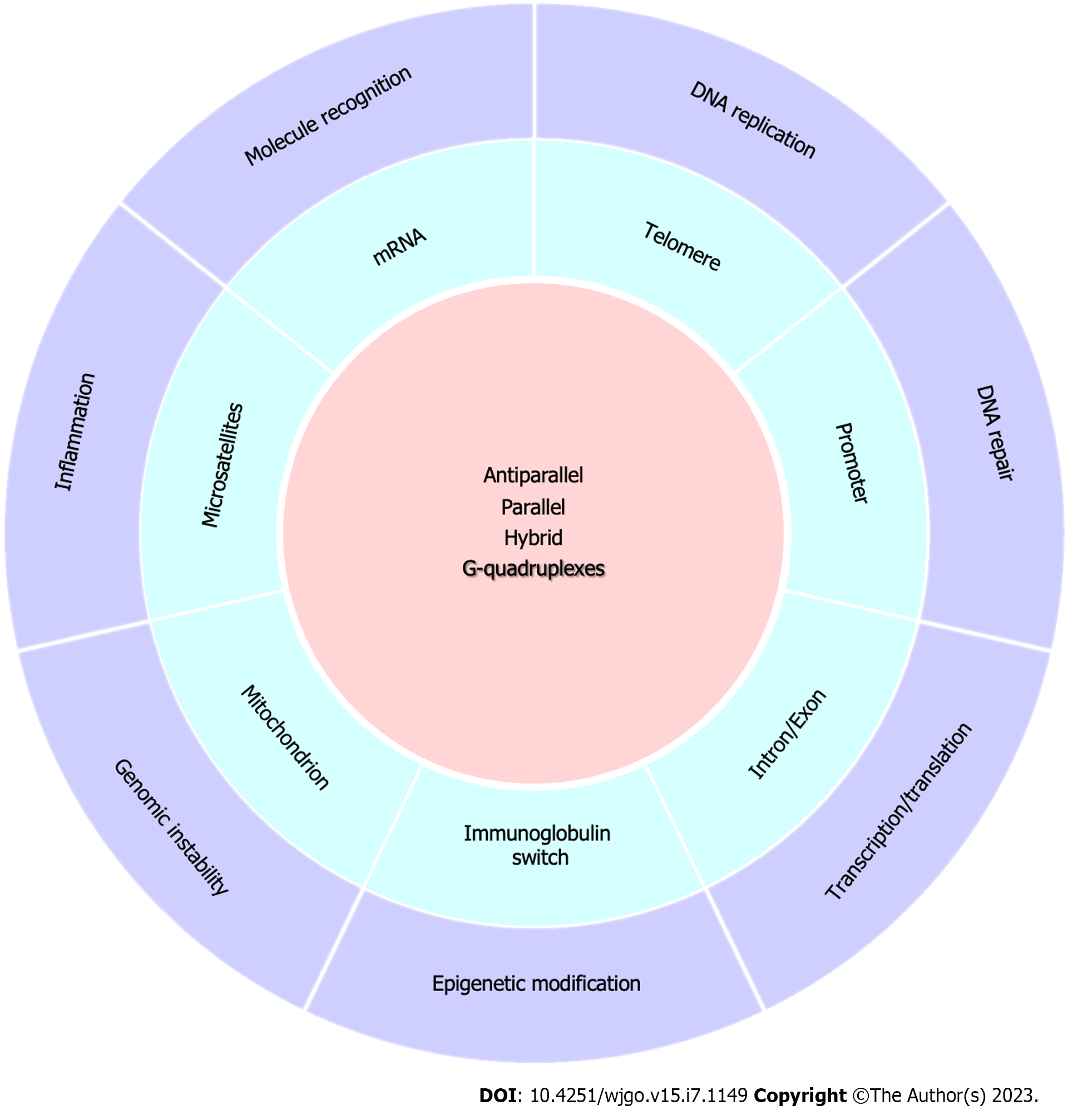
The clinical application value of biological molecules is dependent upon their biological functions, which are affected by intracellular distribution, molecular structure and other properties. Therefore, such molecular characteristics form the basis for clinical application potential, as shown in Figure 1.
Potential G-quadruplex structures in the human genome can be predicted via computer analysis by retrieving the pattern sequences (G≥3N1-7G≥3N1-7G≥3N1-7G≥3)[26]. They can also be formed with less than three guanines contrary to this dogma[27]. With the development of G-quadruplex-specific antibodies, fluorescent probes, sequencing technology, and genomic mapping, G-quadruplex structures are being increasingly detected and visualized in cells[28-33]. Currently, at least 700000 potential G-quadruplex structures have been inferred to exist in humans[34-36]. Telomeric DNA was the first biologically-related G-quadruplex target investigated in detail[37] and was considered to have the highest abundance of potential G-quadruplex structures. The 5000-10000 bp of tandemly repeated sequence (TTAGGG) contained in telomeres can fold into a G-quadruplex to regulate telomere maintenance[38,39]. Maintaining its structural stability can help inhibit the activity of telomerase and thus prevent the unlimited proliferation of tumor cells[40]. In addition to telomeres, genome-wide sequencing analyses have suggested that more than 8000 potential G-quadruplexes are likely enriched in promoter regions spanning 1 kb upstream of the transcription initiation sites in humans[41,42]. In the past, close attention was paid to proto-oncogene promoter G-quadruplexes, including Kirsten rat sarcoma viral oncogene homologue (KRAS)[43], HRAS[44], c-MYC[45], c-KIT[46], RET[47], MST1R[48], and others. G-quadruplexes in promoter regions of carcinoma-related genes were studied as well, such as B-cell lymphoma 2 (BCL2)[49], hypoxia inducible factor 1 subunit alpha (HIF1a)[50], vascular endothelial-derived growth factor (VEGF)[51], platelet-derived growth factor subunit A (PDGFA)[52], PDGF receptor-β (PDGFR-β)[53], human telomerase reverse transcriptase (hTERT)[54], nuclear factor (erythroid-derived 2)-like 2[55], SMARCA4[56], and multidrug resistance protein 1[57]. Recent studies have indicated that G-quadruplexes also exist in promoter regions of MYH7β gene and are associated with various myopathies[58], as well as in CSTB gene, and are related to progressive myoclonus epilepsy type 1[59]. Moreover, there were G-quadruplex-forming sequences (GGGGCC) in intron 1 of the C9orf72 gene, which was the usual hereditary factor of amyotrophic lateral sclerosis and frontotemporal dementia[60]. Similar sequences were also found in other genes, for example, (GGCCT) in the first intron of NOP56 relevant to spinocerebellar ataxia (SCA36)[61], (CCCCATGGTGGTGGCTGGGGACAG) in the coding exon of the PRNP gene indicating Creutzfeldt–Jakob disease[62], TAGGGCGGGAGGGAGGGAA in the first intron of the N-myc gene[63], and (GGGT)4 in human microsatellites[8]. Additionally, abundant potential G-quadruplex formation sites exist in mRNAs (especially in the 5’ UTRs) or microRNAs. For instance, mRNA G-quadruplexes reportedly include VEGF[64], FMR1[65], MMP16[66], transforming growth factor-β (TGFβ2)[10], neuroblastoma RAS viral oncogene homolog (NRAS)[67], insulin-like growth factor 2[68], telomere repeat binding factor 2 (TRF2)[69], PIM1[70], beta-site amyloid precursor protein cleaving enzyme 1[71] and YY1[72]. G-quadruplex structures have recently been explored in miR-92a[73], miR-1229[74] and miR-1587[75]. G-quadruplexes have also been discovered in immunoglobulin switches, microsatellites, and mitochondria genes. However, when and where the potential G-quadruplex structures can actually form and exert corresponding physiological functions in vivo depend on environmental conditions, which require further investigation.
Different from the Watson-Crick base pairing regulation of double-stranded DNA, three to four guanines assemble into a G-quartet by Hoogsteen hydrogen-bonding in a square planar platform. The G-quartets then further stack on top of one another to form G-quadruplexes, which remain stable by monovalent cations in the central ion channel[76]. Because of the different number and spatial arrangement of bases, G-quadruplex structures have obvious polymorphisms. X-ray diffraction and high-field nuclear magnetic resonance spectroscopy are two effective methods for understanding the crystal and solution structures, which can be categorized as intramolecular or intermolecular G-quadruplexes. An intramolecular G-quadruplex is unimolecular and previous studies have confirmed that there are three basic types according to the orientation of the G-quartet: Parallel structure, antiparallel structure, and hybrid structure[77]. These different structures have varying levels of stability, which may affect their respective functions. Because of the restrictions of the external environment and central cation, a G-quadruplex sequence may present multiple configurations. For the human telomeric sequence, crystal or solution structural elucidation revealed that in the presence of K+ solution, the G-quadruplex had parallel, antiparallel, hybrid-2, and hybrid-1 configurations, with an intermediate of two-tetrad[18,78-81]. However, in the presence of Na+ solution, one unfolded state and three G-quadruplex-related configurations are observed, and the structure can interconvert between these forms[82]. KRAS, c-MYC, VEGF, PDGFR-β and HIF1a promoter G-quadruplexes take on parallel structures in K+ solution[50,76], BCL2 promoter G-quadruplexes adopt the hybrid-2 or parallel conformation in K+ solution[76], and c-KIT promoter sequences can form a parallel or antiparallel G-quadruplex[83,84]. G-quadruplex structures present in other sites, such as in mRNAs, also conform to these three basic structural types. For intramolecular G-quadruplexes, more than two unimolecular G-quadruplex sequences can assemble into intermolecular parallel G-quadruplexes[18,58,75].
G-quadruplexes and DNA replication: Current research supports two seemingly opposing views on how G-quadruplexes can influence DNA replication: One view suggests that the G-quadruplex motif is necessary for replication initiation, while the other argues that a G-quadruplex is an obstacle to replication. Evidence supporting the former view is that 70%-90% of replication origins are preceded by a potential G-quadruplex-forming sequence, called the origin G-rich repeated element, which is 250-300 bp upstream of the replication initiation site in the non-nucleosome region[72,85]. Either deleting these elements in several model origins or introducing point mutations that affect G-quadruplex stability may reduce replication initiation activity in cells[86]. In addition, G-quadruplexes can recruit replication activators to play a role in DNA replication[87]. The latter view also has strong evidence, including that small molecular ligands targeting G-quadruplexes can result in DNA damage[88]. Additionally, it was demonstrated that the helicase, chromatin-remodeling protein ATRX, and human CTC1-STN1-TEN1 complex prevented replication defects by unwinding G-quadruplexes[89-92]. There were two possible conclusions regarding this argument. First, a G-quadruplex structure preferentially formed in the firing origin rather than the licensing origin[93,94]. Second, the negative regulation of DNA replication mediated by G-quadruplexes mostly occurred under pathological conditions, such as in the presence of G-quadruplex ligands or absence of ATRX. The negative effects of G-quadruplexes on DNA may be counteracted by unwinding proteins, such as helicase, in wild type cells under undisturbed situation[21].
G-quadruplexes and DNA repair: DNA damage can be triggered by exogenous stimuli, such as physical and chemical factors, or endogenous stress, which includes reactive oxygen species (ROS) production, replicative stress, and the formation of nucleic acid secondary structure. In addition to telomeres, promoters and transcriptional start sites, G-quadruplexes are enriched in DNA double-strand break (DSB) sites during mitosis and meiosis, and G-quadruplex formation may induce DNA damage and negatively impact effective DNA repair mechanisms[22,95]. However, G-quadruplexes can sometimes promote certain repair pathways under specific conditions. There are six main pathways involved with DSB repair over DNA replication: Homologous recombination (HR), nonhomologous end joining, base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), and translesion synthesis (TLS). One model suggested that stabilization of G-quadruplexes can active HR, leading to bypass/repair of G-quadruplex-mediated DNA damage[22]. As a sensor for endogenous oxidative damage of DNA, G-quadruplexes may provide feedback to drive BER to promote genomic stability under oxidative stress[96]. Zoo1 could assist NER function and regulate the selection of DNA repair pathways near G-quadruplex structures[97]. MMR activation was not restricted when G/T and G-quadruplex mismatch were in close proximity[98], and the stable G-quadruplex structure could inhibit the activity of endonuclease of MutL and indirectly interfere with the MMR process[99].
G-quadruplexes and transcription and translation: Potential G-quadruplex forming-sequences are frequently enriched in DNA promoter regions and the 3’ or 5’ UTRs, providing an opportunity for regulation of transcription or translation. For highly expressed cancer-related genes, proteins such as nucleolin and small molecular ligands that promote G-quadruplex formation can induce transcriptional repression. However, proteins that unwind G-quadruplexes such as the nucleoside diphosphate kinase NM23H2, poly ADP-ribose polymerase and RecQ family helicase can lead to transcriptional activation of target genes[100,101]. Moreover, putative G-quadruplex-forming sequences were also found at the docking sites of transcription factors SP1 and c-MYC associated zinc-finger protein (MAZ), which may help recruit SP1 and MAZ and facilitate transcription in cancer progression[102,103]. In noncancerous cells, studies have shown that G-quadruplexes can directly interfere with mitochondrial genome replication, transcription, and respiratory function[104]. Therefore, a G-quadruplex is a key factor that can regulate gene transcription. Similarly, RNA G-quadruplexes can control translation. For instance, oxymatrine inhibited the translation of VEGFA mRNA in human cervical cancer cells by selectively binding to the G-quadruplex structure in VEGFA 5’ UTRs[105]. Additionally, DEAH box polypeptide 36 (DHX36) can bind to 5’ UTRs G-quadruplexes and control translation to promote muscle stem cell regeneration[106]. Thus, G-quadruplexes play significant roles in gene transcription and translation.
G-quadruplexes and epigenetic modifications: C-5 methylation of cytosine by DNA methyltransferase DNMT1, DNMT3A and DNMT3B is a key DNA epigenetic modification in mammalian development and disease. About 90% of CpGs can be highly methylated, but CpG islands (CGIs), found in dense guanine-cytosine-rich regions, largely lack methylation and are universally present in the promoter regions of genes[107]. CGIs can be progressively methylated during certain biological events, such as aging[108] and cancer[109], but the underlying regulatory mechanisms are not fully clear. Studies have shown that G-quadruplex structures are present in CGIs and are closely related to reduce levels of CGIs methylation in the human genome[110]. G4-chromatin immunoprecipitation sequencing (G4-ChIP-seq) analysis indicated that G-quadruplex structures were colocalized with DNMT1 and inhibited methylation by inhibiting activity of this enzyme[110]. Recent studies have shown that the methylation efficiency decreased with increasing G-quadruplex stability, and the degree of methylation can be controlled by adjusting the G-quadruplex topology[111].
In addition to DNA methylation, histone modification is also an important epigenetic regulation. The local conformations and biological functions of G-quadruplexes can be regulated by their specific binding proteins. For example, RNA G-quadruplexes and RNA-binding proteins participate in telomere maintenance and transcriptional regulation through histone modifications. G-quadruplex RNA-binding proteins, such as translocated in liposarcoma/fused in sarcoma and TRF2, can promote the trimethylation of histone H3 at lysine 9 in telomere histones through G-quadruplex telomeric repeat-containing RNA (TERRA)[112,113]. G-quadruplex TERRA possibly regulates methylation and demethylation of histones in telomeric DNA, and can act as a noncompetitive inhibitor to suppress lysine specific histone demethylase-mediated histone demethylation[114]. Polycomb repressive complex 2 (PRC2) interactions with TERRA can catalyze the trimethylation of histone H3 at lysine 27 (H3K27me3), and G-quadruplex RNA can specifically prevent PRC2 from interacting with genes in human and mouse cells to block methylation at H3K27[115]. These mechanisms work together to maintain telomere length and chromatin function.
G-quadruplexes and genomic instability: DNA is vulnerable to damage from various types of endogenous and exogenous stimuli. This can hinder DNA replication and induce genomic instability, which includes point mutations, insertions, deletions, inversions, translocations, expansions/contractions of repeated sequences, gross chromosomal rearrangements, aneuploidy and other characteristics. Such genomic instability is often observed in cancer and can be induced by G-quadruplexes. G-quadruplexes are enriched at regions of base substitutions, insertion-deletion mutations, and chromosome translocation breakpoints that are associated with a variety of human cancers, such as colon cancer, and is the main inducing factor of carcinogenic transformation[116-118]. The instability of potential G-quadruplex-forming sequences increases in a transcription-dependent manner, as transcription can provoke genomic instability of G-quadruplexes by releasing single-stranded DNA, which is easy to fold into secondary structures[117,119].
