Published online Jun 15, 2023. doi: 10.4251/wjgo.v15.i6.988
Peer-review started: January 17, 2023
First decision: February 28, 2023
Revised: March 18, 2023
Accepted: April 12, 2023
Article in press: April 12, 2023
Published online: June 15, 2023
Processing time: 148 Days and 18.2 Hours
Glucocorticoid modulatory element-binding protein 1 (GMEB1), which has been identified as a transcription factor, is a protein widely expressed in various tissues. Reportedly, the dysregulation of GMEB1 is linked to the genesis and development of multiple cancers.
To explore GMEB1’s biological functions in hepatocellular carcinoma (HCC) and figuring out the molecular mechanism.
GMEB1 expression in HCC tissues was analyzed employing the StarBase database. Immunohistochemical staining, Western blotting and quantitative real-time PCR were conducted to examine GMEB1 and Yes-associate protein 1 (YAP1) expression in HCC cells and tissues. Cell counting kit-8 assay, Transwell assay and flow cytometry were utilized to examine HCC cell proliferation, migration, invasion and apoptosis, respectively. The JASPAR database was employed for predicting the binding site of GMEB1 with YAP1 promoter. Dual-luciferase reporter gene assay and chromatin immunoprecipitation-qPCR were conducted to verify the binding relationship of GMEB1 with YAP1 promoter region.
GMEB1 was up-regulated in HCC cells and tissues, and GMEB1 expression was correlated to the tumor size and TNM stage of HCC patients. GMEB1 overexpression facilitated HCC cell multiplication, migration, and invasion, and suppressed the apoptosis, whereas GMEB1 knockdown had the opposite effects. GMEB1 bound to YAP1 promoter region and positively regulated YAP1 expression in HCC cells.
GMEB1 facilitates HCC malignant proliferation and metastasis by promoting the transcription of the YAP1 promoter region.
Core Tip: Glucocorticoid modulatory element-binding protein 1 (GMEB1) was highly expressed in hepatocellular carcinoma (HCC) tissues. Functionally, GMEB1 modulates the malignant biological behaviors of HCC cells. Mechanistically, GMEB1 promotes the expression of Yes-associate protein 1 at transcriptional level. In short, the present study suggested that for HCC, GMEB1 might be a diagnostic biomarker and treatment target.
- Citation: Chen C, Lin HG, Yao Z, Jiang YL, Yu HJ, Fang J, Li WN. Transcription factor glucocorticoid modulatory element-binding protein 1 promotes hepatocellular carcinoma progression by activating Yes-associate protein 1. World J Gastrointest Oncol 2023; 15(6): 988-1004
- URL: https://www.wjgnet.com/1948-5204/full/v15/i6/988.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i6.988
Liver cancer is the third leading cause of cancer-related deaths worldwide, with more than 780000 deaths due to liver cancer in 2018. Approximately 90% of liver cancer cases originate in hepatocytes and are referred to as hepatocellular carcinoma (HCC)[1]. HCC is the fifth most common cancer in males and the ninth most common cancer in females, with an estimated 500000 and 200000 new cases annually in the world, respectively. The pathogenesis of hepatocellular carcinoma is complicated by structural mutations in the proto-oncogene and the addition of exogenous pathogenic factors such as viruses, excessive alcohol consumption, obesity and aflatoxins, which have contributed to the development of HCC[2,3]. Therefore, HCC is one of the most common types of clinical malignancies in the digestive system, and it also ranks fourth among the causes of cancer death globally[4]. Currently, the predominant treatment options for HCC include surgical resection, liver transplantation, radiotherapy and chemotherapy[5]. Nevertheless, most HCC cases have already been in an intermediate to advanced stage at diagnosis, missing the optimal time for surgical treatment[6]. Besides, the high cost and huge shortage of donors greatly limit the clinical application of liver transplantation[7,8]. In this context, it is necessary to identify more reliable early diagnostic markers for HCC diagnosis and to discover more effective new therapeutic targets for clinical intervention, thus providing new insights to improve the prognosis and overall survival rate of HCC patients.
Glucocorticoid modulatory element-binding protein 1 (GMEB1) is a nucleoprotein with a molecular weight of 88 kDa, and can interact with GMEB2 and bind with the glucocorticoid regulatory element (GME) of the tyrosine aminotransferase (TAT) gene promoter sequence, thereby regulating the glucocorticoid receptor transactivation[9]. It has been demonstrated that IL-2 can inhibit glucocorticoid-induced T-cell apoptosis by boosting GMEB1 expression and activating the PI3K/AKT pathway[10], which is a preliminary indication of the anti-apoptotic function of GMEB1. Meanwhile, FOXL2 is an important transcription factor involved in the transcriptional regulation of several target genes, and GMEB1 was found to bind to FOXL2, whose interaction with FOXL2 could regulate the apoptotic process of cells[11]. Additionally, GMEB1 can also bind to pro-caspases and repress its activation and apoptosis[12,13]. Given that tumor development cannot occur without uncontrolled cell proliferation and apoptotic escape, the anti-apoptotic effect of GMEB1 has prompted researchers to explore the relevance of GMEB1 dysregulation to tumorigenesis and development. For example, GMEB1 suppresses CASP8 activation via regulating CFLARL ubiquitination and degradation to repress the cellular apoptosis and thereby promoting malignant progression of non-small cell lung cancer[9]. Additional bioinformatics studies have further shown that high expression of several transcription factors, including GMEB1, is a promising biomarker and/or therapeutic target for prostate cancer[14]. Thus, it becomes clear that GMEB1 can contribute to malignant progression in various tumor types through different mechanisms. Nonetheless, the expression of GMEB1 in HCC and the molecular mechanism to promote the malignant evolution of HCC have not been elucidated.
Yes-associate protein 1 (YAP1) is one of the prime effector proteins downstream of the Hippo pathway, which controls organ size, normal tissue homeostasis and stem cell function through modulating cell proliferation and apoptosis[15]. YAP1 has been proved to play an important role as an oncoprotein in a variety of tumors. For example, in clinical specimens, YAP has been reported to be overexpressed and overactivated after nucleation in prostate, colon, breast and non-small cell lung cancers, as well as ovarian and hepatocellular carcinomas[16-20], significantly contributing to the development and progression of these tumors. More importantly, in studies surrounding the pathogenesis of HCC, there is a growing consensus that abnormalities in the Hippo signaling pathway are closely associated with the development of HCC. The abnormal expression and dysfunction of YAP1, a key protein in the Hippo signaling pathway, is directly related to the malignant progression of HCC[21]. A clinical study including 177 HCC patients further indicated that YAP1 was an independent prognostic marker significantly associated with shorter disease-free and overall survival in hepatocellular carcinoma[22]. Currently, the mechanisms underlying the promotion of tumor progression by YAP1 revolve around the binding of YAP1 to the transcription factor TEAD[23], which in turn initiates the transcription of many genes involved in cell proliferation and survival, including downstream target genes such as BIRC5, CTGF and Cyclin D1[24]. However, as research has continued, the upstream proteins that regulate YAP1 have been further identified. In addition to the classical upstream LATS1/2 that regulates YAP1 expression[25], it has been reported that, for example, the transcription factor FOXM1 promotes the proliferation, migration and invasion of breast cancer cells through promoting YAP1 transcription activation[26]. Furthermore, in HCC, SIX4 can promote cell metastasis by upregulating YAP1 and c-MET[27].
Could GMEB1, a transcription factor closely associated with tumor development, also promote the malignant progression of HCC by regulating the transcription of YAP1 and thus affecting the normal function of the Hippo signaling pathway? With this question in mind, we would like to confirm the potential regulatory relationship between GMEB1 and YAP1 in this study, thus providing a new intervention target for the Hippo signaling pathway, a key signaling pathway in HCC pathogenesis. To this end, we will firstly focus on examining the expression of GMEB1 in human HCC tissues and HCC cell lines, and clarifying the regulatory mechanism of the interaction between GMEB1 and YAP1 in promoting HCC progression in this study. We hope that our study will reveal the crucial role of GMEB1 in the development of HCC, explore the additional biological functions of GMEB1, and provide new therapeutic strategies for the clinical treatment of patients with HCC accompanied with YAP1 overexpression.
With the approval of Zhejiang Xiaoshan Hopital’s Ethics Committee, cancerous tissues and their corresponding para-cancerous tissues from 55 patients with HCC previously admitted to our hospital were selected for this study. Immediately after surgical removal, the tissues were frozen at -196 °C and kept at -80 °C until use. The clinical data and clinicopathological data of all subjects were complete, and none of them underwent preoperative chemotherapy or radiotherapy.
We fixed the cancerous and normal para-cancerous tissues in 10% formaldehyde and embedded them in paraffin. Afterwards, the tissues were cut into 4-μm-thick slices, deparaffinized with xylene, and washed with PBS. The antigen retrieval was induced by heat in PBS (microwave heating, 96 °C, 15 min), and then the tissues were treated with 3% H2O2 for removing endogenous peroxidases. The slices were incubated with citrate buffer to repair the antigen, and blocked with 5% bovine serum albumin (Sangon Biotech Co., Ltd., Shanghai, China) at 4 °C for 30 min. The slices were incubated at 4 °C overnight with the primary antibody GMEB1 (ab240646, 1:100, Abcam, Cambridge, United Kingdom) and then with horseradish peroxidase-conjugated secondary antibody (1:5000, ab6721, Abcam) for 30 min at room temperature. The sections were stained with diaminobenzidine (Sigma-Aldrich, St. Louis, MO, United States) at room temperature for 10 min. Ultimately, the slices were counter-stained with hematoxylin and sealed and fixed with PerMount (BIOS, Beijing, China). Eventually, the slices were observed and photographed under an optical microscope (Olympus Optical Company, Tokyo, Japan).