G-quadruplex and inflammation: G-quadruplexes are correlated with inflammation. Studies have shown that there was a high frequency of potential G-quadruplex formation sequences in the promoter regions of many inflammatory factors, such as tumor necrosis factor, TGF-β, interleukin (IL)-6, IL-12, IL-17, the XC and TAFA family chemokines, and β-chain family cytokines[120]. G-quadruplexes are also distributed in the binding sites of transcription factors involved in inflammatory and immune processes, including nuclear factor nuclear factor kappa B1, interferon regulatory factor 5, transcription factor p65, transcription factor RelB, and nuclear factor of activated T cells 5[120]. In addition, genes containing G-quadruplex structures that can regulate and participate in inflammatory-related processes have been identified through experimental studies[121]. G-quadruplexes can trigger inflammatory reactions by upregulating proinflammatory cytokines, making these structures a marker of increased inflammation and a contributor to inflammatory diseases development[121]. However, another study suggested that G-quadruplexes can interfere with switch-like recombination in B cells to alleviate allergic inflammation[122]. Collectively, this evidence suggests that G-quadruplexes may be a potential target for treating inflammation-related diseases.
Ion and molecule recognition functions of G-quadruplexes: The stability of G-quadruplex structure needs to be maintained by the monovalent cations located in the central ion channel, allowing the G-quadruplex sequence to specifically recognize monovalent cations such as K+ and Na+. In addition, because the specific G-quadruplex-forming sequence can fold into a special conformation and the fluorescence emission of some small molecules is significantly enhanced after binding with G-quadruplexes, G-quadruplexes could be used to identify small molecular ligands (berberine, porphyrin, and more) or proteins (thrombin, nucleolin, and more) that can specifically bind to them[16,57,123,124] or assist imaging. Therefore, G-quadruplexes have been widely used as recognition elements to construct biosensors for detecting targeting ions and molecules, such as tumor biomarkers, in tumor diagnosis, as well as targeting agents or drug carriers of anticancer drug delivery systems for tumor treatment[125,126]. In such applications, the G-quadruplex sequences are also called aptamers.
The above complex biological functions of G-quadruplexes imply that they have broad application prospects for the diagnosis and treatment of gastrointestinal cancers. The role of a G-quadruplex as a recognition element in the molecular diagnosis of gastrointestinal cancers will not be discussed in this review. The application of G-quadruplexes in therapy is mainly discussed from two perspectives: (1) The therapeutic effect of small molecule ligands and biomolecules targeting G-quadruplexes to regulate gene transcription; and (2) The therapeutic effect of G-quadruplex sequences for molecular recognition functions.
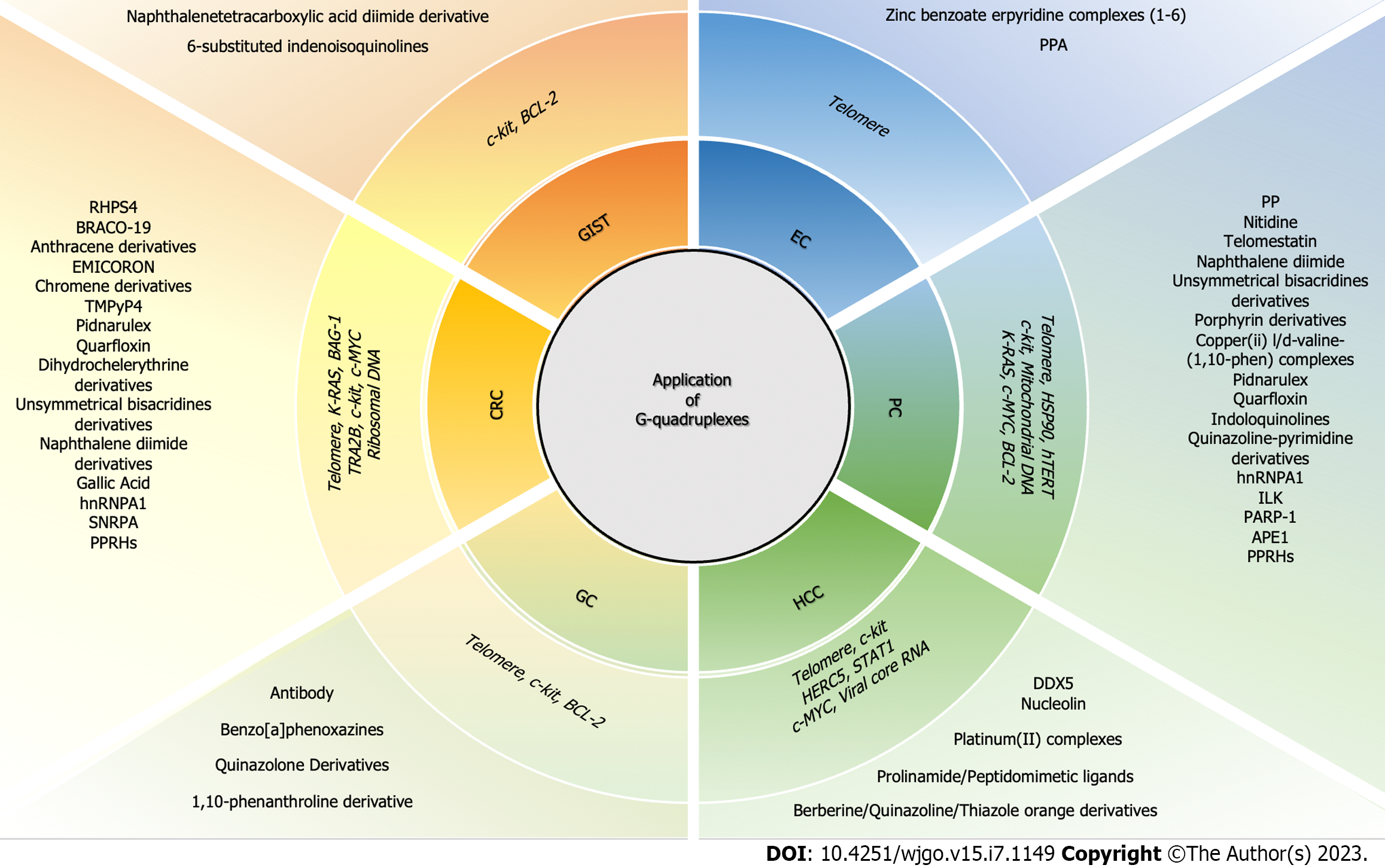
Esophageal cancer: Esophageal cancer (EC) is a gastrointestinal disease with high mortality rates. Surgery is the first choice of treatment for resectable EC cases, but neoadjuvant chemotherapy can improve the 5-year survival rate without increasing postoperative complications. Targeting G-quadruplexes may provide a new perspective for treating EC, although relevant research on this is currently limited. The telomere is an early G-quadruplex target. The G-quadruplex ligand 2,6-bis[3-(N-piperidino) propionamido] anthracene-9,10-dione, which is also considered to be a telomerase inhibitor, can shorten telomeres and exert antiproliferative and proapoptotic effects in both BIC-1 and SEG-1 EC cell lines[127]. A recent study found that zinc benzoate terpyridine complexes (1-6) in combination with G-quadruplex sequence (G2T4G2CAG2GT4G2T) resulted in various degrees of antiproliferative effects in the EC cell line Eca-109[128] (Table 1). Hence, further exploration of a G-quadruplex-related treatment strategy in EC is needed.
| Ligands/biomolecules | Cell lines | Targeting gene/G-quadruplex | Effects on G-quadruplex | Effects on genes | Anticancer phenotypes | Ref. |
| 2,6-bis[3-(N-Piperidino) propionamido] anthrace- ne-9,10-dione | BIC-1, SEG-1 | Telomere | Not detected | Shortened telomeres | Inhibited telomerase activity, arrested cell proliferation, reduced colony number and size, and promoted cell apoptosis | [127] |
| Zinc benzoate erpyridine complexes (1-6) | Eca-109 | G2T4G2CA, G2GT4G2T | Bound with G-quadruplex | Not detected | Inhibited cell proliferation | [128] |
Pancreatic cancer: Pancreatic cancer (PC) is a refractory tumor disease with poor prognosis among cancers. About 97% of PC cases are accompanied by alterations of genes and 90% have KRAS oncogene mutations, which are essential for initiation of pancreatic ductal adenocarcinoma. Because KRAS can drive oncogene addiction, inhibiting gene mutation and downregulating gene expression are reasonable ways to block PC progression. Many attempts have been paid to target KRAS oncogenes, but clinically useful therapies are still limited. The mutant KRAS protein has attracted much attention, causing other approaches involving targeting KRAS transcription to not be fully explored. Additionally, telomere, heat shock protein 90 (HSP90), c-MYC, Bcl-2 and others are important genes that affect cancer cell fate. Therefore, inhibiting the transcription of PC-related genes may be effective. There are G-quadruplex configurations in telomere and the promoter regions of HSP90, KRAS, c-MYC and Bcl-2, that are potential targets. Stable G-quadruplex structure usually acts as an obstacle to gene transcription. Small molecular ligands that stabilize G-quadruplex conformation can exhibit clear anticancer effects in PC.
Naphthalene diimide compounds are part of an important ligand set. A series of tetrasubstituted naphthalene diimide ligands tended to make telomeric G-quadruplexes fold into a parallel conformation, preventing binding of human protection of telomeres 1 and topoisomerase IIIα with telomeric DNA, triggering cytotoxicity in multiple PC cell lines[129]. Tetrasubstituted naphthalene diimide derivatives (compounds 3d) retain high affinity to human telomeric G-quadruplexes, upregulate DNA damage responsive genes such as CDKN1A and DDIT3, downregulate telomere maintenance genes such as POT1 and PARP1, and induce cellular senescence[130]. Tetrasubstituted naphthalene diimide isomer ligands (compounds 2-5) are more inclined to stabilize telomeric G-quadruplex structure and improve antiproliferative potency[131]. Tetrasubstituted naphthalene diimide derivative (MM41) combines with and stabilizes G-quadruplex structure and downregulates expression levels of BCL-2 and KRAS to promote apoptosis and decrease tumor growth of MIA-Pa-Ca2 xenografts[132,133]. Tetrasubstituted naphthalene diimide derivative (CM03) causes DNA damage and promotes the presence of nuclear G-quadruplexes in PANC-1 cells; inhibits expression of GLI4, PLXNA1, PRKCZ and MAPK11; partitions PARD6A, and CBFA2T3 in MIA PaCa-2 and PANC-1 cells; and decreases tumor growth of MIA-Pa-Ca2 xenografts[133-135]. Tetrasubstituted naphthalene diimide derivative (SOP1812) was verified to have antiproliferative activity by combining with hTERT and telomere G-quadruplexes[135]. Another naphthalene diimide derivative (BMSG-SH3) decreases telomerase activity, inhibits HSP90 expression, and reduces tumor growth of MIA-Pa-Ca2 xenografts by 50% through maintaining the stability of telomere and HSP90 promoter G-quadruplex structures[136].
Porphyrin compounds are part of another important ligand set. A cationic alkyl-substituted porphyrin compound C14 binds to the KRAS promoter G-quadruplex, protoxidizes the guanines, suppresses gene expression and eventually leads to growth inhibition of PC cell line PANC-1 under photosensitive conditions[137]. Alkyl cationic porphyrins can promote apoptosis in vitro and restrict metabolism and tumor growth in vivo by targeting G-quadruplexes of KRAS and NRAS mRNAs[138]. Porphyrin derivative octaacetyl and tetrakis can both induce apoptosis and block metastasis by inhibiting epithelial to mesenchymal transition through stabilizing KRAS promoter G-quadruplexes and downregulating KRAS expression levels[139], while porphyrin derivative (5Me) may regulate cell proliferation and cell cycle progression by interacting with telomere, Bcl-2, c-MYC and KRAS G-quadruplexes[140]. Previous studies have shown that TMPyP4 can bind to intermolecular G-quadruplexes to arrest cell proliferation and induce both cellular senescence and apoptosis in MIA PaCa-2 cells[140].
Different from TMPyP4, telomestatin can bind to intramolecular G-quadruplexes and control cell proliferation, senescence and apoptosis in MIA PaCa-2 cells[141]. The benzophenanthridine alkaloid nitidine combines with the KRAS promoter G-quadruplex and stabilizes its structure, further downregulating KRAS expression levels and inducing cytotoxicity in AsPC-1, BxPC-3, MIA PaCa-2, and PANC-1 cells[142]. 4,11-bis(2-Aminoethy-llamino)anthra[2,3-b]furan-5,10-dione(2a),11-bis(2-aminoethylamino)anthra[2,3b]thiophene-5,10-dione (2b) stabilizes KRAS RNA G-quadruplexes, inhibits its translation, and induces apoptosis and growth inhibition of PANC-1 cells[143]. Unsymmetrical bisacridines derivatives can inhibit the proliferation of cancer cells in vitro and in vivo by increasing the stability of telomere, c-MYC and KRAS G-quadruplexes[144]. Copper(ii) l/d-valine-(1,10-phen) complexes (complex 1a, 1b) induce cytotoxicity of BxPC3 and AsPC1 cells from its affinity with telomeric G-quadruplexes[145]. CX-5461, the ligand of telomere, c-MYC and c-kit G-quadruplexes, also exhibits antiproliferative activity, and phase I/II clinical trials of CX-5461 as an anticancer drug have been launched[146,147]. A small molecular fluoroquinolone derivative CX-3543 (quarfloxin) can decrease tumor growth of MIA PaCa-2 xenografts by disrupting nucleolin/G-quadruplex complexes on rDNA and inhibiting rRNA synthesis[148,149]. FDA-approved antihelminthic pyrvinium pamoate inhibits mitochondrial RNA transcription and tumor growth by selectively binding to mitochondrial G-quadruplexes[150]. NSC 317605 and novel indoloquinolines derived from it show KRAS G-quadruplex-dependent cytotoxicity in PC cell lines[151]. Two sets of quinazoline-pyrimidine derivative ligands have been shown to prevent tumor growth via targeting telomere, c-MYC,
Some small molecules can play anticancer roles in PC mainly by stabilizing G-quadruplex structures to inhibit gene transcription. However, in addition, some proteins can promote PC progression by destabilizing G-quadruplexes to support gene transcription. For example, integrin linked kinase (ILK) can stimulate KRAS expression via destabilization of G-quadruplexes mediated by hnRNPA1 in the promoter. This in turn affected ILK expression levels, with transcriptional activation mediated by E2F1. This has been called the KRAS-E2F1-ILK-hnRNPA1 regulatory loop, which can result in aggressive phenotypes in the tumor microenvironment[153-155]. Under oxidative stress conditions, poly (ADP-ribose) polymerase 1 (PARP-1) is recruited and binds to KRAS promoter G-quadruplexes, which favors the recruitment of MAZ and hnRNPA1 to the KRAS promoter by activating a ROS-G-quadruplex-PARP-1 axis. This ultimately results in stimulation of KRAS transcription[156]. Different from this mechanism, the G-quadruplex-binding protein apurinic/apyrimidinic endonuclease 1 can also bind to KRAS G-quadruplexes. However, it maintains the structural stability and recruits MAZ to promote KRAS upregulation in vivo and in vitro[157]. Moreover, polypurine reverse Hoogsteen hairpins (PPRHs) can suppress gene transcription and cell proliferation by promoting the formation of KRAS and c-MYC G-quadruplexes in PC cells[158,159].
In summary, the G-quadruplex targets of PC include KRAS, KRAS mRNA, telomere, HSP90, hTERT, Bcl-2, c-MYC, and mitochondrial G-quadruplexes, the regulatory functions of which involve transcription and translation. Proteins that can promote oncogene transcription through G-quadruplexes are also expected to become potential anticancer targets. All details are described in Table 2.
| Ligands/biomolecules | Cell lines | Targeting gene/G-quadruplex | Effects on G-quadruplex | Effects on genes | Anticancer phenotypes | Ref. |
| Tetrasubstituted naphthalene diimide ligands | PANC-1, MIA PaCa-2, HPAC, BxPc-3 | Telomere | Induced formation of a parallel G-quadruplex | Inhibited the binding of hPOT1 and topoisomerase IIIα to telomeric DNA | Cytotoxicity | [129] |
| Tetrasubstituted naphthalene diimide derivatives (compounds 3d) | MIA PaCa-2 | Telomere | Retained high affinity to human telomeric G-quadruplex | Upregulated some DNA damage responsive genes, downregulated some telomere maintenance genes | Induced cellular senescence but did not inhibit telomerase activity | [130] |
| Naphthalene diimide isomer ligands (compounds 2-5) | MIA PaCa-2, PANC-1 | HSP90 | Stabilized G-quadruplex structure | Not detected | Inhibited cell proliferation | [131] |
| Tetrasubstituted naphthalene diimide derivative (MM41) | MIA PaCa-2 | BCL-2, K-RAS | Bound and stabilized G-quadruplex structure | Downregulated expression of BCL-2, K-RAS | Promoted cell apoptosis, decreased tumor growth of MIA-Pa-Ca2 xenografts | [132] |
| Tetrasubstituted naphthalene diimide derivative (CM03) | MIA PaCa-2, PANC-1 | Not detected | Increased presence of nuclear G-quadruplex | Induces DNA damage, downregulated expression of Gli4, PLXNA1, PRKCZ, MAPK11, PARD6A, CBFA2T3 | Decreased tumor growth of MIA-Pa-Ca2 xenografts | [133-135] |
| Tetrasubstituted naphthalene diimide derivative (SOP1812) | MIA PaCa-2, PANC-1, Capan-1, BXPC-3 | hTERT, telomere | Had affinity with G-quadruplex | Downregulated expression of WNT5B, DVL1, AXIN1, APC2, GLI1, MAPK11, BCL-2, hTERT | Inhibited cell proliferation, reduced MIA PaCa-2 xenograft growth | [135] |
| Tetrasubstituted naphthalene diimide derivative (BMSG-SH3) | MIA PaCa-2 | HSP90 | Stabilized G-quadruplex structure | Not detected | Reduced telomerase activity and HSP90 expression, 50% decreased tumor growth of MIA-Pa-Ca2 xenografts | [136] |
| Cationic alkyl-substituted porphyrin compound C14 | PANC-1 | KRAS | Bound with G-quadruplex and protoxidized the guanines | Downregulated expression of KRAS | Induced cell growth arrest | [137] |
| Alkyl cationic porphyrins | MIA PaCa-2, PANC-1 | KRAS mRNA, NRAS mRNA | Bound G-quadruplex | Downregulated expression of KRAS, NRAS only if photoactivated | Activated apoptosis, reduced the metabolic activity of pancreatic cancer cells and the growth of a PANC-1 xenograft | [138] |
| Porphyrin derivative (Octaacetyl) | PANC-1, MIA PaCa-2 | KRAS | Bound and stabilized G-quadruplex | Downregulated expression of KRAS | Cytotoxicity, induced apoptosis, blocked metastasis by inhibiting epithelial to mesenchymal transition | [139] |
| Porphyrin derivative (Tetrakis) | PANC-1, MIA PaCa-2 | KRAS | Bound and stabilized G-quadruplex | Downregulated expression of KRAS | Cytotoxicity, induced apoptosis, blocked metastasis by inhibiting epithelial to messenchymal transition | [139] |
| Porphyrin derivative (5Me) | PANC-1 | Telomere, Bcl-2, c-MYC, KRAS | Bound and stabilized G-quadruplex | Not detected | Inhibited cell proliferation, arrest G2/M phase cell cycle | [140] |
| TMPyP4 | MIA PaCa-2 | Intermolecular G-quadruplex | Not detected | Shortened telomeres | Cytotoxicity, arrested cell proliferation, induced anaphase bridges, cellular senescence and apoptosis | [141] |
| Telomestatin | MIA PaCa-2 | Intramolecular G-quadruplex | Not detected | Shortened telomeres | Cytotoxicity, arrested cell proliferation, and induced cellular senescence and apoptosis | [141] |
| Nitidine | AsPC-1, BxPC-3, MIA PaCa-2, PANC-1 | KRAS | Bound and stabilized G-quadruplex structure | Downregulated expression of KRAS | Cytotoxicity | [142] |
| 4,11-bis(2-Aminoethy- llamino)anthra[2,3-b]furan-5,10-dione(2a),11-bis(2-aminoethylamino) anthra[2,3b]thiophene-5,10-dione (2b) | PANC-1 | KRAS mRNA | Bound and stabilized G-quadruplex | Inhibited translation of KRAS | Induced apoptosis, inhibited cell growth and colony formation | [143] |
| Unsymmetrical bisacridines derivatives | PANC-1, MIA PaCa-2, BXpC-3, AsPC-1, Capan-2 | Telomere, c-MYC, KRAS | Bound and stabilized G-quadruplex | Not detected | Inhibited cell proliferation, reduced PANC-1 and MIA PaCa-2 xenograft growth in vivo | [144] |
| Copper(ii) l/d-valine-(1,10-phen) complexes (complex 1a, 1b) | BxPC3, AsPC1 | Telomere | Had affinity with G-quadruplex | Not detected | Cytotoxicity | [145] |
| CX-5461 (Pidnarulex) | MIA PaCa-2, PANC-1 | Telomere, c-MYC, c-kit | Bound with G-quadruplex | Not detected | Inhibited cell proliferation | [146,147] |
| CX-3543 (Quarfloxin) | MIA PaCa-2 | Nucleolin/ribosomal DNA G-quadruplex complexes | Disrupts nucleolin/G-quadruplex complexes on ribosomal DNA | Inhibited rRNA synthesis | Inhibited proliferation, inhibited Pol I transcription, induced apoptosis, decreased tumor growth of MIA PaCa-2 xenografts | [148,149] |
| Antihelminthic pyrvinium pamoate | PANC-1, Capan-1, HS766T, CFPAC, MIA PaCa-2 | Mitochondrial DNA | Bound G-quadruplex | Inhibited transcription of mitochondrial RNA | Inhibited cell viability, mitochondrial pathways, tumor growth of MIA PaCa-2 xenografts | [150] |
| NSC 317605 and novel indoloquinolines | AsPc1, PANC1, BxPc3, MIA PaCa-2 | c-MYC, KRAS | Bound and stabilized G-quadruplex | Downregulated expression of KRAS | Cytotoxicity | [151] |
| Quinazoline-pyrimidine derivatives | Tumor-naïve pancreatic stellate cells | Telomere, c-MYC, c-kit, KRAS, BCL-2 | Bound and stabilized G-quadruplex | Not detected | Inhibited tumor growth | [152] |
| hnRNPA1 and integrinlinked kinase | AsPC-1, PANC-1, MIA PaCa-2, Capan-2 | KRAS | Destabilized G-quadruplex | Stimulated transcription of KRAS | Promoted KRAS-E2F1-ILK-hnRNPA1 circuitry, tumor growth and aggressive phenotypes | [153-155] |
| Poly [ADP-ribose] polymerase 1 | PANC-1 | KRAS | Destabilized G-quadruplex | Stimulated transcription of KRAS | Activated a ROS-G-quadruplex-PARP-1 axis | [156] |
| Apurinic/apyrimidinic endonuclease 1 | PANC-1, BxPc3, MIA PaCa-2 | KRAS | Bound and stabilized G-quadruplex | Upregulated expression of KRAS | Did not sensitize pancreatic cancer cells to chemotherapeutic drugs in vitro and in vivo | [157] |
| Polypurine reverse Hoogsteen hairpins | AsPc-1, MIA PaCa-2 | KRAS, c-MYC | Bound and stabilized G-quadruplex | Inhibited transcription of KRAS and c-MYC | Inhibited cell proliferation | [158,159] |
Hepatocellular carcinoma: Different from the genetic pathogenesis of PC, specific mutations in proto-oncogenes that can induce hepatocellular carcinoma (HCC) have not been identified. However, anticancer strategies for HCC involving oncogene G-quadruplexes are still being explored. At present, G-quadruplex ligands targeting c-MYC, c-kit and HERC5 have been synthesized and verified for potential application in HCC treatment. Platinum (II) complexes with tridentate ligands, prolinamide derivatives containing triazole, a series of novel 9-O-substituted-13-octylberberine derivatives and novel 9-N-substituted-13-alkylberberine derivatives were all tested and found to have good antiproliferative activities in HepG2 cells, mainly from their good affinity with c-MYC promoter G-quadruplexes and their improved structural stability[160-163]. A series of thiazole orange derivatives were synthesized to effectively bind to telomeric G-quadruplexes, which can stabilize the structures and exhibit cytotoxicity in HCC cell lines[164]. The peptidomimetic ligands showed high affinity to c-kit1 G-quadruplexes also exhibit antiproliferative and proapoptotic properties in HepG2 cells[165]. A 7,11-disubstituted quinazoline derivative HZ-6d targeting HERC5 G-quadruplexes showed anticancer effects in vivo and in vitro through downregulation of HERC5 expression[166].
Viral hepatitis is a primary cause of HCC. Hepatitis B virus (HBV) and hepatitis C virus (HCV) infections can develop into chronic hepatitis and then cirrhosis, eventually leading to HCC. Therefore, early intervention is an effective strategy for delaying HCC progression. In recent years, G-quadruplexes have become a potential target for antiviral therapy. RNA helicase dead box polypeptide 5 can facilitate mRNA translation of STAT1 by unwinding the RNA G-quadruplex structure at the 5’ end of the 5’ UTR, subsequently stimulating the antiviral effects of interferon-α in HBV-infected hepatoma cells[167]. Additionally, cellular nucleolin can directly interact with viral core RNA G-quadruplexes, thereby suppressing the replication and expression of wild-type HCV[168]. All details are described in Table 3.
| Ligands/biomolecules | Cell lines | Targeting gene/G-quadruplex | Effects on G-quadruplex | Effects on genes | Anticancer phenotypes | Ref. |
| Platinum(II) complexes with tridentate ligands | HepG2 | c-MYC | Bound and stabilized G-quadruplex | Inhibited c-MYC expression | Cytotoxicity | [160] |
| Prolinamide derivatives containing triazole | HepG2 | c-MYC | Bound and stabilized G-quadruplex | Inhibited c-MYC expression | Cytotoxicity | [161] |
| A series of novel 9-O-substituted-13-octylberberine derivatives | HepG2, Sk-Hep-1, Huh-7 | c-MYC | Bound and stabilized G-quadruplex | Not detected | Cytotoxicity, blocked cell cycle, induced apoptosis, inhibited tumor growth of H22 xenografts | [162] |
| Series of novel 9-N-substituted-13-alkylberberine derivatives | HepG2, Sk-Hep-1, Huh-7, Hep3 | c-MYC | Bound and stabilized G-quadruplex | Not detected | Cytotoxicity, blocked cell cycle, induced apoptosis, inhibited tumor growth of H22 xenografts | [163] |
| Thiazole orange derivatives | HepG2 | Telomere | Bound and stabilized G-quadruplex | Not detected | Cytotoxicity | [164] |
| Peptidomimetic ligands | HepG2 | c-kit1 | Had high affinity with G-quadruplex | Not detected | Inhibited cell proliferation, induced apoptosis | [165] |
| A 7, 11-disubstituted quinazoline derivative HZ-6d | HepG2, SMMC-7721 | HERC5 | Bound and stabilized G-quadruplex | Inhibited HERC5 expression | Inhibited cell growth, migration, induced apoptosis, suppressed tumor growth of SMMC-7721 xenografts | [166] |
| DDX5 | HepG2, Huh7, Snu387, Snu423, HepaRG, HepAD38 | STAT1 mRNA | Unwound G-quadruplex | Promoted translation of STAT1 | Upregulated expression of STAT1 and enhanced IFN-α mediated antiviral effects | [167] |
| Nucleolin | Huh7.5.1, Huh7.5 | Viral core RNA, G-quadruplex | Directly interacted with G-quadruplex | Inhibited viral RNA replication | Suppressed wild-type viral replication and expression | [168] |
Gastric cancer: Gastric cancer (GC) ranked third worldwide in malignant tumor mortality rates in 2020. Most patients had late stage disease at diagnosis. For advanced GC, chemotherapy is the preferred option, but the associated adverse effects should not be ignored. It is necessary to seek new methods to treat GC, which could include targeted drug therapies based on G-quadruplexes. Small molecules selectively binding to c-kit, telomere and BCL-2 G-quadruplexes have been found to antagonize GC. For example, benzo[a]phenoxazines and quinazolone derivatives display cytotoxicity effects in HGC-27 cells by interacting with c-kit promoter G-quadruplexes and inhibiting gene transcription, while a 1,10-phenanthroline derivative causes DNA damage, telomere dysfunction, autophagy, and antitumor effects in AGS cells by stabilizing telomere, c-kit and BCL-2 G-quadruplexes[169-171]. Use of G-quadruplex antibody confirmed that the targeting regulation could help suppress GC[172]. All details are described in Table 4.
| Ligands/biomolecules | Cell lines | Targeting gene/G-quadruplex | Effects on G-quadruplex | Effects on genes | Anticancer phenotypes | Ref. |
| Benzo[a]phenoxazines | HGC-27 | c-kit | Bound with G-quadruplex | Inhibited c-MYC transcription | Cytotoxicity | [169] |
| Quinazolone derivatives | HGC-27 | c-kit | Stabilized G-quadruplex | Inhibited c-kit transcription | Cytotoxicity | [170] |
| 3-(4-(1H-imidazo[4,5-f][1,10]phenanthrolin-2-yl)-3-(ptolyl)-1Hpyrazol-1-yl)-N,N-dimethylpropan-1-amine (13d) | AGS | Telomere, c-kit, BCL-2 | Stabilized G-quadruplex | Induced telomere dysfunction, DNA damage response | Inhibited cell proliferation, migration, and invasion, promoted cell apoptosis and autophagy by blocking the Akt/mTOR pathway | [171] |
| G-quadruplex antibody | AGS | G-quadruplex | Not detected | Inhibited transcription of hTERT and BCL-2 | Inhibited cell proliferation, migration, invasion and expression of hTERT and BCL-2, induced apoptosis, blocked cell cycle | [172] |
Colorectal cancer: Colorectal cancer (CRC) is a digestive tract disease with high morbidity and mortality. KRAS mutations are present in about 50% of CRC patients. Gene-targeted therapy is a promising direction for treating CRC. Currently, the potential G-quadruplex gene targets being studied in CRC include telomere, c-MYC, KRAS and c-kit. For the telomeric G-quadruplex ligands, BRACO-19 leads to rapid growth inhibition of flavopiridol-resistant cells[173]; 3,11-difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]-acridinium methosulfate (RHPS4), as well as RHPS4-derivatives, induces DNA damage and antiproliferative activity, stabilizes topoisomerase (TOPO) I, and displays cytotoxic and synergistic anticancer effects with TOPO1 inhibitors in CRC cell lines[174-177]. A series of anthracene derivatives substituted with one or two 4,5-dihydro-1H-imidazol-2-yl-hydrazonic groups stabilize G-quadruplexes to different degrees, inhibit telomerase activity, and mediate cytotoxicity[178]. EMICORON cause telomere damage and block cell proliferation and tumor growth of a patient-derived tumor xenograft model[179,180]. Chromene derivatives, the binders of TERRA G-quadruplexes, have cytotoxic effects in HT29 cells[181]. For the ligands targeting c-MYC G-quadruplexes, TMPyP4-mediated stabilization of the mutated G-quadruplex reinstate c-MYC G-quadruplex structure and inhibit its gene expression[182]. CX3543 (quarfoxin) exhibit proapoptotic and antiproliferative effects by downregulating c-MYC and CCAT1 expression levels in vivo and in vitro[183]. CX-5461 (pidnarulex) induces DNA damage and inhibits tumor growth in vivo by binding to telomere, c-MYC and c-kit G-quadruplexes[184]. Dihydrochelerythrine and its derivatives improve the stability of c-MYC and c-kit G-quadruplexes and inhibit HCT116 cell proliferation[185]. Unsymmetrical bisacridines derivatives stabilize c-MYC and KRAS G-quadruplexes and induce cytotoxicity, apoptosis and senescence in HCT116 cells[144,186]. Additionally, the ligands 7-carboxylate indolo[3,2-b] quinoline tri-alkylamine derivatives targeting KRAS and HSP90A promoter G-quadruplexes also show anti-CRC activity by decreasing KRAS and HSP90A expression levels[187]. 3-[2-(Diethylamino)ethyl]-12-methyl-6-oxo-2,3,6,12-tetrahydro-1Hbenzo[4,5]imidazo [1,2-a] imidazo[1’,2’:1,6]pyrido[2,3-d]pyrimidin-14-ium bromide inhibits cell proliferation by interacting with KRAS G-quadruplexes[188]. In addition to those common cancer-related genes, G-quadruplexes of other functional genes have been shown on anticancer drug research and development. A naphthalene diimides compound T5 was shown to inhibit CRC cell growth by decreasing RNA polymerase I (Pol I)-mediated transcription by targeting ribosomal DNA G-quadruplexes[189]. Thiosugar naphthalene diimide conjugates exhibit cytotoxic effects by targeting telomere, c-MYC and KRAS G-quadruplexes[190]. The natural product gallic acid was found to selectively recognize and stabilize G-quadruplexes of rDNA and c-MYC, inhibit their associated mRNA expression, and subsequently suppress tumor growth in vitro and in vivo[191].