As previously mentioned, the staining signals were scored by the percentage of positive tumor cells and staining intensity[28]. The grading of the positive tumor cell ratio is as follows: 0% (0 point), 0.01%-25% (1 point), 25.01%-50% (2 points), 50.01%-75% (3 points) and ≥ 75% (4 points). The degree of staining intensity: no staining (0 point), weak staining (1 point), medium staining (2 points) and strong staining (3 points). The following formula was adopted to calculate the immunohistochemistry (IHC) score of each section: IHC score = staining intensity × percentage of positively stained tumor cells. The total score is 0-12 points. The score > 4 was defined as high expression, while the score ≤ 4 was defined as low expression.
From the Chinese Center for Type Culture Collection (Wuhan, China), we bought human HCC cell lines (HepG2, HCCLM3, Huh7 and MHCC97H) and human-derived liver cell line (HL-7702). All cells were cultured in RPMI-1640 medium (Thermo Fisher Scientific, Carlsbad, MA, United States) with 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, United States) and 100 μg/mL streptomycin and 100 U/mL penicillin (Invitrogen, Carlsbad, CA, United States) at 37 °C in 5% CO2. From GenePharma Co., Ltd. (Shanghai, China), we purchased pcDNA-GMEB1, pcDNA empty vector (NC), small interfering RNA (siRNA) negative control (si-NC), siRNAs against GMEB1 (si-GMEB1#1 and si-GMEB1#2), pcDNA-YAP1 and siRNA against YAP1 (si-YAP1). HepG2 and Huh7 cells were transfected employing Lipofectamine® 3000 (Invitrogen, Carlsbad, CA, United States) under the supplier’s instructions. At 48 h after transfection, Western blot was conducted to verify the transfection efficiency.
The lentiviral overexpression vector pCDH-CMV-MCS-EF1-Puro and the shRNA vector pLKO.1-puro were purchased from GenScript Biotech. The HCC cell line HepG2 was inoculated in 6-well cell culture plates with 2 mL of medium containing 10% fetal bovine serum 1 d prior to the infection and incubated in a constant temperature incubator at 37 °C with 5% CO2. The lentivirus was transfected when the cell density reached 80%. An appropriate amount of lentivirus was added to the cell culture plate according to the MOI (multiplicity of infection) = 5 and incubated overnight at 37 °C with 5% CO2. Change to fresh medium 24 h after infection. The puromycin was added 36 h after infection to allow the cells infected with the puromycin resistance gene to proliferate sufficiently. 72 h later the puromycin medium was withdrawn and the remaining cells continued to be cultured to obtain stable overexpression or silenced cell lines.
TRIzol reagent (Invitrogen, Carlsbad, CA, United States) was utilized to extract total RNA from tissues and cells, and the RNA purity and concentration were determined by UV absorption method. Subsequently, a PrimeScript-RT Kit (Takara, Shiga, Japan) was employed for synthesizing complementary DNA (cDNA) from 1 µg of total RNA under the manufacturer’s protocol, and then ABI7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, United States) and SYBR® Premix Ex Taq™ (TaKaRa, Dalian, China) were used for quantitative real-time PCR (qRT-PCR). All fluorescence data were converted to relative quantification, and the 2-ΔΔCt method was utilized to calculate the relative expression, with GAPDH as the internal control. GMEB1 primer sequence: forward, 5'-GCA
The cells were harvested and incubated with pre-cooled radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China) for 20 min on ice. We collected the supernatant following centrifuging the cell lysis solution for 20 min at 13000 r/min at 4 °C. Then, a BCA protein quantification kit was utilized to measure the concentration of protein. The protein was isolated by 12% SDS-PAGE, and transferred onto a polyvinylidene difluoride membrane. After being blocked with Tris-buffered saline Tween (TBST) solution containing 3% bovine serum albumin at room temperature for 1 h, the membrane was incubated overnight with diluted primary antibodies at 4 °C: anti-GMEB1 antibody (Abcam, 1:1000, ab240646), anti-GAPDH antibody (1:1000, Abcam, ab8245), and anti-YAP1 antibody (1:1000, Abcam, ab52771). The membrane was washed 5 times with TBST, 3 min for each time. Then, the membrane and the added diluted secondary antibody (1:5000, Abcam, ab6721) were incubated at room temperature for 40 min. The protein bands were observed employing the ECL Western blot kit (Beyotime, Shanghai, China). ImageJ software (NIH, Bethesda, MD, United States) was adopted for analyzing each band’s gray value, and the ratio of the target protein’s gray value to that of GAPDH functioned as the relative protein expression for analysis.
Cell Counting Kit-8 (CCK-8) (Beyotime, Shanghai, China) was employed for detecting cell proliferation. HepG2 and Huh7 cells during logarithmic growth were taken and inoculated into 96-well plates (5 × 103 cells/well), which were incubated in a 37 °C, 5% CO2 incubator for 1, 2 and 3 days, respectively. After that, 10 μL of CCK-8 reagent was added to each well and incubated for 2 h at 37 °C. We discarded the supernatant, and used an automatic microplate reader (Bio-Rad, Hercules, CA, United States) to determine the absorbance (OD) value at 450 nm of each well.
Matrigel (BD Biosciences, Franklin Lakes, NJ, United States) preserved at -20 °C was melted at 4 °C overnight and mixed with serum-free medium. 100 μL of the medium was pipetted into the upper Transwell chamber, which was then let stand for 5 h at 37 °C. HepG2 and Huh7 cells were collected to prepare single-cell suspension with a density of 5 × 105 cells/mL. 500 μL of 10% fetal bovine serum-containing medium was added to the lower compartment, and 200 μL of cell suspension was added to the top compartment. The Transwell chamber was incubated at 37 °C for 48 h. We wiped off the remaining cells and Matrigel carefully. The migrated cells in the lower compartment were fixed with 4% formaldehyde at room temperature for 30 min and stained with crystal violet for 20 min at room temperature. The cells in 5 random views were photographed and counted under the inverted microscope (Nikon, Tokyo, Japan). In the migration assay, Matrigel was not added to the top compartment of Transwell, and the rest was as same as that in the invasion assay.
Cell apoptosis was detected via the Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (Southern Biotechnology, Birmingham, Al, United States). Huh7 and HepG2 cells 48 h after transfection were trypsinized and collected and inoculated into 6-well plates. The cell density was adjusted to 2 × 104 cells/well, and the culturing was continued for 24 h. After washing with precooled PBS twice, the cells were re-suspended in 1×Binding buffer. We added 5 μL of PI and 5 μL of Annexin V-FITC to the cell suspension, which was subsequently mixed fully and incubated for 15 min at room temperature away from light. Afterwards, under the requirements of the instructions, the apoptosis was detected by the flow cytometer (BD Biosciences, San Jose, CA, United States) within 1 h.
We employed the JASPR database (http://jaspar.genereg.net/) for predicting the binding sites of GMEB1 to YAP1 promoter region. The target fragments of mutant YAP1 and wild-type YAP1 were established and inserted into the pGL3 vector (Promega, Madison, WI, United States) to construct pGL3-YAP1-mutant (YAP1-MUT) and pGL3-YAP1-wild type (YAP1-WT) reporter vectors. YAP1-MUT or YAP1-WT and pcDNA-GMEB1 overexpression plasmids or si-GMEB1#1 were cotransfected into HepG2 and Huh7 cells. The luciferase activity was measured 48 h after transfection with the Dual-Luciferase Reporter Assay system (Promega, Madison, WI, United States) under the instructions.
A EZ-ChIPTM kit (Millipore, Billerica, MA, United States) was adopted for performing chromatin immunoprecipitation (ChIP) assay. About 1 × 107 HepG2 and Huh7 cells were cross-linked at 37 °C for 10 min in 1% formaldehyde, and reacted with 125 mmol/L glycine solution at room temperature for 5 min. The mixture was sonicated on ice, 15 s each time, and 15 times at 15-second intervals. Following that, the sonicated product was centrifuged for 10 min at 4 °C at 12000 g. The supernatant was harvested, divided into two tubes, and was incubated with negative control IgG antibody (Abcam, ab6721) or GMEB1 antibody (Abcam, ab240646) at 4 °C overnight. Next, protein agarose/agarose gel was utilized to precipitate DNA-protein complexes which were then centrifuged for 5 min at 12000 g at 4 °C. Subsequently, the cross-linking was reversed at 65 °C overnight, and DNA fragments were recovered after extraction and purification by phenol/chloroform. Eventually, the binding of GMEB1 with YAP1 promoter region and YAP1 specific primers was detected via qRT-PCR.
5-6 wk C57BL/6 mice (purchased from Beijing Viton Lever Laboratory Animal Technology Co., Ltd.) were housed under pathogenic conditions at 26-28 °C and 50-65% humidity. All animal experiments conformed to ethical norms and were approved by the animal ethics committee of our institution. To construct subcutaneous tumors, sh-NC (shRNA negative control), sh-GMEB1, oe-NC (overexpression negative control), oe-YAP1, sh-GMEB1+oe-YAP1 cells were suspended in 100 µL PBS diluted in Matrigel matrix gel (356234, Corning) in quantities of 1 × 106 (PBS:Matrigel = 2:1), cells were injected subcutaneously into the right axilla of mice (n = 5/group) and tumor size was measured every 3 days. Tumor volume was calculated using the formula: V = (L × W2)/2, where V is the volume (mm3), L is the long diameter and W is the short diameter. Transplanted tumors were taken from Balb/c nude mice after 30 d for subsequent experiments.
To examine the effect on metastatic capacity, sh-NC (shRNA negative control), sh-GMEB1, oe-NC, oe-YAP1, sh-GMEB1+ oe-YAP1 cells were injected into nude mice (n = 5/group) via tail vein at an amount of 1 × 106, respectively. Mice were executed 60 days after injection, and number of metastatic foci on the lungs of each group were counted.
SPSS22.0 statistical software was employed to carry out statistical analysis of the data. All experiments were independently repeated three times. The results of in vitro experiments were presented as mean ± SD, and the results of all in vivo experiments were expressed as mean ± SEM. Comparisons between groups were conducted using Student's t test or one-way ANOVA. Pearson correlation analysis was utilized for evaluate the correlation between GMEB1 and YAP1 expression in HCC tissues. The relationship between GMEB1 expression and the clinicopathological characteristics of HCC patients was assessed by the χ2 test. P < 0.05 was considered a statistically significant difference between groups.
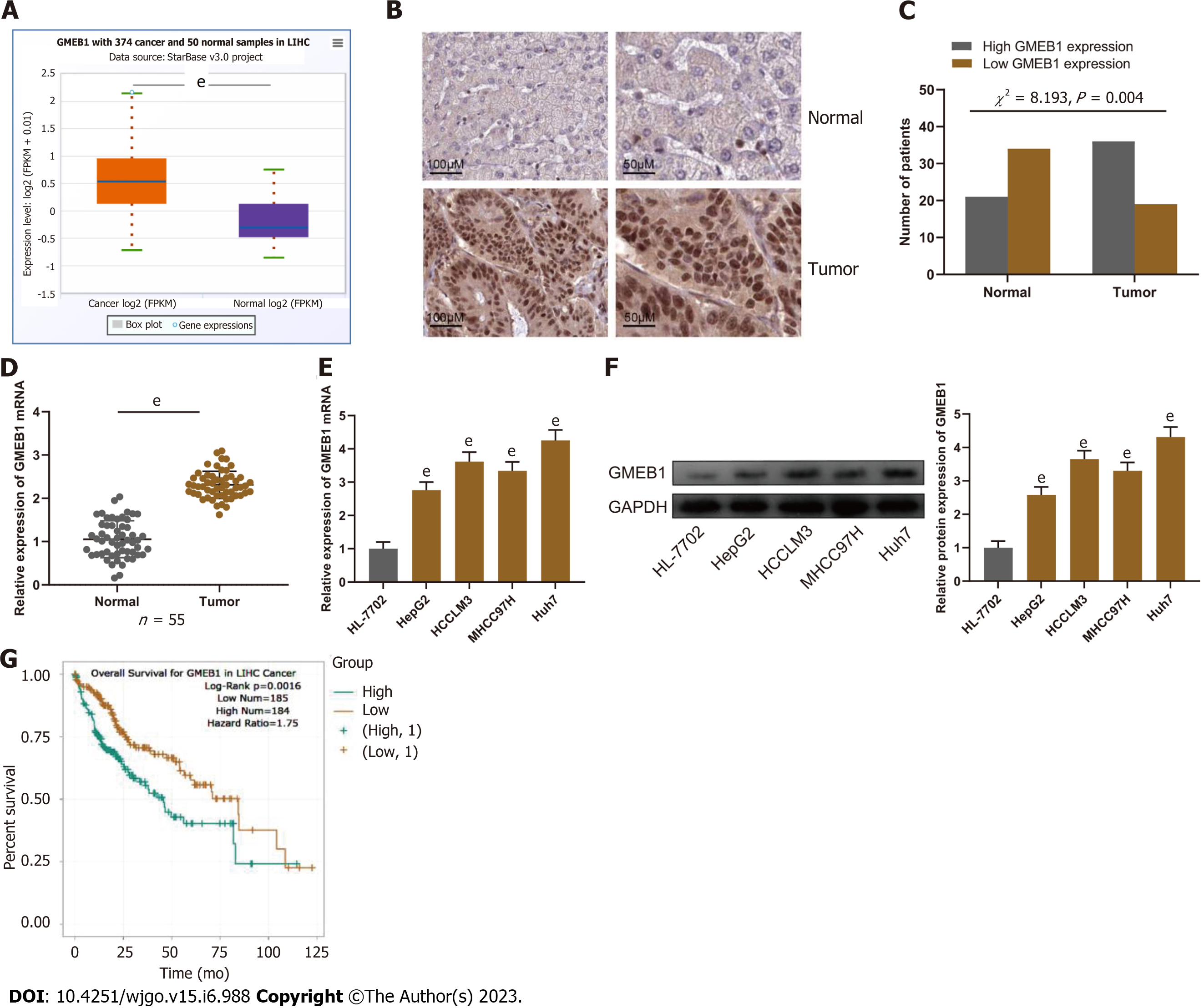
To investigate GMEB1 expression in HCC, we analyzed GMEB1 expression in HCC through the StarBase database (http://starbase.sysu.edu.cn/), and it was unveiled that GMEB1 is aberrantly upregulated in HCC tissue samples (Figure 1A). Furthermore, we performed another database analysis of GMEB1 expression level in different subtypes of hepatocellular carcinoma (https://ualcan.path.uab.edu/), and the results revealed that GMEB1 expression was still significantly higher in HCC than in normal liver tissues (Supplementary Figure 1A). Interestingly, GMEB1 expression was further upregulated with the increasing tumor stage (Supplementary Figure 1B), tentatively suggesting a correlation between high GMEB1 expression and poorer liver cancer stage. Subsequently, we validated results from the database on clinical samples. IHC was adopted to detect GMEB1 expression in 55 selected HCC patients’ cancer tissues and corresponding para-cancer tissues. Statistical analysis manifested that GMEB1 expression in the cancerous tissues of HCC patients was significantly higher as opposed to the para-cancer tissues (Figure 1B). Additionally, qRT-PCR analysis found that GMEB1 mRNA was markedly upregulated in HCC tissues and cells (HepG2, HCCLM3, MHCC97H and Huh7) as against para-cancerous tissues or the HL-7702 cell line (Figure 1C and D). Western blot analysis manifested that relative to HL-7702 cells, the GMEB1 protein level in HCC cells was remarkably elevated (Figure 1E).
Due to the high expression of GMEB1 in HCC tissues, we further explored the association between GMEB1 expression level and patient prognosis through the database analysis. The results from StarBase database suggested that high GMEB1 expression was linked to HCC patients’ short overall survival (Figure 1F). At the same time, the overall survival of patients with the same stage of hepatocellular carcinoma was significantly longer in patients with low GMEB1 expression compared to those with high GMEB1 expression (Supplementary Figure 1C). Chi-square test revealed that high GMEB1 expression was related to advanced TNM stage and large tumor size in HCC patients (Table 1). These results indicated that GMEB1 is highly expressed in HCC tissues and that upregulation of GMEB1 expression is strongly associated with an exacerbation of the malignant phenotype and poor prognosis. Also, this section provisionally suggested that the upregulation of GMEB1 may play a role in the malignant progression of hepatocellular carcinoma.