The functional protein or oligonucleotide molecules regulating CRC progression based on special G-quadruplexes have also been explored. For example, hnRNPA1 destabilizes TRA2B promoter G-quadruplexes and stimulates its mRNA and protein expression levels, which facilitates proliferation of HCT116 cells[192]. Small nuclear ribonucleoprotein polypeptide A consistently modulates translation of BAG-1 and inhibits HCT116 cell proliferation[193,194]. PPRHs induces c-MYC G-quadruplexes and inhibits proliferation of SW480 cells[159].
The LMNAV6 promoter region forms multiple G-quadruplexes, which increases its transcriptional activity, promotes Lamin A/C protein expression, and induces CRC cell proliferation[195]. FLJ39051, a highly expressed long noncoding RNA in CRC, contains G-quadruplexes. It combines with the RNA helicase DHX36 and promotes CRC cell migration[196]. At present, small molecular ligands or proteins targeting LMNAV6 and FLJ39051 G-quadruplexes have not been reported. All details are described in Table 5.
| Ligands/biomolecules | Cell lines | Targeting gene/G-quadruplex | Effects on G-quadruplex | Effects on genes | Anticancer phenotypes | Ref. |
| BRACO-19 | HCT116, Flavopiridol-resistant HCT116 | Telomere | Stabilized G-quadruplex | Not detected | Rapid inhibition of cell growth | [173] |
| RHPS4 (3,11-difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium methosulfate) and RHPS4-derivatives | HT29, HCT116 | Telomere | Bound with G-quadruplex | Induced DNA damage | Stabilized TOPO1, cytotoxicity, inhibited cell proliferation, had synergistic anticancer effects with TOPO1 inhibitors | [174-177] |
| A series of anthracene derivatives substituted with one or two 4,5-dihydro-1H-imidazol-2-yl-hydrazonic groups | LoVo | Telomere | Induced G-quadruplex structures, bound and stabilized G-quadruplex | Induced DNA damage | Cytotoxicity, telomerase inhibition | [178] |
| EMICORON | HT29, HCT116, A90 colon epithelial tumor cell line | Telomere | Bound with G-quadruplex | Increased telomere damage | Cytotoxicity, inhibited cell proliferation and tumor growth of patient-derived tumor xenograft | [179,180] |
| Chromene derivatives | HT29 | Telomere RNA | Bound with G-quadruplex | Not detected | Cytotoxicity | [181] |
| TMPyP4 | SW480, SW620 | c-MYC | Stabilized the mutated G-quadruplex structure | Inhibited c-MYC expression | Silenced c-MYC expression | [182] |
| CX-3543 (quarfloxin) | HT29 | c-MYC | Not detected | Inhibited c-MYC expression | Reduced CCAT1 expression, promoted cell apoptosis, inhibited cell proliferation and tumor growth of HT29 xenografts | [183] |
| CX-5461 (pidnarulex) | HT-29, DLD-1, CT26 | Telomere, c-MYC, c-kit | Bound with G-quadruplex | Caused DNA damage | Inhibited tumor growth of CT26 xenografts | [184] |
| Dihydrochelerythrine and its derivatives | HCT116 | c-MYC, c-kit | Stabilized G-quadruplex | Not detected | Inhibited cell proliferation | [185] |
| Unsymmetrical bisacridines derivatives | HCT116 | c-MYC, KRAS | Bound and stabilized G-quadruplex | Not detected | Induced cytotoxicity, apoptosis and senescence | [144,186] |
| 7-carboxylate indolo[3,2-b] quinoline tri-alkylamine derivatives | HCT116, SW620 | KRAS, HSP90A | Stabilized G-quadruplex | Decreased KRAS and HSP90 mRNA expression, and KRAS transcription | Inhibited cell proliferation and protein expression of KRAS and HSP90A, promoted apoptosis | [187] |
3-[2-(Diethylamino)ethyl]-12-methyl-6-oxo-2,3,6,12-tetrahydro-1H-benzo[4,5]imidazo[1,2- a]imidazo[1’,2’:1,6]pyrido[2,3-d]pyrimidin-14-ium bromide | HCT116 | KRAS | Bound and stabilized G-quadruplex | Decreased KRAS mRNA expression | Inhibited cell proliferation | [188] |
| Naphthalene diimides compound T5 | Colorectal cancer cell | rDNA | Had high affinity with G-quadruplex | Impaired RNA Pol I elongation, inhibited Pol I transcription | Inhibited cell growth by inducing a rapid inhibition of Pol I transcription, nucleolus disruption, proteasome-dependent Pol I catalytic subunit A degradation and autophagy | [189] |
| Thiosugar naphthalene diimide conjugates | HT29 | Telomere, c-MYC, KRAS | Bound and stabilized G-quadruplex | Not detected | Cytotoxicity | [190] |
| Gallic acid | SW480 SW620 | rDNA, c-MYC | Bound and stabilized G-quadruplex | Inhibited expression of rDNA and c-MYC | Cytotoxicity, inhibited tumor growth of SW480 xenografts | [191] |
| HnRNPA1 | HCT116 | TRA2B promoter | Destabilized G-quadruplex | Stimulated TRA2B transcription | Promoted cell proliferation and expression of TRA2B | [192] |
| SNRPA | HCT116 | BAG-1 mRNA | Bound with G-quadruplex | Inhibited translation of BAG-1 | Inhibited cell proliferation | [193,194] |
| PPRHs | SW480 | c-MYC | Bound and stabilized G-quadruplex | Inhibited transcription of c-MYC | Inhibited cell proliferation | [159] |
Gastrointestinal stromal tumors: Gastrointestinal stromal tumors (GISTs) are soft tissue sarcomas originating from Cajal interstitial cells. They mostly frequently occur in the stomach, small intestine, and colorectum, but rarely occur in the esophagus, mesentery, omentum and retroperitoneum. GISTs are characterized by aberrant expression of c-kit oncogene, CD117 and CD34. The kinase inhibitor imatinib is an effective drug, but resistance to imatinib induced by active-site mutations has become a practical challenge that cannot be fully addressed by second and third-generation inhibitors. There are two G-quadruplex-forming sequences (c-kit1 positioned between -12 and -33 bp, c-kit2 positioned between -64 and -83 bp) upstream of the transcription initiation sites of the human c-kit promoter. There are also potential binding sites for transcription factors SP1 and AP2, providing an opportunity for c-kit-targeted therapy. A series of 6-substituted indenoisoquinolines and N,N’-Bis[2-(pyrrolidin-1-yl)ethylamino]-2,6-bis[2-(pyrrolidin-1-yl)ethylamino]-1,4,5,8-naphthalenetetracarboxylic acid diimide have been confirmed to stabilize c-kit promoter G-quadruplexes, mediate cytotoxicity, and downregulate c-kit protein expression levels in GIST cell lines[197,198]. The latter can also stabilize BCL-2 promoter and mRNA G-quadruplexes to promote cytotoxicity and inhibit BCL-2 protein expression[198]. All details are described in Table 6.
| Ligands/biomolecules | Cell lines | Targeting gene/G-quadruplex | Effects on G-quadruplex | Effects on genes | Anticancer phenotypes | Ref. |
| 6-Substituted indenoisoquinolines | GIST882 | c-kit | Stabilized G-quadruplex | Inhibited c-kit transcription | Cytotoxicity, inhibited expression of c-kit protein | [196] |
| N,N’-Bis(2-(pyrrolidin-1-yl)ethylamino)-2,6-bis(2-(pyrrolidin-1-yl)ethylamino)-1,4,5,8-naphthalenetetracarboxylic acid diimide | GIST882, GIST48, GIST62 | c-kit, BCL-2, BCL-2 mRNA | Stabilized G-quadruplex | Not detected | Cytotoxicity, inhibited expression of c-kit and BCL-2 proteins | [197,198] |
In summary, targeting G-quadruplexes of cancer-related genes in cancer cells and inducing cytotoxic effects by regulating gene transcription may be an effective strategy for preventing and treating various gastrointestinal cancers. An overview of the advancement of potential drugs that target G-quadruplexes in gastrointestinal cancers is shown as Figure 2.
As anticancer agents: In addition to acting as a target of ligands or proteins, G-quadruplexes can serve as anticancer agents. They can recognize specific biomacromolecules with a high degree of specificity, regulate their biological function, and interfere with cancer progression. The G-quadruplex formed by the G-rich sequence T-22AG can competitively bind to nuclear protein, inhibit its combination with KRAS G-quadruplex, and thus inhibit gene transcription and proliferation of Panc-1 cells[199]. AS1411 was an earlier discovered G-quadruplex sequence with antiproliferative activity by targeting nucleolin in a variety of cancer cells, such as PC, GC and CRC[200]. The sequences TBA and its derivatives exhibit antiproliferative effects in HCT 116p53−/− cells via the G-quadruplex structure; the target of which may be uL3[201]. Similarly, the G-quadruplex sequences INT-B (T30175) and its derivatives, along with d(GGGT)4 and its analogs also inhibit HCT 116p53−/− cell proliferation, but the specific target remains unclear[202,203]. All details are described in Table 7.
| Tumor model | G-quadruplex name | Sequence (5’-3’) | Protein target | Cells | Anticancer phenotype | Ref. |
| Pancreatic cancer | T-22AG | GGAGGGGGAGAAGGGAGAAGGG | Nuclear protein | Panc-1 | Reduces cell growth | [199] |
| AS1411 | GGTGGTGGTGGTTGTGGTGGTGGTGG | Nucleolin | PANC-1 | Inhibited cell proliferation | [200] | |
| Gastric cancer | AS1411 | GGTGGTGGTGGTTGTGGTGGTGGTGG | Nucleolin | KATOIIIe, HGC27 | Inhibited cell proliferation | [200] |
| Colorectal cancer | AS1411 | GGTGGTGGTGGTTGTGGTGGTGGTGG | Nucleolin | HCC 2998, HT-29, KM12, HCT-116, SW620, HCT-15, LS174T | Inhibited cell proliferation | [200] |
| TBA | GGTTGGTGTGGTTGG | uL3 | HCT 116p53-/- | Impaired ribosomal RNA processing, leading to the accumulation of pre-ribosomal RNAs, arrested cells in the G2/M phase and induced early apoptosis | [201] | |
| L-TBA | GGTTGGTGTGGTTGG | |||||
| LQ1 | GGTTGGTGTGGTTGG | |||||
| LQ2 | GGTTGGGTGTGGTTGG | |||||
| LQ3 | GGTTGGGTGTGGTTGG | |||||
| INT-B (T30175) | GTGGTGGGTGGGTGGGT | Not detected | HCT 116p53-/- | Inhibited cell proliferation | [202] | |
| INT-BS2 | GSGGTGGGTGGGTGGGT | |||||
| INT-BS5 | GTGGSGGGTGGGTGGGT | |||||
| INT-BS9 | GTGGTGGGSGGGTGGGT | |||||
| INT-BS13 | GTGGTGGGTGGGSGGGT | |||||
| INT-BS17 | GTGGTGGGTGGGTGGGS | |||||
| TT-INT-B | TTGTGGTGGGTGGGTGGGT | |||||
| Qnat | GGGTGGGTGGGTGGGT | Not detected | HCT 116p53-/- | Inhibited cell proliferation | [203] | |
| QS4 | GGGSGGGTGGGTGGGT | |||||
| QS8 | GGGTGGGSGGGTGGGT | |||||
| QS12 | GGGTGGGTGGGSGGGT | |||||
| QS16 | GGGTGGGTGGGTGGGS |
As an assistant in anticancer agents: G-quadruplex sequences mainly have two functions. In the research and development of anticancer drugs, G-quadruplex sequences were often used as the carriers of drugs or targeted agents in delivery systems to improve the delivery efficiency and targeting of anticancer drugs[204]. The carried molecules have included such chemotherapeutic drugs as paclitaxel, docetaxel, doxorubicin, triptolide, epirubicin, gemcitabine, thymoquinone, TMPyP4 and 5-fluorouracil, targeting cancers like HCC, PC and CRC[205,206]. As an important component of anticancer agents, G-quadruplex sequences can also help produce or improve anticancer efficacy. For example, the G-quadruplex dependent intracellular self-assembly device can continuously produce ROS to enhance antitumor effects of 5-aminolevulinic-acid in EC cells[207]. The parallel G-quadruplex configurations boost the cellular uptake of 5-fluoro-20-deoxyuridine oligomers, which stimulate cytotoxicity in 5-fluorouracil resistant CRC cells[208].
Although many small molecular ligands or biomolecules targeting G-quadruplexes have been found to have anticancer activity in vitro and in vivo, it is still uncertain whether they can achieve such effects in humans. Unfortunately, many promising drugs have not passed clinical trials in the past, as biological systems are complex and many internal and external factors can affect drug effectiveness. The application of G-quadruplex targets in the treatment of gastrointestinal cancers will also face some challenges.
At first, G-quadruplexes were simply described as an obstacle to the transcription of cancer-related genes, leading to increased efforts to design and develop small molecule ligands as anticancer drugs targeting G-quadruplex structure, which attracted widespread attention[209]. However, evidence has shown that G-quadruplexes can regulate gene transcription at multiple levels, including through epigenetic modification and chromatin structure[210]. Because of the complexity of gene expression regulation, G-quadruplexes can play dual roles in gene transcription: Blocking polymerase to inhibit gene transcription; and recruiting transcription factors to promote gene transcription[117]. Under certain conditions, G-quadruplexes can trigger opposing effects on the same target[211]. The regulation of translation by RNA G-quadruplexes also has two sides. G-quadruplexes can prevent ribosome entry under conditions of cap-dependent translation, but can also prompt ribosome entry under conditions of cap-independent translation[212]. With both DNA G-quadruplex-mediated regulation of transcription and RNA G-quadruplex-mediated regulation of translation, the final effects depend on the specific environment. Further research is required to investigate if ligands targeting G-quadruplexes in the tumor microenvironment can result in the predicted anticancer effects and if they can affect normal cells.
Chromatin and DNA modifications: The formation of G-quadruplexes in vivo is the result of the comprehensive action of various factors within its cell environment, including chromatin. Although a previous study indicated that transcriptional activation increased the instability of potential G-quadruplex-forming sequences, ChIP-seq research confirmed that promoter G-quadruplex formation preceded transcription rather than depending on transcription. Additionally, chromatin compaction led to a loss of RNA polymerase II (Pol II) and promoter G-quadruplexes[213]. Different types of DNA modifications can directly influence the formation of G-quadruplexes. For example, the stability and kinetic associations of G-quadruplex structures were increased by cytosine methylation (in addition with 5mC), which did not directly act on the Hoogsteen bonding[214]. Guanine bases in nucleic acids can be oxidized to 8-oxo-7,8-dihydroguanine (8-oxoguanine), which can destroy the G-quadruplex structure in cancers[215]. Oncogene promoter regions are prone to hypomethylation, while those of tumor suppressor genes are prone to hypermethylation. These factors may indirectly impact the formation of G-quadruplexes and the resulting regulatory effects.
G-quadruplex-binding proteins: G-quadruplexes play various regulatory functions by interacting with proteins. G-quadruplex-binding proteins indirectly participate in biological processes such as DNA replication, gene transcription and telomere maintenance via G-quadruplexes. The influence of binding proteins on the formation of G-quadruplexes mainly involves two aspects: Unfolding G-quadruplex structures and stabilizing G-quadruplex structures. Helicases are important binding proteins that can unwind G-quadruplexes and interfere with their regulatory functions. Such proteins are mainly classified as canonical helicases, including the RecQ-like and DEAD box or DEAH box helicase families. In vitro, these three helicases have been reported to bind to the 3’ tail of the DNA substrate and subsequently repetitively catalyze 3’-5’ unfolding of G-quadruplexes in an ATP-independent manner[216]. In addition, nonhelicase binding proteins, such as G-rich RNA sequence binding factor 1 and cellular nucleic acid-binding protein, can sequester the unfolded G-quadruplex form[216-218]. In contrast, there are also binding proteins that can support the G-quadruplex structure, such as nucleolin and RNA-binding protein 4[168,219]. Additionally, RNA-binding proteins are important influencing factors of RNA G-quadruplexes. G-quadruplex-binding proteins are also potential targets for cancer treatment because their effects contribute to G-quadruplex functions[220]. Significantly, the specific effects of these binding proteins on the targeted G-quadruplexes depend on their specific intracellular environments.