| Characteristic | Number | GMEB1 expression | χ2 | P value | ||
| High (n = 29) | Low (n = 26) | |||||
| Age (yr) | ||||||
| < 60 | 25 | 14 | 11 | |||
| ≥ 60 | 30 | 15 | 15 | 0.197 | 0.657 | |
| Gender | ||||||
| Male | 23 | 17 | 13 | |||
| Female | 32 | 12 | 13 | 0.419 | 0.517 | |
| Alcoholism | ||||||
| Negative | 24 | 15 | 9 | |||
| Positive | 31 | 14 | 17 | 1.632 | 0.201 | |
| HBV | ||||||
| Negative | 28 | 16 | 12 | |||
| Positive | 27 | 13 | 14 | 0.446 | 0.504 | |
| Tumor size (cm) | ||||||
| ≤ 3 | 18 | 12 | 6 | |||
| > 3 | 37 | 10 | 27 | 7.928 | 0.005b | |
| TNM stage | ||||||
| I-II | 20 | 11 | 9 | |||
| III-IV | 35 | 7 | 28 | 7.081 | 0.007b | |
| Differentiation | ||||||
| Well/moderate | 28 | 13 | 15 | |||
| Poor | 27 | 16 | 11 | 0.980 | 0.322 | |
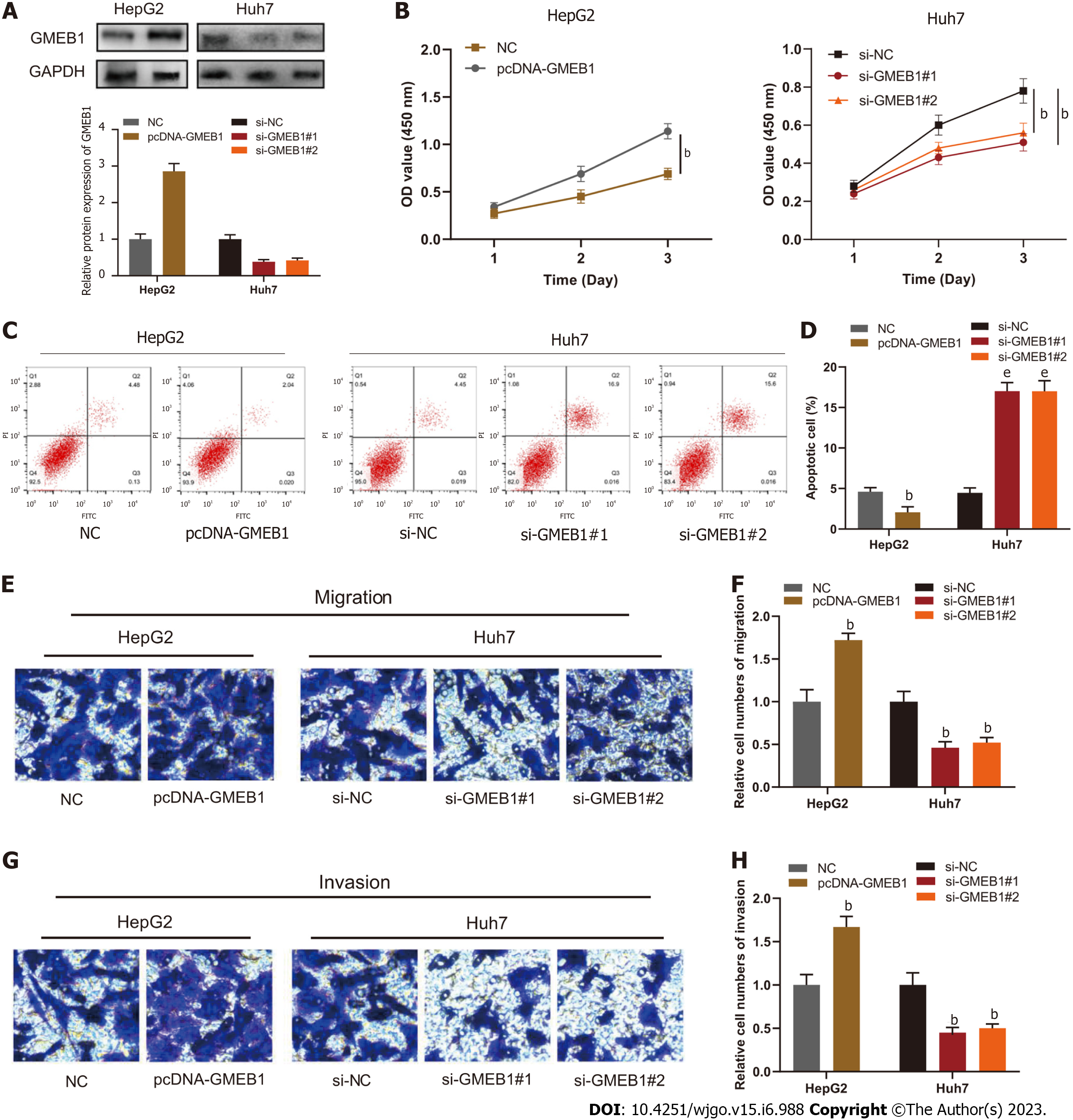
To delve into the biological function of GMEB1 in HCC cells, we transfected HepG2 cells with pcDNA-GMEB1 to construct the GMEB1 overexpression cell model, and transfected Huh7 cells with si-GMEB1#1 or si-GMEB1#2 to construct the GMEB1 knockdown cell model. Western blot analysis showed that the plasmids were successfully transfected into cells, which was reflected in the results that GMEB1 was successfully overexpressed in HepG2 cells while GMEB1 was effectively silenced and down-regulated in Huh7 cells (Figure 2A). Next, to investigate the effect of GMEB1 on the proliferation of hepatocellular carcinoma cells, CCK8 assay was performed to assess the alteration in cell viability and proliferation rate following changes in GMEB1 expression levels. Results from CCK-8 assay demonstrated that overexpression of GMEB1 significantly promoted the proliferation of HepG2 cells while GMEB1 knockdown dramatically inhibited the proliferation of Huh7 cells compared to the si-NC group (Figure 2B). Furthermore, we continued to examine the effect of altered GMEB1 expression levels on cellular apoptosis in different groups by flow cytometry, and analysis of the PI/Annexin V double staining results showed that the percentage of apoptotic cells increased substantially after GMEB1 silencing, while the percentage of apoptotic cells in the overexpression group decreased markedly compared to the control group, suggesting that GMEB1 could enhance the anti-apoptotic ability of cells and promote cell survival (Figure 2C and D). The above results indicated that GMEB1 had a remarkable promotion effect on the growth of hepatocellular carcinoma cells, which was mainly achieved through the dual effect on inhibiting apoptosis and promoting proliferation.
In addition to promoting proliferation, we'd also like to know whether GMEB1 could alter the migration and invasion ability of hepatocellular carcinoma cells as well. The UALCAN database was firstly employed to analyze the correlation between GMEB1 and nodal metastasis status. As we can see in Supplementary Figure 1D, with the increase of nodal metastasis status, the expression level of GMEB1 was boosted as well. In order to further verify this conjecture, we conducted Transwell assay. The results indicated that overexpression of GMEB1 significantly promoted the migration and invasion of HepG2 cells, while knockdown of GMEB1 strongly inhibited the motor ability of Huh7 cells, suggesting that GMEB1 also plays an important role in promoting the migration and invasion ability of hepatocellular carcinoma cells and increasing the metastatic potential of tumors (Figure 2E-H).
Taken together, we can easily infer that GMEB1 can enhance the proliferation and anti-apoptotic ability of tumor cells, thus promoting unlimited proliferation of tumor. In addition, the enhancement of the migration and invasion ability of tumor cells further increases the possibility of tumor metastasis.
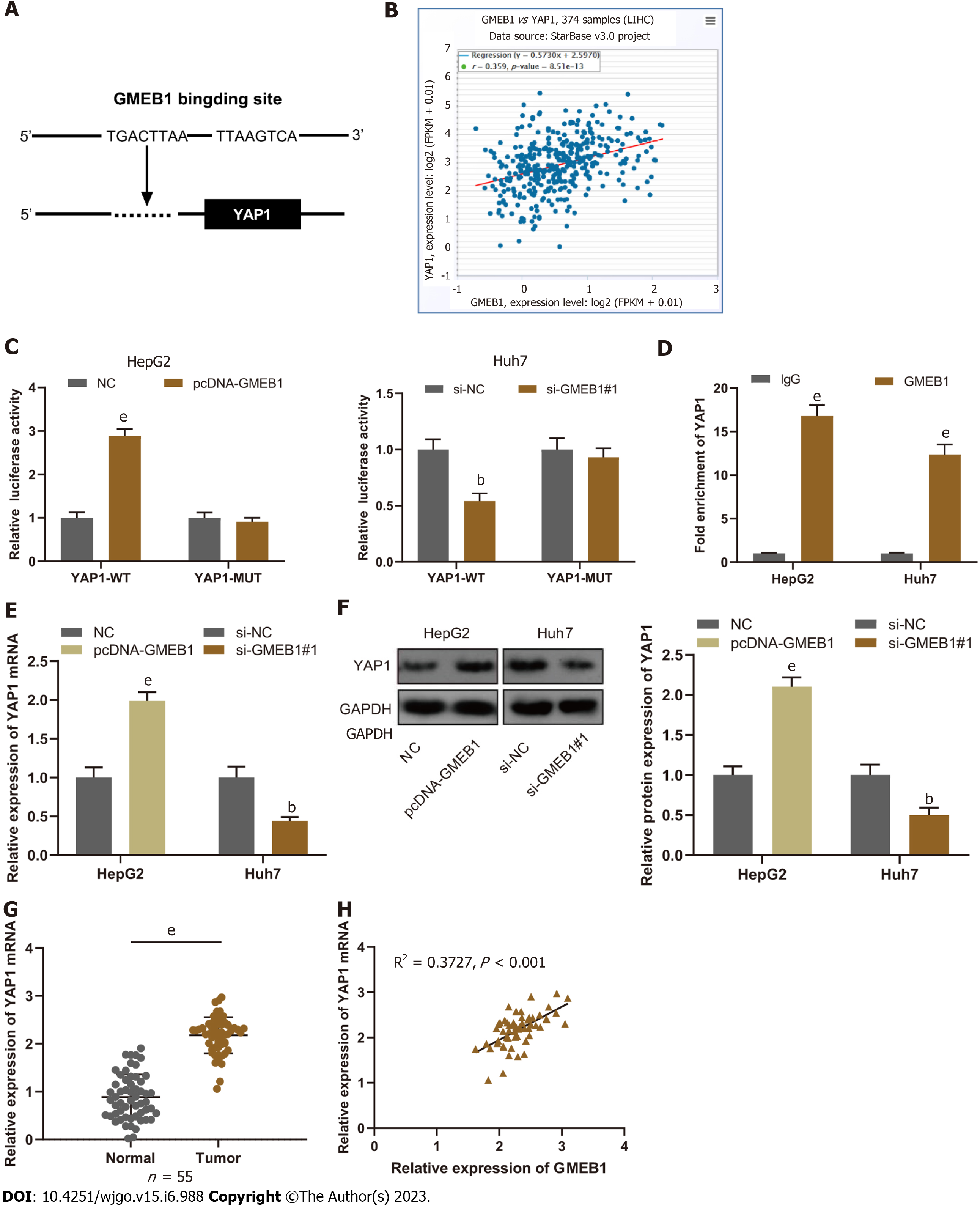
The next question is how GMEB1 promotes malignant proliferation and invasion of hepatocellular carcinoma cells and what are the downstream target genes, which we will then explore in more depth. Firstly, we analyzed the JASPAR databases (http://jaspar.genereg.net/) and found that GMEB1 may bind to three sites in the promoter region of YAP1 (Figure 3A). The StarBase database (http://starbase.sysu.edu.cn/) showed that GMEB1 and YAP1 expression are positively correlated in hepatic cancer tissue samples (Figure 3B). To verify the relationship between GMEB1 and YAP1, we then constructed wild-type plasmids with YAP1 promoter binding region and plasmids with mutations in the predicted binding sites, and then transfected cells with plasmids separately and collected them for dual luciferase reporter gene assays. The results revealed that overexpression of GMEB1 enhanced the luciferase activity of YAP1-WT in HepG2 cells, whereas GMEB1 knockdown reduced the luciferase activity of YAP1-WT in Huh7 cells (Figure 3C); neither GMEB1 overexpression nor knockdown had any significant effect on the luciferase activity of YAP1-MUT (Figure 3C), suggesting that GMEB1 can specifically bind to the YAP1 promoter region, and the binding site predicted by the database is the region where GMEB1 binds to the YAP1 promoter, and GMEB1 can positively regulate YAP1 expression at the transcriptional level. Moreover, ChIP-qPCR analysis demonstrated that GMEB1 was markedly enriched in the promoter region of YAP1 compared to the IgG control (Figure 3D), further confirming that GMEB1 directly regulates the transcriptional activation of YAP1. To this end, we examined the effects of altered GMEB1 expression on YAP1 at the mRNA and protein levels, respectively, and found that overexpression of GMEB1 promoted YAP1 expression in HepG2 cells, whereas knockdown of GMEB1 inhibited YAP1 expression in Huh7 cells (Figure 3E and F). In addition, we also examined the expression of YAP1 in HCC tissues. qRT-PCR results illustrated that the expression of YAP1 mRNA was much higher in HCC tissues than in para-cancerous tissues (Figure 3G); Pearson analysis indicated that GMEB1 mRNA was positively correlated with YAP1 mRNA expression in HCC tissues (Figure 3H).
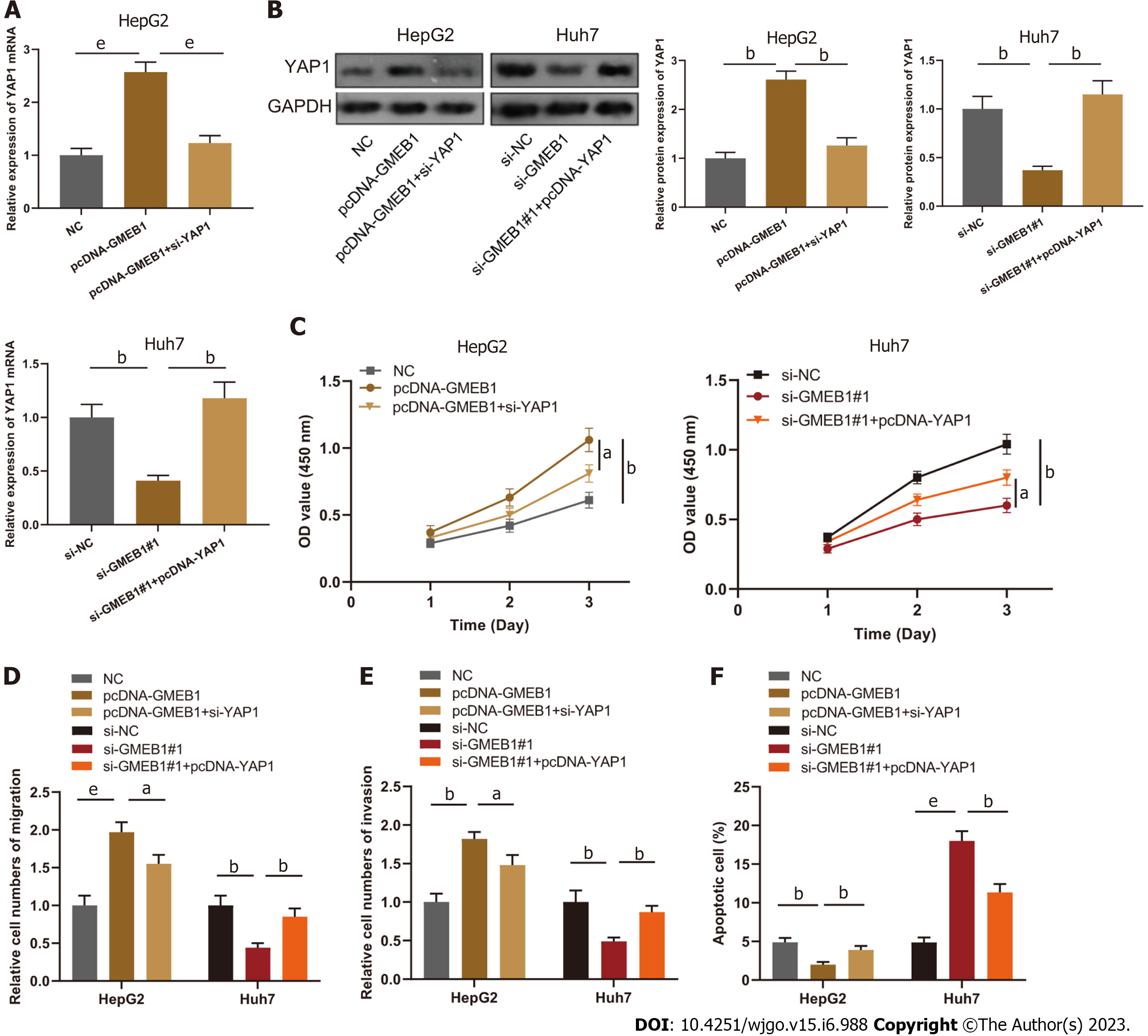
To verify the effect of GMEB1/YAP1 regulatory circuit on proliferation, migration, invasion and apoptosis of HCC cells, we co-transfected HepG2 cells with pcDNA-GMEB1 and si-YAP1; co-transfected Huh7 cells with si-GMEB1#1 and pcDNA-YAP1, respectively. qRT-PCR and Western blot assays illustrated that GMEB1 overexpression and YAP1 protein silencing were successful (Figure 4A and B). The results of CCK-8 assay, Transwell and flow cytometry demonstrated that the promotion of proliferation, migration and invasion as well as the anti-apoptotic effect of GMEB1 overexpression on HepG2 cells was reversed by the knockdown of YAP1; on the other hand, the upregulation of YAP1 also reversed the effect of knockdown of GMEB1 on the proliferation, migration, invasion and anti-apoptotic effect of YAP1 on Huh7 cells (Figure 4C-F).