Inflammatory cytokines: Inflammatory cytokines produced during inflammation reactions can support the production of ROS and nitrogen species (RONS), which may cause DNA damage. RONS can remove an electron from DNA bases and generate an electron hole, then transfer it to a base with a lower ionization potential. Guanine has the lowest ionization energy among the four DNA bases, making it particularly vulnerable to oxidative damage[221]. The most significant oxidative damage involves hydroxyl free radicals interacting with guanine to induce 8-oxoguanine, which can pair with adenine bases, and induce a G>T conversion during replication[3]. The degree of DNA damage depends on the position of the oxidized guanines and G-quartets.
At present, the design of small molecules is mainly based on the specific G-quadruplex configurations. As mentioned previously, there are three basic configurations for G-quadruplexes, and different nucleic acid sequences may form the same G-quadruplex configuration. Therefore, one small molecule ligand may have similar binding stabilities with the G-quadruplex structures of different genes, which may reduce the targeting of gene therapy. For example, berberine can combine with the parallel structures of the KRAS and c-MYC promoters[6,16]. This inhibits KRAS and c-MYC expression and induces cytotoxicity in various cancer cells that express these oncogenes. Further work is needed to determine if this drug can cause negative effects in normal cells.
G-quadruplexes are widely distributed throughout the human genome and are key aspects of gene transcription and translation regulation. Therefore, G-quadruplexes can be the drug targets against multiple human diseases, such as viral infection[222], bacterial infection[223], muscular atrophy[60] and cancer, especially gastrointestinal cancers. However, there are some uncertainties with this application that should be explored further. Firstly, G-quadruplex-mediated regulation of transcription and translation in gastrointestinal tissues require more investigation, especially during tumorigenesis. The development of high-throughput sequencing and single nucleotide polymorphism detection may provide new opportunities to establish specific gene therapy strategies for gastrointestinal cancers based on G-quadruplexes. Secondly, the transcriptional activation function of G-quadruplexes is needed in some normal physiological processes, raising the concern that anticancer therapies targeting G-quadruplexes may interfere with normal cellular activities. Fully understanding the roles of G-quadruplexes in different biological processes, especially in various diseases, is helpful for addressing this challenge. Thirdly, G-quadruplexes can both inhibit and promote gene transcription and translation, with the final effects depending on the intracellular environment. This ultimately directly affects the treatment outcome. With the progress of molecular diagnosis technology, it may be necessary to specifically evaluate the patient’s internal environment before treatment. Fourthly, small molecule ligands and biomolecules may simultaneously target genes with the same G-quadruplex configurations, resulting in a need for improved selectivity or targeting. Fifthly, the formation of a G-quadruplex is affected by a variety of biological factors. Whether these factors can interfere with a G-quadruplex-targeted therapy requires further study. Sixthly, clinical trials are needed to verify the efficacy of such small molecule ligands and biomolecules.
In addition to telomeres, G-quadruplexes are widely present in the promoter regions of oncogenes as well as cancerous genes, and can regulate various biological processes, especially gene transcription and translation, laying a good foundation for G-quadruplexes to become anticancer targets from the perspective of gene regulation. Multiple genes regulating EC, PC, HCC, GC, CRC and GIST have been found to contain G-quadruplex structures, including the key regulatory gene KRAS for PC and CRC, and c-kit for GC and GIST. Many small molecular ligands or biomolecules based on the G-quadruplex of these genes have been designed, synthesized, or discovered, and preclinical studies have shown that these molecules have good anticancer effects. Therefore, G-quadruplexes as targets against gastrointestinal cancers have broad application prospects. However, due to the diversity of G-quadruplex functions and the complexity of the biological internal environment, the application of G-quadruplex as a target of anticancer drugs still faces some challenges, which requires further exploration and research. We hope this work will provide references for anticancer strategies based on G-quadruplex targets in gastrointestinal cancers.
| 1. | Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022;12:31-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 664] [Cited by in RCA: 5961] [Article Influence: 1490.3] [Reference Citation Analysis (2)] |
| 2. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 48671] [Article Influence: 3244.7] [Reference Citation Analysis (12)] |
| 3. | Stein M, Eckert KA. Impact of G-Quadruplexes and Chronic Inflammation on Genome Instability: Additive Effects during Carcinogenesis. Genes (Basel). 2021;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 4. | GELLERT M, LIPSETT MN, DAVIES DR. Helix formation by guanylic acid. Proc Natl Acad Sci U S A. 1962;48:2013-2018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1125] [Cited by in RCA: 1221] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 5. | Tan J, Lan L. The DNA secondary structures at telomeres and genome instability. Cell Biosci. 2020;10:47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 6. | Wang KB, Liu Y, Li J, Xiao C, Wang Y, Gu W, Li Y, Xia YZ, Yan T, Yang MH, Kong LY. Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine. Nat Commun. 2022;13:6016. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 35] [Reference Citation Analysis (1)] |
| 7. | Monsen RC, DeLeeuw LW, Dean WL, Gray RD, Chakravarthy S, Hopkins JB, Chaires JB, Trent JO. Long promoter sequences form higher-order G-quadruplexes: an integrative structural biology study of c-Myc, k-Ras and c-Kit promoter sequences. Nucleic Acids Res. 2022;50:4127-4147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 8. | Ogloblina AM, Bannikova VA, Khristich AN, Oretskaya TS, Yakubovskaya MG, Dolinnaya NG. Parallel G-Quadruplexes Formed by Guanine-Rich Microsatellite Repeats Inhibit Human Topoisomerase I. Biochemistry (Mosc). 2015;80:1026-1038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 9. | Cheong C, Moore PB. Solution structure of an unusually stable RNA tetraplex containing G- and U-quartet structures. Biochemistry. 1992;31:8406-8414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 211] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 10. | Agarwala P, Pandey S, Ekka MK, Chakraborty D, Maiti S. Combinatorial role of two G-quadruplexes in 5' UTR of transforming growth factor β2 (TGFβ2). Biochim Biophys Acta Gen Subj. 2019;1863:129416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 11. | Scionti F, Juli G, Rocca R, Polerà N, Nadai M, Grillone K, Caracciolo D, Riillo C, Altomare E, Ascrizzi S, Caparello B, Cerra M, Arbitrio M, Richter SN, Artese A, Alcaro S, Tagliaferri P, Tassone P, Di Martino MT. TERRA G-quadruplex stabilization as a new therapeutic strategy for multiple myeloma. J Exp Clin Cancer Res. 2023;42:71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 12. | Khan S, Singh A, Nain N, Kukreti S. Alkali cation-mediated topology displayed by an exonic G-rich sequence of TRPA1 gene. J Biomol Struct Dyn. 2022;1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 13. | Sugimoto W, Kinoshita N, Nakata M, Ohyama T, Tateishi-Karimata H, Nishikata T, Sugimoto N, Miyoshi D, Kawauchi K. Intramolecular G-quadruplex-hairpin loop structure competition of a GC-rich exon region in the TMPRSS2 gene. Chem Commun (Camb). 2021;58:48-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 14. | Ghafouri-Fard S, Abak A, Baniahmad A, Hussen BM, Taheri M, Jamali E, Dinger ME. Interaction between non-coding RNAs, mRNAs and G-quadruplexes. Cancer Cell Int. 2022;22:171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 15. | Liu W, Zhu BC, Liu LY, Xia XY, Mao ZW. G-quadruplex structural transition driven by a platinum compound. Nucleic Acids Res. 2022;50:7816-7828. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 16. | Dickerhoff J, Brundridge N, McLuckey SA, Yang D. Berberine Molecular Recognition of the Parallel MYC G-Quadruplex in Solution. J Med Chem. 2021;64:16205-16212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 17. | Beseiso D, Chen EV, McCarthy SE, Martin KN, Gallagher EP, Miao J, Yatsunyk LA. The first crystal structures of hybrid and parallel four-tetrad intramolecular G-quadruplexes. Nucleic Acids Res. 2022;50:2959-2972. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 18. | Criscuolo A, Napolitano E, Riccardi C, Musumeci D, Platella C, Montesarchio D. Insights into the Small Molecule Targeting of Biologically Relevant G-Quadruplexes: An Overview of NMR and Crystal Structures. Pharmaceutics. 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 19. | Kosiol N, Juranek S, Brossart P, Heine A, Paeschke K. G-quadruplexes: a promising target for cancer therapy. Mol Cancer. 2021;20:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 362] [Article Influence: 72.4] [Reference Citation Analysis (0)] |
| 20. | Mellor C, Perez C, Sale JE. Creation and resolution of non-B-DNA structural impediments during replication. Crit Rev Biochem Mol Biol. 2022;57:412-442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 21. | Bryan TM. Mechanisms of DNA Replication and Repair: Insights from the Study of G-Quadruplexes. Molecules. 2019;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 22. | Linke R, Limmer M, Juranek SA, Heine A, Paeschke K. The Relevance of G-Quadruplexes for DNA Repair. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 23. | Jansson-Fritzberg LI, Sousa CI, Smallegan MJ, Song JJ, Gooding AR, Kasinath V, Rinn JL, Cech TR. DNMT1 inhibition by pUG-fold quadruplex RNA. RNA. 2023;29:346-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 24. | Ogasawara S. Transcription Driven by Reversible Photocontrol of Hyperstable G-Quadruplexes. ACS Synth Biol. 2018;7:2507-2513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 25. | Wu CG, Spies M. G-quadruplex recognition and remodeling by the FANCJ helicase. Nucleic Acids Res. 2016;44:8742-8753. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 87] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 26. | Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908-2916. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1262] [Cited by in RCA: 1424] [Article Influence: 67.8] [Reference Citation Analysis (0)] |
| 27. | Asamitsu S, Takeuchi M, Ikenoshita S, Imai Y, Kashiwagi H, Shioda N. Perspectives for Applying G-Quadruplex Structures in Neurobiology and Neuropharmacology. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 28. | Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Plückthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci U S A. 2001;98:8572-8577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 470] [Cited by in RCA: 502] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 29. | Oganesian L, Bryan TM. Physiological relevance of telomeric G-quadruplex formation: a potential drug target. Bioessays. 2007;29:155-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 257] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 30. | Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem. 2013;5:182-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1430] [Cited by in RCA: 1700] [Article Influence: 130.8] [Reference Citation Analysis (0)] |
| 31. | Di Antonio M, Ponjavic A, Radzevičius A, Ranasinghe RT, Catalano M, Zhang X, Shen J, Needham LM, Lee SF, Klenerman D, Balasubramanian S. Single-molecule visualization of DNA G-quadruplex formation in live cells. Nat Chem. 2020;12:832-837. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 228] [Cited by in RCA: 282] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 32. | Henderson A, Wu Y, Huang YC, Chavez EA, Platt J, Johnson FB, Brosh RM Jr, Sen D, Lansdorp PM. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2017;45:6252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 33. | Zhang S, Sun H, Chen H, Li Q, Guan A, Wang L, Shi Y, Xu S, Liu M, Tang Y. Direct visualization of nucleolar G-quadruplexes in live cells by using a fluorescent light-up probe. Biochim Biophys Acta Gen Subj. 2018;1862:1101-1106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 34. | Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901-2907. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 739] [Cited by in RCA: 800] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 35. | Hänsel-Hertsch R, Spiegel J, Marsico G, Tannahill D, Balasubramanian S. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat Protoc. 2018;13:551-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 230] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 36. | Chambers VS, Marsico G, Boutell JM, Di Antonio M, Smith GP, Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol. 2015;33:877-881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 913] [Cited by in RCA: 953] [Article Influence: 86.6] [Reference Citation Analysis (0)] |
| 37. | Henderson E, Hardin CC, Walk SK, Tinoco I Jr, Blackburn EH. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell. 1987;51:899-908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 491] [Cited by in RCA: 542] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 38. | Rhodes D, Lipps HJ. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627-8637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 961] [Cited by in RCA: 1142] [Article Influence: 103.8] [Reference Citation Analysis (0)] |
| 39. | Moye AL, Porter KC, Cohen SB, Phan T, Zyner KG, Sasaki N, Lovrecz GO, Beck JL, Bryan TM. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat Commun. 2015;6:7643. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 218] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 40. | Spiegel J, Adhikari S, Balasubramanian S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020;2:123-136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 469] [Cited by in RCA: 598] [Article Influence: 99.7] [Reference Citation Analysis (0)] |
| 41. | Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406-413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1163] [Cited by in RCA: 1084] [Article Influence: 54.2] [Reference Citation Analysis (0)] |