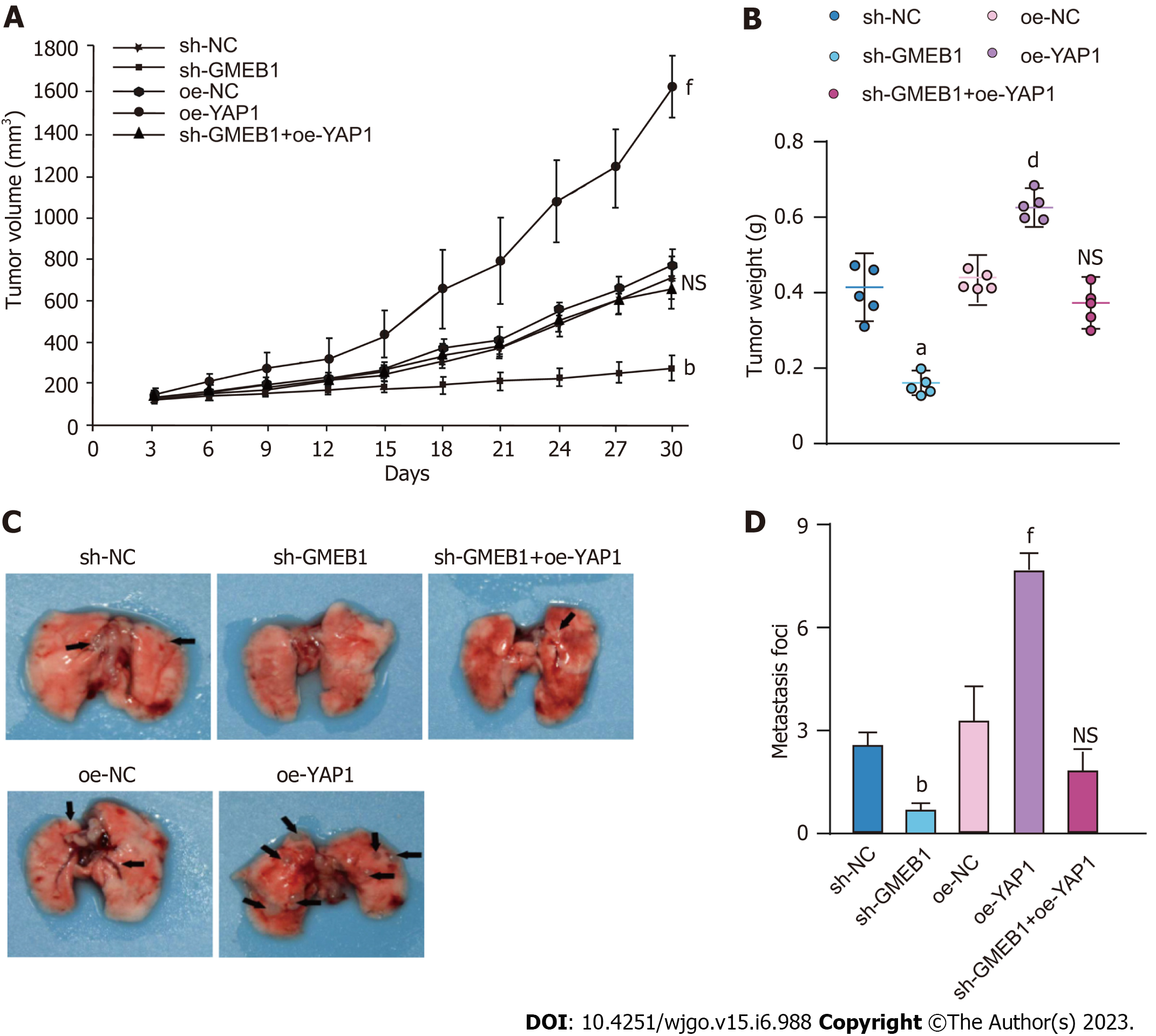
To further validate this effect, we then constructed sh-GMEB1, oe-YAP1 and sh-GMEB1+oeYAP1 stable expression cell lines by lentiviral infection and investigated the effect of the GMEB1/YAP1 regulatory loop on tumor proliferation by Cell-derived xenografts (CDX) model in nude mice. The results revealed that sh-GMEB1 remarkably slowed down the proliferation rate of HepG2-derived xenografts, whereas overexpression of YAP1 significantly enhanced the proliferation ability of tumors compared with the oe-NC group. More interestingly, silencing GMEB1 followed by overexpression of YAP1 markedly reversed the proliferation inhibitory effect of silencing GMEB1 (Figure 5A and B). The above data suggested that the GMEB1/YAP1-regulated signaling axis can effectively increase the level of malignant proliferation of tumor. Subsequently, we examined whether the number of HCC metastasis foci forming in the lung would be altered by tail vein injection of the above cells. As we can see, silencing of GMEB1 significantly reduced the number of metastatic foci, whereas overexpression of YAP1 did the opposite. Furthermore, silencing of GMEB1 followed by overexpression of YAP1 also reversed the inhibitory effect of GMEB1 silencing on the metastatic ability, with no significant difference from the control group (Figure 5C and D). Taken together, these data further confirmed our findings at the cellular level from the perspective of CDX mice model and emphasized the positive regulation of the GMEB1/YAP1 signaling axis on the malignant proliferation and metastasis of hepatocellular carcinoma.
Transcription factors are a major type of DNA-binding protein that regulates gene expression via binding with specific DNA sequences in gene promoter regions, thereby affecting cell growth, differentiation and apoptosis[29,30]. Many studies have shown that transcription factors can partake in regulating HCC occurrence and development. For instance, the transcription factor FOXN3 is downregulated in HCC and can inhibit cancer cell proliferation via negatively regulating E2F5 expression[31]; the transcription factor ATF3 suppresses HCC cell metastasis and proliferation by upregulating CYR61 expression[32]; the transcription factor KLF5 boosts HCC cell multiplication and metastasis by activating the PI3K/AKT/Snail signal pathway[33]. GMEB1 is a glucocorticoid regulatory element-binding protein, a ubiquitous multifunctional DNA-binding protein, and a transcription factor, which features prominently in the modulation of transcription after steroid hormone activation[34,35]. For the first time, this study discovered that GMEB1 was up-regulated in HCC tissues and cells. High GMEB1 expression was correlated to the advanced TNM stage, relatively large tumor size, and relatively short overall survival time of HCC patients. In-vitro functional research indicated that GMEB1 overexpression facilitated HCC cell multiplication, migration and invasion, and repressed the apoptosis, yet knocking down GMEB1 resulted in the opposite effects. This demonstrates that GMEB1 plays a vital role in the malignant progression of HCC.
In the further mechanism study, we mainly focused on the exploration for the downstream target gene of GMEB1, and finally determined that YAP1 was a new downstream target gene of GMEB1 through database prediction and experimental verification.YAP1, located on human chromosome 11q22, is a transcriptional coactivator of the Hippo pathway, and it modulates the transcription of downstream target genes and intracellular signal transduction through phosphorylation, thus maintaining the balance of cell multiplication and apoptosis[36]. Some studies have found that YAP1 is targeted and regulated by multiple microRNAs (miRs), such as miR-506[37], miR-200a[38], miR-139-5p[39] and miR-27b-3p[40]. Reportedly, YAP1 is abnormally high-expressed in various tumor tissues, and participates in facilitating tumor cell multiplication, invasion and migration and the activation of multiple signaling pathways[36-38,41,42]. For example, YAP1 is up-regulated in bladder carcinoma tissues and can interact with mTOR protein to promote bladder carcinoma cell proliferation[43]. The increase in YAP1 expression is connected with the poor prognosis of BC patients, and inhibits PTEN expression to boost BC cell multiplication and repress the apoptosis[44]. In HCC, YAP1 can facilitate cell proliferation, invasion and EMT process in vitro[45]. In our study, we first found that GMEB1 could bind with the YAP1 promoter region and could positively modulate YAP1 expression in HCC cells. Also, there was a positive correlation between YAP1 and GMEB1 expression in HCC tissues. Moreover, alteration of YAP1 expression could mediate GMEB1-induced HCC cell multiplication, migration, invasion and apoptosis.
The crucial role of the Hippo signaling pathway, with YAP/TAZ as the main effector proteins, in the development of hepatocellular carcinoma has been confirmed and highlighted by numerous studies[46], however, no inhibitors directly targeting core proteins of the Hippo signaling pathway have been successfully marketed, making direct intervention of the Hippo signaling pathway difficult and preventing patients with excessive Hippo activation from benefiting more from conventional therapies. For this reason, a number of studies have begun to attempt to interfere with the Hippo signaling pathway through the inhibition of indirectly regulated proteins. These include PFI-2, a highly selective inhibitor of SETD7 methyltransferase, which inhibits YAP nuclear translocation and Hippo pathway activation through direct interaction with SETD7[47], and Ki-16425, a competitive inhibitor of LPA1/2/3, which inhibits Hippo signaling by blocking LPA receptor-induced YAP/TAZ dephosphorylation[48]. The discovery of our study further enriches the range of target options for indirect intervention in the Hippo signaling pathway, thus providing new candidate proteins for inhibitor development.
In conclusion, our study revealed for the first time that GMEB1 is highly expressed in both HCC tissues and cells. Furthermore, we confirmed that YAP1 is a novel transcriptional target gene of GMEB1 and demonstrated that GMEB1/YAP1 regulatory axis has a key role in promoting HCC cell proliferation, migration and invasion and anti-apoptosis. As this suggests that, on the one hand, GMEB1 can be a candidate molecular marker for accurate diagnosis of early-stage liver cancer; on the other hand, targeting GMEB1 may be a potential therapeutic approach to improve the efficiency of HCC treatment. However, our study also has some limitations. Firstly, we only used the CDX animal model to verify the regulation of GMEB1 on the malignant progression of HCC; we lacked samples from clinical sources for further corroboration. In addition, we lacked follow-up studies on patients so as to further corroborate the effect of GMEB1 on the prognosis of hepatocellular carcinoma. In subsequent studies, we will continue to refine our findings in the follow-up study and continue to obtain more reliable experimental conclusions.
Most hepatocellular carcinoma (HCC) cases have already been in an intermediate to advanced stage at diagnosis, missing the optimal time for surgical treatment. Besides, the high cost and huge shortage of donors greatly limit the clinical application of liver transplantation. Therefore, the lack of reliable diagnostic markers and interventions makes the overall survival rate of HCC patients hardly improved, and the prognosis of patients is generally poor.
Mechanistic understanding of proliferation promotion, increased metastatic potential and cellular defense against apoptosis can inform novel therapeutic strategies in HCC treatment. We would like to clarify the biological functions and clinical significance of glucocorticoid modulatory element-binding protein 1 (GMEB1) in HCC to provide more reliable biomarkers and target candidates for HCC diagnosis and treatment.
Due to the lack of reliable early diagnosis biomarkers and clinical intervention targets, patients with HCC are often diagnosed in the middle and late stages with poor prognosis. This study aims to find new molecular biomarkers with high reference value and effective candidate targets for the treatment of HCC.
GMEB1 expression level was detected through bioinformatics analysis first and further validation was conducted in HCC clinical tissue samples by using immunohistochemistry and qPCR; immunoblotting and qPCR were used to detect the expression of GMEB1 in HCC cell lines. Cell counting kit-8 assay, Transwell and flow cytometry were performed to assess the alteration on proliferation and metastasis potential of HCC cells after changes on GMEB1 expression level in HCC cell lines. The binding site of GMEB1 to Yes-associate protein 1 (YAP1) promoter was predicted by bioinformatics analysis and verified by dual luciferase reporter gene assay and ChIP-qPCR.