| 42. | Marsico G, Chambers VS, Sahakyan AB, McCauley P, Boutell JM, Antonio MD, Balasubramanian S. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 2019;47:3862-3874. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 259] [Cited by in RCA: 305] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 43. | Marquevielle J, Robert C, Lagrabette O, Wahid M, Bourdoncle A, Xodo LE, Mergny JL, Salgado GF. Structure of two G-quadruplexes in equilibrium in the KRAS promoter. Nucleic Acids Res. 2020;48:9336-9345. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 44. | Cogoi S, Xodo LE. G4 DNA in ras genes and its potential in cancer therapy. Biochim Biophys Acta. 2016;1859:663-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 45. | Li ML, Yuan JM, Yuan H, Wu BH, Huang SL, Li QJ, Ou TM, Wang HG, Tan JH, Li D, Chen SB, Huang ZS. Design, Synthesis, and Evaluation of New Sugar-Substituted Imidazole Derivatives as Selective c-MYC Transcription Repressors Targeting the Promoter G-Quadruplex. J Med Chem. 2022;65:12675-12700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 46. | Yang Y, Yang Y, Wang S, Li H, Chen DDY. Detecting the formation of human c-KIT oncogene promoter G-Quadruplex by Taylor dispersion analysis. Talanta. 2021;233:122533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 47. | Tong X, Lan W, Zhang X, Wu H, Liu M, Cao C. Solution structure of all parallel G-quadruplex formed by the oncogene RET promoter sequence. Nucleic Acids Res. 2011;39:6753-6763. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 70] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 48. | Robert C, Marquevielle J, Salgado GF. The Promoter Region of the Proto-Oncogene MST1R Contains the Main Features of G-Quadruplexes Formation. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 49. | Dai J, Chen D, Jones RA, Hurley LH, Yang D. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34:5133-5144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 295] [Cited by in RCA: 311] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 50. | Lombardo CM, Welsh SJ, Strauss SJ, Dale AG, Todd AK, Nanjunda R, Wilson WD, Neidle S. A novel series of G-quadruplex ligands with selectivity for HIF-expressing osteosarcoma and renal cancer cell lines. Bioorg Med Chem Lett. 2012;22:5984-5988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 51. | Stein M, Hile SE, Weissensteiner MH, Lee M, Zhang S, Kejnovský E, Kejnovská I, Makova KD, Eckert KA. Variation in G-quadruplex sequence and topology differentially impacts human DNA polymerase fidelity. DNA Repair (Amst). 2022;119:103402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 52. | Qin Y, Rezler EM, Gokhale V, Sun D, Hurley LH. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007;35:7698-7713. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 170] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 53. | Chen JN, He YD, Liang HT, Cai TT, Chen Q, Zheng KW. Regulation of PDGFR-β gene expression by targeting the G-vacancy bearing G-quadruplex in promoter. Nucleic Acids Res. 2021;49:12634-12643. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 54. | Monsen RC, DeLeeuw L, Dean WL, Gray RD, Sabo TM, Chakravarthy S, Chaires JB, Trent JO. The hTERT core promoter forms three parallel G-quadruplexes. Nucleic Acids Res. 2020;48:5720-5734. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 55. | Waller ZA, Howell LA, Macdonald CJ, O'Connell MA, Searcey M. Identification and characterisation of a G-quadruplex forming sequence in the promoter region of nuclear factor (erythroid-derived 2)-like 2 (Nrf2). Biochem Biophys Res Commun. 2014;447:128-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 56. | Navarro A, Benabou S, Eritja R, Gargallo R. Influence of pH and a porphyrin ligand on the stability of a G-quadruplex structure within a duplex segment near the promoter region of the SMARCA4 gene. Int J Biol Macromol. 2020;159:383-393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 57. | Joshi S, Singh A, Kukreti S. Porphyrin induced structural destabilization of a parallel DNA G-quadruplex in human MRP1 gene promoter. J Mol Recognit. 2022;35:e2950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 58. | Singh A, Joshi S, Kukreti S. Cationic porphyrins as destabilizer of a G-quadruplex located at the promoter of human MYH7 β gene. J Biomol Struct Dyn. 2020;38:4801-4816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 59. | Borel C, Migliavacca E, Letourneau A, Gagnebin M, Béna F, Sailani MR, Dermitzakis ET, Sharp AJ, Antonarakis SE. Tandem repeat sequence variation as causative cis-eQTLs for protein-coding gene expression variation: the case of CSTB. Hum Mutat. 2012;33:1302-1309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 60. | Geng Y, Liu C, Cai Q, Luo Z, Miao H, Shi X, Xu N, Fung CP, Choy TT, Yan B, Li N, Qian P, Zhou B, Zhu G. Crystal structure of parallel G-quadruplex formed by the two-repeat ALS- and FTD-related GGGGCC sequence. Nucleic Acids Res. 2021;49:5881-5890. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 61. | Kobayashi H, Abe K, Matsuura T, Ikeda Y, Hitomi T, Akechi Y, Habu T, Liu W, Okuda H, Koizumi A. Expansion of intronic GGCCTG hexanucleotide repeat in NOP56 causes SCA36, a type of spinocerebellar ataxia accompanied by motor neuron involvement. Am J Hum Genet. 2011;89:121-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 219] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 62. | Mead S, Webb TE, Campbell TA, Beck J, Linehan JM, Rutherfoord S, Joiner S, Wadsworth JD, Heckmann J, Wroe S, Doey L, King A, Collinge J. Inherited prion disease with 5-OPRI: phenotype modification by repeat length and codon 129. Neurology. 2007;69:730-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 51] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 63. | Trajkovski M, da Silva MW, Plavec J. Unique structural features of interconverting monomeric and dimeric G-quadruplexes adopted by a sequence from the intron of the N-myc gene. J Am Chem Soc. 2012;134:4132-4141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 64. | Bilgen E, Persil Çetinkol Ö. Doxorubicin exhibits strong and selective association with VEGF Pu(22) G-quadruplex. Biochim Biophys Acta Gen Subj. 2020;1864:129720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 65. | Blice-Baum AC, Mihailescu MR. Biophysical characterization of G-quadruplex forming FMR1 mRNA and of its interactions with different fragile X mental retardation protein isoforms. RNA. 2014;20:103-114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 66. | Morris MJ, Wingate KL, Silwal J, Leeper TC, Basu S. The porphyrin TmPyP4 unfolds the extremely stable G-quadruplex in MT3-MMP mRNA and alleviates its repressive effect to enhance translation in eukaryotic cells. Nucleic Acids Res. 2012;40:4137-4145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 121] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 67. | Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5' UTR of the NRAS proto-oncogene modulates translation. Nat Chem Biol. 2007;3:218-221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 712] [Cited by in RCA: 661] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 68. | Christiansen J, Kofod M, Nielsen FC. A guanosine quadruplex and two stable hairpins flank a major cleavage site in insulin-like growth factor II mRNA. Nucleic Acids Res. 1994;22:5709-5716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 69] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 69. | Gomez D, Guédin A, Mergny JL, Salles B, Riou JF, Teulade-Fichou MP, Calsou P. A G-quadruplex structure within the 5'-UTR of TRF2 mRNA represses translation in human cells. Nucleic Acids Res. 2010;38:7187-7198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 161] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 70. | Arora A, Suess B. An RNA G-quadruplex in the 3' UTR of the proto-oncogene PIM1 represses translation. RNA Biol. 2011;8:802-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 82] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 71. | Fisette JF, Montagna DR, Mihailescu MR, Wolfe MS. A G-rich element forms a G-quadruplex and regulates BACE1 mRNA alternative splicing. J Neurochem. 2012;121:763-773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 87] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 72. | Huang W, Smaldino PJ, Zhang Q, Miller LD, Cao P, Stadelman K, Wan M, Giri B, Lei M, Nagamine Y, Vaughn JP, Akman SA, Sui G. Yin Yang 1 contains G-quadruplex structures in its promoter and 5'-UTR and its expression is modulated by G4 resolvase 1. Nucleic Acids Res. 2012;40:1033-1049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 90] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 73. | Xi M, Li Y, Zhou J. Exploration of the formation and structure characteristics of a miR-92a promoter G-quadruplex by ESI-MS and CD. Talanta. 2020;211:120708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 74. | Imperatore JA, Then ML, McDougal KB, Mihailescu MR. Characterization of a G-Quadruplex Structure in Pre-miRNA-1229 and in Its Alzheimer's Disease-Associated Variant rs2291418: Implications for miRNA-1229 Maturation. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 75. | Li F, Tan W, Chen H, Zhou J, Xu M, Yuan G. Up- and downregulation of mature miR-1587 function by modulating its G-quadruplex structure and using small molecules. Int J Biol Macromol. 2019;121:127-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 76. | Chen L, Dickerhoff J, Sakai S, Yang D. DNA G-Quadruplex in Human Telomeres and Oncogene Promoters: Structures, Functions, and Small Molecule Targeting. Acc Chem Res. 2022;55:2628-2646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 93] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 77. | Ma Y, Iida K, Nagasawa K. Topologies of G-quadruplex: Biological functions and regulation by ligands. Biochem Biophys Res Commun. 2020;531:3-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 84] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 78. | Liu Y, Cheng D, Ge M, Lin W. The Truncated Human Telomeric Sequence forms a Hybrid-Type Intramolecular Mixed Parallel/antiparallel G-quadruplex Structure in K(+) Solution. Chem Biol Drug Des. 2016;88:122-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 79. | Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723-2735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 957] [Cited by in RCA: 940] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 80. | Dai J, Carver M, Punchihewa C, Jones RA, Yang D. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927-4940. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 415] [Cited by in RCA: 455] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 81. | Zhang Z, Dai J, Veliath E, Jones RA, Yang D. Structure of a two-G-tetrad intramolecular G-quadruplex formed by a variant human telomeric sequence in K+ solution: insights into the interconversion of human telomeric G-quadruplex structures. Nucleic Acids Res. 2010;38:1009-1021. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 147] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 82. | Noer SL, Preus S, Gudnason D, Aznauryan M, Mergny JL, Birkedal V. Folding dynamics and conformational heterogeneity of human telomeric G-quadruplex structures in Na+ solutions by single molecule FRET microscopy. Nucleic Acids Res. 2016;44:464-471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 67] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 83. | Mazzini S, Gargallo R, Musso L, De Santis F, Aviñó A, Scaglioni L, Eritja R, Di Nicola M, Zunino F, Amatulli A, Dallavalle S. Stabilization of c-KIT G-Quadruplex DNA Structures by the RNA Polymerase I Inhibitors BMH-21 and BA-41. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 84. | Kotar A, Rigo R, Sissi C, Plavec J. Two-quartet kit* G-quadruplex is formed via double-stranded pre-folded structure. Nucleic Acids Res. 2019;47:2641-2653. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 85. | Cayrou C, Ballester B, Peiffer I, Fenouil R, Coulombe P, Andrau JC, van Helden J, Méchali M. The chromatin environment shapes DNA replication origin organization and defines origin classes. Genome Res. 2015;25:1873-1885. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 141] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 86. | Valton AL, Hassan-Zadeh V, Lema I, Boggetto N, Alberti P, Saintomé C, Riou JF, Prioleau MN. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 2014;33:732-746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 166] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 87. | Prorok P, Artufel M, Aze A, Coulombe P, Peiffer I, Lacroix L, Guédin A, Mergny JL, Damaschke J, Schepers A, Cayrou C, Teulade-Fichou MP, Ballester B, Méchali M. Involvement of G-quadruplex regions in mammalian replication origin activity. Nat Commun. 2019;10:3274. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 137] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 88. | Rodriguez R, Miller KM, Forment JV, Bradshaw CR, Nikan M, Britton S, Oelschlaegel T, Xhemalce B, Balasubramanian S, Jackson SP. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol. 2012;8:301-310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 488] [Cited by in RCA: 587] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 89. | Vannier JB, Sandhu S, Petalcorin MI, Wu X, Nabi Z, Ding H, Boulton SJ. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science. 2013;342:239-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 175] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 90. | Drosopoulos WC, Kosiyatrakul ST, Schildkraut CL. BLM helicase facilitates telomere replication during leading strand synthesis of telomeres. J Cell Biol. 2015;210:191-208. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 138] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 91. | Dyer MA, Qadeer ZA, Valle-Garcia D, Bernstein E. ATRX and DAXX: Mechanisms and Mutations. Cold Spring Harb Perspect Med. 2017;7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 157] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 92. | Zhang M, Wang B, Li T, Liu R, Xiao Y, Geng X, Li G, Liu Q, Price CM, Liu Y, Wang F. Mammalian CST averts replication failure by preventing G-quadruplex accumulation. Nucleic Acids Res. 2019;47:5243-5259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 66] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 93. | Bidula S, Brázda V. Genomic Analysis of Non-B Nucleic Acids Structures in SARS-CoV-2: Potential Key Roles for These Structures in Mutability, Translation, and Replication? Genes (Basel). 2023;14. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 94. | Sugimoto N, Maehara K, Yoshida K, Ohkawa Y, Fujita M. Genome-wide analysis of the spatiotemporal regulation of firing and dormant replication origins in human cells. Nucleic Acids Res. 2018;46:6683-6696. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 64] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 95. | Galati E, Bosio MC, Novarina D, Chiara M, Bernini GM, Mozzarelli AM, García-Rubio ML, Gómez-González B, Aguilera A, Carzaniga T, Todisco M, Bellini T, Nava GM, Frigè G, Sertic S, Horner DS, Baryshnikova A, Manzari C, D'Erchia AM, Pesole G, Brown GW, Muzi-Falconi M, Lazzaro F. VID22 counteracts G-quadruplex-induced genome instability. Nucleic Acids Res. 2021;49:12785-12804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 96. | Lee HT, Bose A, Lee CY, Opresko PL, Myong S. Molecular mechanisms by which oxidative DNA damage promotes telomerase activity. Nucleic Acids Res. 2017;45:11752-11765. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 97. | De Magis A, Götz S, Hajikazemi M, Fekete-Szücs E, Caterino M, Juranek S, Paeschke K. Zuo1 supports G4 structure formation and directs repair toward nucleotide excision repair. Nat Commun. 2020;11:3907. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 98. | Pavlova AV, Monakhova MV, Ogloblina AM, Andreeva NA, Laptev GY, Polshakov VI, Gromova ES, Zvereva MI, Yakubovskaya MG, Oretskaya TS, Kubareva EA, Dolinnaya NG. Responses of DNA Mismatch Repair Proteins to a Stable G-Quadruplex Embedded into a DNA Duplex Structure. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 99. | Pavlova AV, Savitskaya VY, Dolinnaya NG, Monakhova MV, Litvinova AV, Kubareva EA, Zvereva MI. G-Quadruplex Formed by the Promoter Region of the hTERT Gene: Structure-Driven Effects on DNA Mismatch Repair Functions. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 100. | Lejault P, Mitteaux J, Sperti FR, Monchaud D. How to untie G-quadruplex knots and why? Cell Chem Biol. 2021;28:436-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 101. | Voter AF, Qiu Y, Tippana R, Myong S, Keck JL. A guanine-flipping and sequestration mechanism for G-quadruplex unwinding by RecQ helicases. Nat Commun. 2018;9:4201. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 102. | Da Ros S, Nicoletto G, Rigo R, Ceschi S, Zorzan E, Dacasto M, Giantin M, Sissi C. G-Quadruplex Modulation of SP1 Functional Binding Sites at the KIT Proximal Promoter. Int J Mol Sci. 2020;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 103. | Cogoi S, Paramasivam M, Membrino A, Yokoyama KK, Xodo LE. The KRAS promoter responds to Myc-associated zinc finger and poly(ADP-ribose) polymerase 1 proteins, which recognize a critical quadruplex-forming GA-element. J Biol Chem. 2010;285:22003-22016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 121] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 104. | Falabella M, Kolesar JE, Wallace C, de Jesus D, Sun L, Taguchi YV, Wang C, Wang T, Xiang IM, Alder JK, Maheshan R, Horne W, Turek-Herman J, Pagano PJ, St Croix CM, Sondheimer N, Yatsunyk LA, Johnson FB, Kaufman BA. G-quadruplex dynamics contribute to regulation of mitochondrial gene expression. Sci Rep. 2019;9:5605. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 105. | Hu XX, Wang SQ, Gan SQ, Liu L, Zhong MQ, Jia MH, Jiang F, Xu Y, Xiao CD, Shen XC. A Small Ligand That Selectively Binds to the G-quadruplex at the Human Vascular Endothelial Growth Factor Internal Ribosomal Entry Site and Represses the Translation. Front Chem. 2021;9:781198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 106. | Chen X, Yuan J, Xue G, Campanario S, Wang D, Wang W, Mou X, Liew SW, Umar MI, Isern J, Zhao Y, He L, Li Y, Mann CJ, Yu X, Wang L, Perdiguero E, Chen W, Xue Y, Nagamine Y, Kwok CK, Sun H, Muñoz-Cánoves P, Wang H. Translational control by DHX36 binding to 5'UTR G-quadruplex is essential for muscle stem-cell regenerative functions. Nat Commun. 2021;12:5043. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 57] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 107. | Angeloni A, Bogdanovic O. Sequence determinants, function, and evolution of CpG islands. Biochem Soc Trans. 2021;49:1109-1119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 60] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 108. | Xiao FH, Wang HT, Kong QP. Dynamic DNA Methylation During Aging: A "Prophet" of Age-Related Outcomes. Front Genet. 2019;10:107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 109. | Zhao L, Wu X, Zheng J, Dong D. DNA methylome profiling of circulating tumor cells in lung cancer at single base-pair resolution. Oncogene. 2021;40:1884-1895. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 110. | Mao SQ, Ghanbarian AT, Spiegel J, Martínez Cuesta S, Beraldi D, Di Antonio M, Marsico G, Hänsel-Hertsch R, Tannahill D, Balasubramanian S. DNA G-quadruplex structures mold the DNA methylome. Nat Struct Mol Biol. 2018;25:951-957. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 183] [Cited by in RCA: 208] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 111. | Matsumoto S, Tateishi-Karimata H, Sugimoto N. DNA methylation is regulated by both the stability and topology of G-quadruplex. Chem Commun (Camb). 