GMEB1 was abnormally highly expressed in HCC, whose expression correlates with some clinicopathological features of HCC patients, such as tumor size and TNM stage. According to in vitro results, overexpression of GEMB1 promotes the malignant proliferation and metastasis of HCC cell lines, while knockdown of GMEB1 suppresses the degree of malignancy. In mechanism, GEMB1 positively regulates the expression of YAP1 in transcription level, which finally promote the progression of HCC.
GMEB1 is a new oncogenic factor of HCC, which promotes the malignant proliferation and metastasis of HCC by promoting YAP1 transcriptional activation.
This study reveals that GMEB1, a member of KDWK gene family to modulate the transactivation of the glucocorticoid receptor, has a previously unrecognized role in HCC progression, suggesting that it may be a promising biomarker for early-stage HCC diagnosis and it is possible to intervene in anti-tumor therapy against HCC by inhibiting GMEB1-mediated tumor proliferation, metastasis and anti-apoptosis effect.
| 1. | de Castria TB, Khalil DN, Harding JJ, O'Reilly EM, Abou-Alfa GK. Tremelimumab and durvalumab in the treatment of unresectable, advanced hepatocellular carcinoma. Future Oncol. 2022;18:3769-3782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 25] [Reference Citation Analysis (0)] |
| 2. | Hernández-Aquino E, Muriel P. Beneficial effects of naringenin in liver diseases: Molecular mechanisms. World J Gastroenterol. 2018;24:1679-1707. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 194] [Cited by in RCA: 251] [Article Influence: 31.4] [Reference Citation Analysis (15)] |
| 3. | Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133-1145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1163] [Cited by in RCA: 1234] [Article Influence: 88.1] [Reference Citation Analysis (2)] |
| 4. | Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Abate D, Abbasi N, Abbastabar H, Abd-Allah F, Abdel-Rahman O, Abdelalim A, Abdoli A, Abdollahpour I, Abdulle ASM, Abebe ND, Abraha HN, Abu-Raddad LJ, Abualhasan A, Adedeji IA, Advani SM, Afarideh M, Afshari M, Aghaali M, Agius D, Agrawal S, Ahmadi A, Ahmadian E, Ahmadpour E, Ahmed MB, Akbari ME, Akinyemiju T, Al-Aly Z, AlAbdulKader AM, Alahdab F, Alam T, Alamene GM, Alemnew BTT, Alene KA, Alinia C, Alipour V, Aljunid SM, Bakeshei FA, Almadi MAH, Almasi-Hashiani A, Alsharif U, Alsowaidi S, Alvis-Guzman N, Amini E, Amini S, Amoako YA, Anbari Z, Anber NH, Andrei CL, Anjomshoa M, Ansari F, Ansariadi A, Appiah SCY, Arab-Zozani M, Arabloo J, Arefi Z, Aremu O, Areri HA, Artaman A, Asayesh H, Asfaw ET, Ashagre AF, Assadi R, Ataeinia B, Atalay HT, Ataro Z, Atique S, Ausloos M, Avila-Burgos L, Avokpaho EFGA, Awasthi A, Awoke N, Ayala Quintanilla BP, Ayanore MA, Ayele HT, Babaee E, Bacha U, Badawi A, Bagherzadeh M, Bagli E, Balakrishnan S, Balouchi A, Bärnighausen TW, Battista RJ, Behzadifar M, Bekele BB, Belay YB, Belayneh YM, Berfield KKS, Berhane A, Bernabe E, Beuran M, Bhakta N, Bhattacharyya K, Biadgo B, Bijani A, Bin Sayeed MS, Birungi C, Bisignano C, Bitew H, Bjørge T, Bleyer A, Bogale KA, Bojia HA, Borzì AM, Bosetti C, Bou-Orm IR, Brenner H, Brewer JD, Briko AN, Briko NI, Bustamante-Teixeira MT, Butt ZA, Carreras G, Carrero JJ, Carvalho F, Castro C, Castro F, Catalá-López F, Cerin E, Chaiah Y, Chanie WF, Chattu VK, Chaturvedi P, Chauhan NS, Chehrazi M, Chiang PP, Chichiabellu TY, Chido-Amajuoyi OG, Chimed-Ochir O, Choi JJ, Christopher DJ, Chu DT, Constantin MM, Costa VM, Crocetti E, Crowe CS, Curado MP, Dahlawi SMA, Damiani G, Darwish AH, Daryani A, das Neves J, Demeke FM, Demis AB, Demissie BW, Demoz GT, Denova-Gutiérrez E, Derakhshani A, Deribe KS, Desai R, Desalegn BB, Desta M, Dey S, Dharmaratne SD, Dhimal M, Diaz D, Dinberu MTT, Djalalinia S, Doku DT, Drake TM, Dubey M, Dubljanin E, Duken EE, Ebrahimi H, Effiong A, Eftekhari A, El Sayed I, Zaki MES, El-Jaafary SI, El-Khatib Z, Elemineh DA, Elkout H, Ellenbogen RG, Elsharkawy A, Emamian MH, Endalew DA, Endries AY, Eshrati B, Fadhil I, Fallah Omrani V, Faramarzi M, Farhangi MA, Farioli A, Farzadfar F, Fentahun N, Fernandes E, Feyissa GT, Filip I, Fischer F, Fisher JL, Force LM, Foroutan M, Freitas M, Fukumoto T, Futran ND, Gallus S, Gankpe FG, Gayesa RT, Gebrehiwot TT, Gebremeskel GG, Gedefaw GA, Gelaw BK, Geta B, Getachew S, Gezae KE, Ghafourifard M, Ghajar A, Ghashghaee A, Gholamian A, Gill PS, Ginindza TTG, Girmay A, Gizaw M, Gomez RS, Gopalani SV, Gorini G, Goulart BNG, Grada A, Ribeiro Guerra M, Guimaraes ALS, Gupta PC, Gupta R, Hadkhale K, Haj-Mirzaian A, Hamadeh RR, Hamidi S, Hanfore LK, Haro JM, Hasankhani M, Hasanzadeh A, Hassen HY, Hay RJ, Hay SI, Henok A, Henry NJ, Herteliu C, Hidru HD, Hoang CL, Hole MK, Hoogar P, Horita N, Hosgood HD, Hosseini M, Hosseinzadeh M, Hostiuc M, Hostiuc S, Househ M, Hussen MM, Ileanu B, Ilic MD, Innos K, Irvani SSN, Iseh KR, Islam SMS, Islami F, Jafari Balalami N, Jafarinia M, Jahangiry L, Jahani MA, Jahanmehr N, Jakovljevic M, James SL, Javanbakht M, Jayaraman S, Jee SH, Jenabi E, Jha RP, Jonas JB, Jonnagaddala J, Joo T, Jungari SB, Jürisson M, Kabir A, Kamangar F, Karch A, Karimi N, Karimian A, Kasaeian A, Kasahun GG, Kassa B, Kassa TD, Kassaw MW, Kaul A, Keiyoro PN, Kelbore AG, Kerbo AA, Khader YS, Khalilarjmandi M, Khan EA, Khan G, Khang YH, Khatab K, Khater A, Khayamzadeh M, Khazaee-Pool M, Khazaei S, Khoja AT, Khosravi MH, Khubchandani J, Kianipour N, Kim D, Kim YJ, Kisa A, Kisa S, Kissimova-Skarbek K, Komaki H, Koyanagi A, Krohn KJ, Bicer BK, Kugbey N, Kumar V, Kuupiel D, La Vecchia C, Lad DP, Lake EA, Lakew AM, Lal DK, Lami FH, Lan Q, Lasrado S, Lauriola P, Lazarus JV, Leigh J, Leshargie CT, Liao Y, Limenih MA, Listl S, Lopez AD, Lopukhov PD, Lunevicius R, Madadin M, Magdeldin S, El Razek HMA, Majeed A, Maleki A, Malekzadeh R, Manafi A, Manafi N, Manamo WA, Mansourian M, Mansournia MA, Mantovani LG, Maroufizadeh S, Martini SMS, Mashamba-Thompson TP, Massenburg BB, Maswabi MT, Mathur MR, McAlinden C, McKee M, Meheretu HAA, Mehrotra R, Mehta V, Meier T, Melaku YA, Meles GG, Meles HG, Melese A, Melku M, Memiah PTN, Mendoza W, Menezes RG, Merat S, Meretoja TJ, Mestrovic T, Miazgowski B, Miazgowski T, Mihretie KMM, Miller TR, Mills EJ, Mir SM, Mirzaei H, Mirzaei HR, Mishra R, Moazen B, Mohammad DK, Mohammad KA, Mohammad Y, Darwesh AM, Mohammadbeigi A, Mohammadi H, Mohammadi M, Mohammadian M, Mohammadian-Hafshejani A, Mohammadoo-Khorasani M, Mohammadpourhodki R, Mohammed AS, Mohammed JA, Mohammed S, Mohebi F, Mokdad AH, Monasta L, Moodley Y, Moosazadeh M, Moossavi M, Moradi G, Moradi-Joo M, Moradi-Lakeh M, Moradpour F, Morawska L, Morgado-da-Costa J, Morisaki N, Morrison SD, Mosapour A, Mousavi SM, Muche AA, Muhammed OSS, Musa J, Nabhan AF, Naderi M, Nagarajan AJ, Nagel G, Nahvijou A, Naik G, Najafi F, Naldi L, Nam HS, Nasiri N, Nazari J, Negoi I, Neupane S, Newcomb PA, Nggada HA, Ngunjiri JW, Nguyen CT, Nikniaz L, Ningrum DNA, Nirayo YL, Nixon MR, Nnaji CA, Nojomi M, Nosratnejad S, Shiadeh MN, Obsa MS, Ofori-Asenso R, Ogbo FA, Oh IH, Olagunju AT, Olagunju TO, Oluwasanu MM, Omonisi AE, Onwujekwe OE, Oommen AM, Oren E, Ortega-Altamirano DDV, Ota E, Otstavnov SS, Owolabi MO, P A M, Padubidri JR, Pakhale S, Pakpour AH, Pana A, Park EK, Parsian H, Pashaei T, Patel S, Patil ST, Pennini A, Pereira DM, Piccinelli C, Pillay JD, Pirestani M, Pishgar F, Postma MJ, Pourjafar H, Pourmalek F, Pourshams A, Prakash S, Prasad N, Qorbani M, Rabiee M, Rabiee N, Radfar