2022;58:12459-12462. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 112. | Takahama K, Takada A, Tada S, Shimizu M, Sayama K, Kurokawa R, Oyoshi T. Regulation of telomere length by G-quadruplex telomere DNA- and TERRA-binding protein TLS/FUS. Chem Biol. 2013;20:341-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 147] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 113. | Mei Y, Deng Z, Vladimirova O, Gulve N, Johnson FB, Drosopoulos WC, Schildkraut CL, Lieberman PM. TERRA G-quadruplex RNA interaction with TRF2 GAR domain is required for telomere integrity. Sci Rep. 2021;11:3509. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 73] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 114. | Porro A, Feuerhahn S, Lingner J. TERRA-reinforced association of LSD1 with MRE11 promotes processing of uncapped telomeres. Cell Rep. 2014;6:765-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 108] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 115. | Beltran M, Tavares M, Justin N, Khandelwal G, Ambrose J, Foster BM, Worlock KB, Tvardovskiy A, Kunzelmann S, Herrero J, Bartke T, Gamblin SJ, Wilson JR, Jenner RG. G-tract RNA removes Polycomb repressive complex 2 from genes. Nat Struct Mol Biol. 2019;26:899-909. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 116. | Katapadi VK, Nambiar M, Raghavan SC. Potential G-quadruplex formation at breakpoint regions of chromosomal translocations in cancer may explain their fragility. Genomics. 2012;100:72-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 117. | Kim N. The Interplay between G-quadruplex and Transcription. Curr Med Chem. 2019;26:2898-2917. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 114] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 118. | Georgakopoulos-Soares I, Morganella S, Jain N, Hemberg M, Nik-Zainal S. Noncanonical secondary structures arising from non-B DNA motifs are determinants of mutagenesis. Genome Res. 2018;28:1264-1271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 110] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 119. | Williams JD, Fleetwood S, Berroyer A, Kim N, Larson ED. Sites of instability in the human TCF3 (E2A) gene adopt G-quadruplex DNA structures in vitro. Front Genet. 2015;6:177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 120. | Bidula S. Analysis of putative G-quadruplex forming sequences in inflammatory mediators and their potential as targets for treating inflammatory disorders. Cytokine. 2021;142:155493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 121. | Wang X, Chen S, Zhao Z, Chen F, Huang Y, Guo X, Lei L, Wang W, Luo Y, Yu H, Wang J. Genomic G-quadruplex folding triggers a cytokine-mediated inflammatory feedback loop to aggravate inflammatory diseases. iScience. 2022;25:105312. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 122. | Dalloul Z, Chenuet P, Dalloul I, Boyer F, Aldigier JC, Laffleur B, El Makhour Y, Ryffel B, Quesniaux VFJ, Togbé D, Mergny JL, Cook-Moreau J, Cogné M. G-quadruplex DNA targeting alters class-switch recombination in B cells and attenuates allergic inflammation. J Allergy Clin Immunol. 2018;142:1352-1355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 123. | Zhu BC, He J, Xia XY, Jiang J, Liu W, Liu LY, Liang BB, Yao HG, Ke Z, Xia W, Mao ZW. Solution structure of a thrombin binding aptamer complex with a non-planar platinum(ii) compound. Chem Sci. 2022;13:8371-8379. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 124. | Santos T, Salgado GF, Cabrita EJ, Cruz C. Nucleolin: a binding partner of G-quadruplex structures. Trends Cell Biol. 2022;32:561-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 125. | Liu J, Yan L, He S, Hu J. Engineering DNA quadruplexes in DNA nanostructures for biosensor construction. Nano Res. 2022;15:3504-3513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 126. | Zhang C, Fu S, Zhang F, Han M, Wang X, Du J, Zhang H, Li W. Affibody Modified G-quadruplex DNA Micelles Incorporating Polymeric 5-Fluorodeoxyuridine for Targeted Delivery of Curcumin to Enhance Synergetic Therapy of HER2 Positive Gastric Cancer. Nanomaterials (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 127. | Shammas MA, Koley H, Beer DG, Li C, Goyal RK, Munshi NC. Growth arrest, apoptosis, and telomere shortening of Barrett's-associated adenocarcinoma cells by a telomerase inhibitor. Gastroenterology. 2004;126:1337-1346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 46] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 128. | Jiang J, Li J, Liu C, Liu R, Liang X, Zhou Y, Pan L, Chen H, Ma Z. Study on the substitution effects of zinc benzoate terpyridine complexes on photoluminescence, antiproliferative potential and DNA binding properties. J Biol Inorg Chem. 2020;25:311-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 129. | Hampel SM, Sidibe A, Gunaratnam M, Riou JF, Neidle S. Tetrasubstituted naphthalene diimide ligands with selectivity for telomeric G-quadruplexes and cancer cells. Bioorg Med Chem Lett. 2010;20:6459-6463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 83] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 130. | Micco M, Collie GW, Dale AG, Ohnmacht SA, Pazitna I, Gunaratnam M, Reszka AP, Neidle S. Structure-based design and evaluation of naphthalene diimide G-quadruplex ligands as telomere targeting agents in pancreatic cancer cells. J Med Chem. 2013;56:2959-2974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 149] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 131. | Mpima S, Ohnmacht SA, Barletta M, Husby J, Pett LC, Gunaratnam M, Hilton ST, Neidle S. The influence of positional isomerism on G-quadruplex binding and anti-proliferative activity of tetra-substituted naphthalene diimide compounds. Bioorg Med Chem. 2013;21:6162-6170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 132. | Ohnmacht SA, Marchetti C, Gunaratnam M, Besser RJ, Haider SM, Di Vita G, Lowe HL, Mellinas-Gomez M, Diocou S, Robson M, Šponer J, Islam B, Pedley RB, Hartley JA, Neidle S. A G-quadruplex-binding compound showing anti-tumour activity in an in vivo model for pancreatic cancer. Sci Rep. 2015;5:11385. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 94] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 133. | Marchetti C, Zyner KG, Ohnmacht SA, Robson M, Haider SM, Morton JP, Marsico G, Vo T, Laughlin-Toth S, Ahmed AA, Di Vita G, Pazitna I, Gunaratnam M, Besser RJ, Andrade ACG, Diocou S, Pike JA, Tannahill D, Pedley RB, Evans TRJ, Wilson WD, Balasubramanian S, Neidle S. Targeting Multiple Effector Pathways in Pancreatic Ductal Adenocarcinoma with a G-Quadruplex-Binding Small Molecule. J Med Chem. 2018;61:2500-2517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 126] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 134. | Ahmed AA, Marchetti C, Ohnmacht SA, Neidle S. A G-quadruplex-binding compound shows potent activity in human gemcitabine-resistant pancreatic cancer cells. Sci Rep. 2020;10:12192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 135. | Ahmed AA, Angell R, Oxenford S, Worthington J, Williams N, Barton N, Fowler TG, O'Flynn DE, Sunose M, McConville M, Vo T, Wilson WD, Karim SA, Morton JP, Neidle S. Asymmetrically Substituted Quadruplex-Binding Naphthalene Diimide Showing Potent Activity in Pancreatic Cancer Models. ACS Med Chem Lett. 2020;11:1634-1644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 136. | Gunaratnam M, de la Fuente M, Hampel SM, Todd AK, Reszka AP, Schätzlein A, Neidle S. Targeting pancreatic cancer with a G-quadruplex ligand. Bioorg Med Chem. 2011;19:7151-7157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 54] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 137. | Faudale M, Cogoi S, Xodo LE. Photoactivated cationic alkyl-substituted porphyrin binding to g4-RNA in the 5'-UTR of KRAS oncogene represses translation. Chem Commun (Camb). 2012;48:874-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 55] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 138. | Ferino A, Nicoletto G, D'Este F, Zorzet S, Lago S, Richter SN, Tikhomirov A, Shchekotikhin A, Xodo LE. Photodynamic Therapy for ras-Driven Cancers: Targeting G-Quadruplex RNA Structures with Bifunctional Alkyl-Modified Porphyrins. J Med Chem. 2020;63:1245-1260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 139. | Pattanayak R, Barua A, Das A, Chatterjee T, Pathak A, Choudhury P, Sen S, Saha P, Bhattacharyya M. Porphyrins to restrict progression of pancreatic cancer by stabilizing KRAS G-quadruplex: In silico, in vitro and in vivo validation of anticancer strategy. Eur J Pharm Sci. 2018;125:39-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 140. | Chilakamarthi U, Koteshwar D, Jinka S, Vamsi Krishna N, Sridharan K, Nagesh N, Giribabu L. Novel Amphiphilic G-Quadruplex Binding Synthetic Derivative of TMPyP4 and Its Effect on Cancer Cell Proliferation and Apoptosis Induction. Biochemistry. 2018;57:6514-6527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 141. | Liu W, Sun D, Hurley LH. Binding of G-quadruplex-interactive agents to distinct G-quadruplexes induces different biological effects in MiaPaCa cells. Nucleosides Nucleotides Nucleic Acids. 2005;24:1801-1815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 142. | Kaiser CE, Van Ert NA, Agrawal P, Chawla R, Yang D, Hurley LH. Insight into the Complexity of the i-Motif and G-Quadruplex DNA Structures Formed in the KRAS Promoter and Subsequent Drug-Induced Gene Repression. J Am Chem Soc. 2017;139:8522-8536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 147] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 143. | Miglietta G, Cogoi S, Marinello J, Capranico G, Tikhomirov AS, Shchekotikhin A, Xodo LE. RNA G-Quadruplexes in Kirsten Ras (KRAS) Oncogene as Targets for Small Molecules Inhibiting Translation. J Med Chem. 2017;60:9448-9461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 72] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 144. | Paluszkiewicz E, Horowska B, Borowa-Mazgaj B, Peszyńska-Sularz G, Paradziej-Łukowicz J, Augustin E, Konopa J, Mazerska Z. Design, synthesis and high antitumor potential of new unsymmetrical bisacridine derivatives towards human solid tumors, specifically pancreatic cancers and their unique ability to stabilize DNA G-quadruplexes. Eur J Med Chem. 2020;204:112599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 145. | Arjmand F, Sharma S, Parveen S, Toupet L, Yu Z, Cowan JA. Copper(ii) l/d-valine-(1,10-phen) complexes target human telomeric G-quadruplex motifs and promote site-specific DNA cleavage and cellular cytotoxicity. Dalton Trans. 2020;49:9888-9899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 146. | Bossaert M, Pipier A, Riou JF, Noirot C, Nguyên LT, Serre RF, Bouchez O, Defrancq E, Calsou P, Britton S, Gomez D. Transcription-associated topoisomerase 2α (TOP2A) activity is a major effector of cytotoxicity induced by G-quadruplex ligands. Elife. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 147. | Xu H, Di Antonio M, McKinney S, Mathew V, Ho B, O'Neil NJ, Santos ND, Silvester J, Wei V, Garcia J, Kabeer F, Lai D, Soriano P, Banáth J, Chiu DS, Yap D, Le DD, Ye FB, Zhang A, Thu K, Soong J, Lin SC, Tsai AH, Osako T, Algara T, Saunders DN, Wong J, Xian J, Bally MB, Brenton JD, Brown GW, Shah SP, Cescon D, Mak TW, Caldas C, Stirling PC, Hieter P, Balasubramanian S, Aparicio S. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat Commun. 2017;8:14432. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 360] [Cited by in RCA: 424] [Article Influence: 47.1] [Reference Citation Analysis (0)] |
| 148. | Drygin D, Siddiqui-Jain A, O'Brien S, Schwaebe M, Lin A, Bliesath J, Ho CB, Proffitt C, Trent K, Whitten JP, Lim JK, Von Hoff D, Anderes K, Rice WG. Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res. 2009;69:7653-7661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 455] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 149. | Xu H, Hurley LH. A first-in-class clinical G-quadruplex-targeting drug. The bench-to-bedside translation of the fluoroquinolone QQ58 to CX-5461 (Pidnarulex). Bioorg Med Chem Lett. 2022;77:129016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 57] [Reference Citation Analysis (0)] |
| 150. | Schultz CW, McCarthy GA, Nerwal T, Nevler A, DuHadaway JB, McCoy MD, Jiang W, Brown SZ, Goetz A, Jain A, Calvert VS, Vishwakarma V, Wang D, Preet R, Cassel J, Summer R, Shaghaghi H, Pommier Y, Baechler SA, Pishvaian MJ, Golan T, Yeo CJ, Petricoin EF, Prendergast GC, Salvino J, Singh PK, Dixon DA, Brody JR. The FDA-Approved Anthelmintic Pyrvinium Pamoate Inhibits Pancreatic Cancer Cells in Nutrient-Depleted Conditions by Targeting the Mitochondria. Mol Cancer Ther. 2021;20:2166-2176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 151. | Psaras AM, Carty RK, Miller JT, Tumey LN, Brooks TA. Indoloquinoline-Mediated Targeted Downregulation of KRAS through Selective Stabilization of the Mid-Promoter G-Quadruplex Structure. Genes (Basel). 2022;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 152. | Bhuma N, Chand K, Andréasson M, Mason J, Das RN, Patel AK, Öhlund D, Chorell E. The effect of side chain variations on quinazoline-pyrimidine G-quadruplex DNA ligands. Eur J Med Chem. 2023;248:115103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 153. | Chu PC, Yang MC, Kulp SK, Salunke SB, Himmel LE, Fang CS, Jadhav AM, Shan YS, Lee CT, Lai MD, Shirley LA, Bekaii-Saab T, Chen CS. Regulation of oncogenic KRAS signaling via a novel KRAS-integrin-linked kinase-hnRNPA1 regulatory loop in human pancreatic cancer cells. Oncogene. 2016;35:3897-3908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (1)] |
| 154. | Cogoi S, Rapozzi V, Cauci S, Xodo LE. Critical role of hnRNP A1 in activating KRAS transcription in pancreatic cancer cells: A molecular mechanism involving G4 DNA. Biochim Biophys Acta Gen Subj. 2017;1861:1389-1398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 155. | Chu PC, Kulp SK, Bekaii-Saab T, Chen CS. Targeting integrin-linked kinase to suppress oncogenic KRAS signaling in pancreatic cancer. Small GTPases. 2018;9:452-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 156. | Cinque G, Ferino A, Pedersen EB, Xodo LE. Role of Poly [ADP-ribose] Polymerase 1 in Activating the Kirsten ras (KRAS) Gene in Response to Oxidative Stress. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 157. | Pramanik S, Chen Y, Song H, Khutsishvili I, Marky LA, Ray S, Natarajan A, Singh PK, Bhakat KK. The human AP-endonuclease 1 (APE1) is a DNA G-quadruplex structure binding protein and regulates KRAS expression in pancreatic ductal adenocarcinoma cells. Nucleic Acids Res. 2022;50:3394-3412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 158. | Psaras AM, Valiuska S, Noé V, Ciudad CJ, Brooks TA. Targeting KRAS Regulation with PolyPurine Reverse Hoogsteen Oligonucleotides. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 159. | Valiuska S, Psaras AM, Noé V, Brooks TA, Ciudad CJ. Targeting MYC Regulation with Polypurine Reverse Hoogsteen Oligonucleotides. Int J Mol Sci. 2022;24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 160. | Wang P, Leung CH, Ma DL, Yan SC, Che CM. Structure-based design of platinum(II) complexes as c-myc oncogene down-regulators and luminescent probes for G-quadruplex DNA. Chemistry. 2010;16:6900-6911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 100] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 161. | Chauhan A, Paladhi S, Debnath M, Dash J. Selective recognition of c-MYC G-quadruplex DNA using prolinamide derivatives. Org Biomol Chem. 2016;14:5761-5767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 162. | Chen J, Duan Y, Yu X, Zhong J, Bai J, Li NG, Zhu Z, Xu J. Development of novel 9-O-substituted-13-octylberberine derivatives as potential anti-hepatocellular carcinoma agents. J Enzyme Inhib Med Chem. 2022;37:2423-2433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 163. | Chen J, Duan Y, Yang K, Wang J, Yan J, Gu C, Wang S, Zhu Z, Liu EH, Xu J. Design, synthesis and biological evaluation of novel 9-N-substituted-13-alkylberberine derivatives from Chinese medicine as anti-hepatocellular carcinoma agents. Bioorg Med Chem. 2023;79:117156. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 164. | Jin J, Hou J, Long W, Zhang X, Lu YJ, Li D, Zhang K, Wong WL. Synthesis of fluorescent G-quadruplex DNA binding ligands for the comparison of terminal group effects in molecular interaction: Phenol versus methoxybenzene. Bioorg Chem. 2020;99:103821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 165. | Chauhan A, Paladhi S, Debnath M, Mandal S, Das RN, Bhowmik S, Dash J. A small molecule peptidomimetic that binds to c-KIT1 G-quadruplex and exhibits antiproliferative properties in cancer cells. Bioorg Med Chem. 2014;22:4422-4429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 166. | Wang Y, Ding Q, Xu T, Li CY, Zhou DD, Zhang L. HZ-6d targeted HERC5 to regulate p53 ISGylation in human hepatocellular carcinoma. Toxicol Appl Pharmacol. 2017;334:180-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 167. | Sun J, Wu G, Pastor F, Rahman N, Wang WH, Zhang Z, Merle P, Hui L, Salvetti A, Durantel D, Yang D, Andrisani O. RNA helicase DDX5 enables STAT1 mRNA translation and interferon signalling in hepatitis B virus replicating hepatocytes. Gut. 2022;71:991-1005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 168. | Bian WX, Xie Y, Wang XN, Xu GH, Fu BS, Li S, Long G, Zhou X, Zhang XL. Binding of cellular nucleolin with the viral core RNA G-quadruplex structure suppresses HCV replication. Nucleic Acids Res. 2019;47:56-68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 169. | McLuckie KI, Waller ZA, Sanders DA, Alves D, Rodriguez R, Dash J, McKenzie GJ, Venkitaraman AR, Balasubramanian S. G-quadruplex-binding benzo[a]phenoxazines down-regulate c-KIT expression in human gastric carcinoma cells. J Am Chem Soc. 2011;133:2658-2663. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 140] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 170. | Ma X, Awadasseid A, Zhou K, Wang X, Shen C, Zhao X, Cheng M, Zhang W. A 1,10-phenanthroline derivative selectively targeting telomeric G-quadruplex induces cytoprotective autophagy, causing apoptosis of gastric cancer cells. Life Sci. 2021;287:120095. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 171. | Wang X, Zhou CX, Yan JW, Hou JQ, Chen SB, Ou TM, Gu LQ, Huang ZS, Tan JH. Synthesis and Evaluation of Quinazolone Derivatives as a New Class of c-KIT G-Quadruplex Binding Ligands. ACS Med Chem Lett. 2013;4:909-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 172. | Zhang YQ, Pei JH, Shi SS, Zheng J, Wang JM, Guo XS, Cui GY, Wang XY, Zhang HP, Hu WQ. G-quadruplex antibody attenuates human gastric cancer cell proliferation and promotes apoptosis through hTERT/telomerase pathway. Eur Rev Med Pharmacol Sci. 2018;22:2614-2623. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 173. | Incles CM, Schultes CM, Kelland LR, Neidle S. Acquired cellular resistance to flavopiridol in a human colon carcinoma cell line involves up-regulation of the telomerase catalytic subunit and telomere elongation. Sensitivity of resistant cells to combination treatment with a telomerase inhibitor. Mol Pharmacol. 2003;64:1101-1108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 174. | Salvati E, Leonetti C, Rizzo A, Scarsella M, Mottolese M, Galati R, Sperduti I, Stevens MF, D'Incalci M, Blasco M, Chiorino G, Bauwens S, Horard B, Gilson E, Stoppacciaro A, Zupi G, Biroccio A. Telomere damage induced by the G-quadruplex ligand RHPS4 has an antitumor effect. J Clin Invest. 2007;117:3236-3247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 199] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 175. | Rizzo A, Salvati E, Porru M, D'Angelo C, Stevens MF, D'Incalci M, Leonetti C, Gilson E, Zupi G, Biroccio A. Stabilization of quadruplex DNA perturbs telomere replication leading to the activation of an ATR-dependent ATM signaling pathway. Nucleic Acids Res. 2009;37:5353-5364. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 135] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 176. | Biroccio A, Porru M, Rizzo A, Salvati E, D'Angelo C, Orlandi A, Passeri D, Franceschin M, Stevens MF, Gilson E, Beretta G, Zupi G, Pisano C, Zunino F, Leonetti C. DNA damage persistence as determinant of tumor sensitivity to the combination of Topo I inhibitors and telomere-targeting agents. Clin Cancer Res. 2011;17:2227-2236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 177. | Rizzo A, Iachettini S, Zizza P, Cingolani C, Porru M, Artuso S, Stevens M, Hummersone M, Biroccio A, Salvati E, Leonetti C. Identification of novel RHPS4-derivative ligands with improved toxicological profiles and telomere-targeting activities. J Exp Clin Cancer Res. 2014;33:81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 178. | Folini M, Pivetta C, Zagotto G, De Marco C, Palumbo M, Zaffaroni N, Sissi C. Remarkable interference with telomeric function by a G-quadruplex selective bisantrene regioisomer. Biochem Pharmacol. 2010;79:1781-1790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 179. | Franceschin M, Rizzo A, Casagrande V, Salvati E, Alvino A, Altieri A, Ciammaichella A, Iachettini S, Leonetti C, Ortaggi G, Porru M, Bianco A, Biroccio A. Aromatic core extension in the series of N-cyclic bay-substituted perylene G-quadruplex ligands: increased telomere damage, antitumor activity, and strong selectivity for neoplastic over healthy cells. ChemMedChem. 2012;7:2144-2154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 180. | Porru M, Artuso S, Salvati E, Bianco A, Franceschin M, Diodoro MG, Passeri D, Orlandi A, Savorani F, D'Incalci M, Biroccio A, Leonetti C. Targeting G-Quadruplex DNA Structures by EMICORON Has a Strong Antitumor Efficacy against Advanced Models of Human Colon Cancer. Mol Cancer Ther. 2015;14:2541-2551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 181. | Rocca R, Scionti F, Nadai M, Moraca F, Maruca A, Costa G, Catalano R, Juli G, Di Martino MT, Ortuso F, Alcaro S, Tagliaferri P, Tassone P, Richter SN, Artese A. Chromene Derivatives as Selective TERRA G-Quadruplex RNA Binders with Antiproliferative Properties. Pharmaceuticals (Basel). 2022;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 182. | Thakur RK, Kumar P, Halder K, Verma A, Kar A, Parent JL, Basundra R, Kumar A, Chowdhury S. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res. 2009;37:172-183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 157] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 183. | Yao YX, Xu BH, Zhang Y. CX-3543 Promotes Cell Apoptosis through Downregulation of CCAT1 in Colon Cancer Cells. Biomed Res Int. 2018;2018:9701957. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 184. | Chung SY, Chang YC, Hsu DS, Hung YC, Lu ML, Hung YP, Chiang NJ, Yeh CN, Hsiao M, Soong J, Su Y, Chen MH. A G-quadruplex stabilizer, CX-5461 combined with two immune checkpoint inhibitors enhances in vivo therapeutic efficacy by increasing PD-L1 expression in colorectal cancer. Neoplasia. 2023;35:100856. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 185. | Malhotra R, Rarhi C, Diveshkumar KV, Barik R, D'cunha R, Dhar P, Kundu M, Chattopadhyay S, Roy S, Basu S, Pradeepkumar PI, Hajra S. Dihydrochelerythrine and its derivatives: Synthesis and their application as potential G-quadruplex DNA stabilizing agents. Bioorg Med Chem. 2016;24:2887-2896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 186. | Pawłowska M, Kulesza J, Augustin E. c-Myc Protein Level Affected by Unsymmetrical Bisacridines Influences Apoptosis and Senescence Induced in HCT116 Colorectal and H460 Lung Cancer Cells. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 187. | Brito H, Martins AC, Lavrado J, Mendes E, Francisco AP, Santos SA, Ohnmacht SA, Kim NS, Rodrigues CM, Moreira R, Neidle S, Borralho PM, Paulo A. Targeting KRAS Oncogene in Colon Cancer Cells with 7-Carboxylate Indolo[3,2-b]quinoline Tri-Alkylamine Derivatives. PLoS One. 2015;10:e0126891. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 188. | D'Aria F, D'Amore VM, Di Leva FS, Amato J, Caterino M, Russomanno P, Salerno S, Barresi E, De Leo M, Marini AM, Taliani S, Da Settimo F, Salgado GF, Pompili L, Zizza P, Shirasawa S, Novellino E, Biroccio A, Marinelli L, Giancola C. Targeting the KRAS oncogene: Synthesis, physicochemical and biological evaluation of novel G-Quadruplex DNA binders. Eur J Pharm Sci. 2020;149:105337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 189. | Sanchez-Martin V, Schneider DA, Ortiz-Gonzalez M, Soriano-Lerma A, Linde-Rodriguez A, Perez-Carrasco V, Gutierrez-Fernandez J, Cuadros M, González C, Soriano M, Garcia-Salcedo JA. Targeting ribosomal G-quadruplexes with naphthalene-diimides as RNA polymerase I inhibitors for colorectal cancer treatment. Cell Chem Biol. 2021;28:1590-1601.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 190. | Belmonte-Reche E, Benassi A, Peñalver P, Cucchiarini A, Guédin A, Mergny JL, Rosu F, Gabelica V, Freccero M, Doria F, Morales JC. Thiosugar naphthalene diimide conjugates: G-quadruplex ligands with antiparasitic and anticancer activity. Eur J Med Chem. 2022;232:114183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 191. | Sanchez-Martin V, Plaza-Calonge MDC, Soriano-Lerma A, Ortiz-Gonzalez M, Linde-Rodriguez A, Perez-Carrasco V, Ramirez-Macias I, Cuadros M, Gutierrez-Fernandez J, Murciano-Calles J, Rodríguez-Manzaneque JC, Soriano M, Garcia-Salcedo JA. Gallic Acid: A Natural Phenolic Compound Exerting Antitumoral Activities in Colorectal Cancer via Interaction with G-Quadruplexes. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 192. | Nishikawa T, Kuwano Y, Takahara Y, Nishida K, Rokutan K. HnRNPA1 interacts with G-quadruplex in the TRA2B promoter and stimulates its transcription in human colon cancer cells. Sci Rep. 2019;9:10276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 193. | Bolduc F, Turcotte MA, Perreault JP. The Small Nuclear Ribonucleoprotein Polypeptide A (SNRPA) binds to the G-quadruplex of the BAG-1 5'UTR. Biochimie. 2020;176:122-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 194. | Jodoin R, Carrier JC, Rivard N, Bisaillon M, Perreault JP. G-quadruplex located in the 5'UTR of the BAG-1 mRNA affects both its cap-dependent and cap-independent translation through global secondary structure maintenance. Nucleic Acids Res. 2019;47:10247-10266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 195. | Nishikawa T, Kuwano Y, Nakata M, Rokutan K, Nishida K. Multiple G-quadruplexes in the LMNA promoter regulate LMNA variant 6 transcription and promote colon cancer cell growth. Biochim Biophys Acta Gene Regul Mech. 2021;1864:194746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 196. | Matsumura K, Kawasaki Y, Miyamoto M, Kamoshida Y, Nakamura J, Negishi L, Suda S, Akiyama T. The novel G-quadruplex-containing long non-coding RNA GSEC antagonizes DHX36 and modulates colon cancer cell migration. Oncogene. 2017;36:1191-1199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 88] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 197. | Bejugam M, Gunaratnam M, Müller S, Sanders DA, Sewitz S, Fletcher JA, Neidle S, Balasubramanian S. Targeting the c-Kit Promoter G-quadruplexes with 6-Substituted Indenoisoquinolines. ACS Med Chem Lett. 2010;1:306-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 56] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 198. | Gunaratnam M, Collie GW, Reszka AP, Todd AK, Parkinson GN, Neidle S. A naphthalene diimide G-quadruplex ligand inhibits cell growth and down-regulates BCL-2 expression in an imatinib-resistant gastrointestinal cancer cell line. Bioorg Med Chem. 2018;26:2958-2964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 199. | Cogoi S, Quadrifoglio F, Xodo LE. G-rich oligonucleotide inhibits the binding of a nuclear protein to the Ki-ras promoter and strongly reduces cell growth in human carcinoma pancreatic cells. Biochemistry. 2004;43:2512-2523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 200. | Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp Mol Pathol. 2009;86:151-164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 703] [Cited by in RCA: 647] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 201. | Pecoraro A, Virgilio A, Esposito V, Galeone A, Russo G, Russo A. uL3 Mediated Nucleolar Stress Pathway as a New Mechanism of Action of Antiproliferative G-quadruplex TBA Derivatives in Colon Cancer Cells. Biomolecules. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 202. | Virgilio A, Benigno D, Pecoraro A, Russo A, Russo G, Esposito V, Galeone A. Exploring New Potential Anticancer Activities of the G-Quadruplexes Formed by [(GTG(2)T(G(3)T)(3)] and Its Derivatives with an Abasic Site Replacing Single Thymidine. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 203. | Virgilio A, Pecoraro A, Benigno D, Russo A, Russo G, Esposito V, Galeone A. Antiproliferative Effects of the Aptamer d(GGGT)(4) and Its Analogues with an Abasic-Site Mimic Loop on Different Cancer Cells. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 204. | Lopes-Nunes J, Oliveira PA, Cruz C. G-Quadruplex-Based Drug Delivery Systems for Cancer Therapy. Pharmaceuticals (Basel). 2021;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 205. | Bates PJ, Reyes-Reyes EM, Malik MT, Murphy EM, O'Toole MG, Trent JO. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: Uses and mechanisms. Biochim Biophys Acta Gen Subj. 2017;1861:1414-1428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 239] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 206. | Park JY, Cho YL, Chae JR, Moon SH, Cho WG, Choi YJ, Lee SJ, Kang WJ. Gemcitabine-Incorporated G-Quadruplex Aptamer for Targeted Drug Delivery into Pancreas Cancer. Mol Ther Nucleic Acids. 2018;12:543-553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 70] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 207. | Shi J, Nie W, Zhao X, Yang X, Cheng H, Zhou T, Zhang Y, Zhang K, Liu J. An Intracellular Self-Assembly-Driven Uninterrupted ROS Generator Augments 5-Aminolevulinic-Acid-Based Tumor Therapy. Adv Mater. 2022;34:e2201049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 208. | Clua A, Fàbrega C, García-Chica J, Grijalvo S, Eritja R. Parallel G-quadruplex Structures Increase Cellular Uptake and Cytotoxicity of 5-Fluoro-2'-deoxyuridine Oligomers in 5-Fluorouracil Resistant Cells. Molecules. 2021;26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 209. | Awadasseid A, Ma X, Wu Y, Zhang W. G-quadruplex stabilization via small-molecules as a potential anti-cancer strategy. Biomed Pharmacother. 2021;139:111550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 76] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 210. | Robinson J, Raguseo F, Nuccio SP, Liano D, Di Antonio M. DNA G-quadruplex structures: more than simple roadblocks to transcription? Nucleic Acids Res. 2021;49:8419-8431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 209] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 211. | Rigo R, Palumbo M, Sissi C. G-quadruplexes in human promoters: A challenge for therapeutic applications. Biochim Biophys Acta Gen Subj. 2017;1861:1399-1413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 96] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 212. | Kharel P, Becker G, Tsvetkov V, Ivanov P. Properties and biological impact of RNA G-quadruplexes: from order to turmoil and back. Nucleic Acids Res. 2020;48:12534-12555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 119] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 213. | Shen J, Varshney D, Simeone A, Zhang X, Adhikari S, Tannahill D, Balasubramanian S. Promoter G-quadruplex folding precedes transcription and is controlled by chromatin. Genome Biol. 2021;22:143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 214. | Stevens AJ, de Jong L, Kennedy MA. The Dynamic Regulation of G-Quadruplex DNA Structures by Cytosine Methylation. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 215. | Hahm JY, Park J, Jang ES, Chi SW. 8-Oxoguanine: from oxidative damage to epigenetic and epitranscriptional modification. Exp Mol Med. 2022;54:1626-1642. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 179] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 216. | Varshney D, Spiegel J, Zyner K, Tannahill D, Balasubramanian S. The regulation and functions of DNA and RNA G-quadruplexes. Nat Rev Mol Cell Biol. 2020;21:459-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 698] [Cited by in RCA: 884] [Article Influence: 147.3] [Reference Citation Analysis (0)] |
| 217. | Hensen F, Potter A, van Esveld SL, Tarrés-Solé A, Chakraborty A, Solà M, Spelbrink JN. Mitochondrial RNA granules are critically dependent on mtDNA replication factors Twinkle and mtSSB. Nucleic Acids Res. 2019;47:3680-3698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 218. | David AP, Pipier A, Pascutti F, Binolfi A, Weiner AMJ, Challier E, Heckel S, Calsou P, Gomez D, Calcaterra NB, Armas P. CNBP controls transcription by unfolding DNA G-quadruplex structures. Nucleic Acids Res. 2019;47:7901-7913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 219. | Niu K, Zhang X, Song Q, Feng Q. G-Quadruplex Regulation of VEGFA mRNA Translation by RBM4. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (1)] |
| 220. | Shu H, Zhang R, Xiao K, Yang J, Sun X. G-Quadruplex-Binding Proteins: Promising Targets for Drug Design. Biomolecules. 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 53] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 221. | Fleming AM, Burrows CJ. Interplay of Guanine Oxidation and G-Quadruplex Folding in Gene Promoters. J Am Chem Soc. 2020;142:1115-1136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 117] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 222. | Ruggiero E, Richter SN. Viral G-quadruplexes: New frontiers in virus pathogenesis and antiviral therapy. Annu Rep Med Chem. 2020;54:101-131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 51] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 223. | Zheng BX, Yu J, Long W, Chan KH, Leung AS, Wong WL. Structurally diverse G-quadruplexes as the noncanonical nucleic acid drug target for live cell imaging and antibacterial study. Chem Commun (Camb). 2023;59:1415-1433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Jiang L, China; Uhlmann D, Germany; Wong WL, China S-Editor: Wang JJ L-Editor: A P-Editor: Zhang XD