A, Rafiei A, Rahim F, Rahimi M, Rahman MA, Rajati F, Rana SM, Raoofi S, Rath GK, Rawaf DL, Rawaf S, Reiner RC, Renzaho AMN, Rezaei N, Rezapour A, Ribeiro AI, Ribeiro D, Ronfani L, Roro EM, Roshandel G, Rostami A, Saad RS, Sabbagh P, Sabour S, Saddik B, Safiri S, Sahebkar A, Salahshoor MR, Salehi F, Salem H, Salem MR, Salimzadeh H, Salomon JA, Samy AM, Sanabria J, Santric Milicevic MM, Sartorius B, Sarveazad A, Sathian B, Satpathy M, Savic M, Sawhney M, Sayyah M, Schneider IJC, Schöttker B, Sekerija M, Sepanlou SG, Sepehrimanesh M, Seyedmousavi S, Shaahmadi F, Shabaninejad H, Shahbaz M, Shaikh MA, Shamshirian A, Shamsizadeh M, Sharafi H, Sharafi Z, Sharif M, Sharifi A, Sharifi H, Sharma R, Sheikh A, Shirkoohi R, Shukla SR, Si S, Siabani S, Silva DAS, Silveira DGA, Singh A, Singh JA, Sisay S, Sitas F, Sobngwi E, Soofi M, Soriano JB, Stathopoulou V, Sufiyan MB, Tabarés-Seisdedos R, Tabuchi T, Takahashi K, Tamtaji OR, Tarawneh MR, Tassew SG, Taymoori P, Tehrani-Banihashemi A, Temsah MH, Temsah O, Tesfay BE, Tesfay FH, Teshale MY, Tessema GA, Thapa S, Tlaye KG, Topor-Madry R, Tovani-Palone MR, Traini E, Tran BX, Tran KB, Tsadik AG, Ullah I, Uthman OA, Vacante M, Vaezi M, Varona Pérez P, Veisani Y, Vidale S, Violante FS, Vlassov V, Vollset SE, Vos T, Vosoughi K, Vu GT, Vujcic IS, Wabinga H, Wachamo TM, Wagnew FS, Waheed Y, Weldegebreal F, Weldesamuel GT, Wijeratne T, Wondafrash DZ, Wonde TE, Wondmieneh AB, Workie HM, Yadav R, Yadegar A, Yadollahpour A, Yaseri M, Yazdi-Feyzabadi V, Yeshaneh A, Yimam MA, Yimer EM, Yisma E, Yonemoto N, Younis MZ, Yousefi B, Yousefifard M, Yu C, Zabeh E, Zadnik V, Moghadam TZ, Zaidi Z, Zamani M, Zandian H, Zangeneh A, Zaki L, Zendehdel K, Zenebe ZM, Zewale TA, Ziapour A, Zodpey S, Murray CJL. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2019;5:1749-1768. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1723] [Cited by in RCA: 1834] [Article Influence: 262.0] [Reference Citation Analysis (0)] |
| 5. | Kirstein MM, Wirth TC. [Multimodal treatment of hepatocellular carcinoma]. Internist (Berl). 2020;61:164-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 6. | Zhou ZF, Peng F, Li JY, Ye YB. Intratumoral IL-12 Gene Therapy Inhibits Tumor Growth In A HCC-Hu-PBL-NOD/SCID Murine Model. Onco Targets Ther. 2019;12:7773-7784. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 7. | Dageforde LA, Fowler KJ, Chapman WC. Liver transplantation for hepatocellular carcinoma: current update on treatment and allocation. Curr Opin Organ Transplant. 2017;22:128-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 8. | O'Rourke JM, Shetty S, Shah T, Perera MTPR. Liver transplantation for hepatocellular carcinoma: pushing the boundaries. Transl Gastroenterol Hepatol. 2019;4:1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 9. | An W, Yao S, Sun X, Hou Z, Lin Y, Su L, Liu X. Glucocorticoid modulatory element-binding protein 1 (GMEB1) interacts with the de-ubiquitinase USP40 to stabilize CFLAR(L) and inhibit apoptosis in human non-small cell lung cancer cells. J Exp Clin Cancer Res. 2019;38:181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 10. | Kawabe K, Lindsay D, Braitch M, Fahey AJ, Showe L, Constantinescu CS. IL-12 inhibits glucocorticoid-induced T cell apoptosis by inducing GMEB1 and activating PI3K/Akt pathway. Immunobiology. 2012;217:118-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 11. | L'Hôte D, Georges A, Todeschini AL, Kim JH, Benayoun BA, Bae J, Veitia RA. Discovery of novel protein partners of the transcription factor FOXL2 provides insights into its physiopathological roles. Hum Mol Genet. 2012;21:3264-3274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 12. | Tsuruma K, Nakagawa T, Morimoto N, Minami M, Hara H, Uehara T, Nomura Y. Glucocorticoid modulatory element-binding protein 1 binds to initiator procaspases and inhibits ischemia-induced apoptosis and neuronal injury. J Biol Chem. 2006;281:11397-11404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 13. | Nakagawa T, Tsuruma K, Uehara T, Nomura Y. GMEB1, a novel endogenous caspase inhibitor, prevents hypoxia- and oxidative stress-induced neuronal apoptosis. Neurosci Lett. 2008;438:34-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 14. | Jiang M, Cheng Y, Wang D, Lu Y, Gu S, Wang C, Huang Y, Li Y. Transcriptional network modulated by the prognostic signature transcription factors and their long noncoding RNA partners in primary prostate cancer. EBioMedicine. 2021;63:103150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 15. | Shibata M, Ham K, Hoque MO. A time for YAP1: Tumorigenesis, immunosuppression and targeted therapy. Int J Cancer. 2018;143:2133-2144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 139] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 16. | Zagiel B, Melnyk P, Cotelle P. Progress with YAP/TAZ-TEAD inhibitors: a patent review (2018-present). Expert Opin Ther Pat. 2022;32:899-912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (1)] |
| 17. | Hao B, Chen X, Cao Y. Yes-associated protein 1 promotes the metastasis of U251 glioma cells by upregulating Jagged-1 expression and activating the Notch signal pathway. Exp Ther Med. 2018;16:1411-1416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 18. | Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, Schinzel AC, Sood S, Rosenbluh J, Kim JW, Zwang Y, Roberts TM, Root DE, Jacks T, Hahn WC. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 539] [Cited by in RCA: 647] [Article Influence: 53.9] [Reference Citation Analysis (0)] |
| 19. | Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747-2761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2041] [Cited by in RCA: 2547] [Article Influence: 141.5] [Reference Citation Analysis (0)] |
| 20. | Sheen-Chen SM, Huang CY, Tsai CH, Liu YW, Wu SC, Huang CC, Eng HL, Chan YC, Ko SF, Tang RP. Yes-associated protein is not an independent prognostic marker in breast cancer. Anticancer Res. 2012;32:3321-3325. [PubMed] |
| 21. | Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, Mu D, Lucito R, Powers S, Lowe SW. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253-1267. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 904] [Cited by in RCA: 913] [Article Influence: 45.7] [Reference Citation Analysis (0)] |
| 22. | Xu MZ, Yao TJ, Lee NP, Ng IO, Chan YT, Zender L, Lowe SW, Poon RT, Luk JM. Yes-associated protein is an independent prognostic marker in hepatocellular carcinoma. Cancer. 2009;115:4576-4585. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 402] [Cited by in RCA: 424] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 23. | Xie Q, Chen J, Feng H, Peng S, Adams U, Bai Y, Huang L, Li J, Huang J, Meng S, Yuan Z. YAP/TEAD-mediated transcription controls cellular senescence. Cancer Res. 2013;73:3615-3624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 138] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 24. | Piccolo FM, Kastan NR, Haremaki T, Tian Q, Laundos TL, De Santis R, Beaudoin AJ, Carroll TS, Luo JD, Gnedeva K, Etoc F, Hudspeth AJ, Brivanlou AH. Role of YAP in early ectodermal specification and a Huntington's Disease model of human neurulation. Elife. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 25. | Kastan N, Gnedeva K, Alisch T, Petelski AA, Huggins DJ, Chiaravalli J, Aharanov A, Shakked A, Tzahor E, Nagiel A, Segil N, Hudspeth AJ. Small-molecule inhibition of Lats kinases may promote Yap-dependent proliferation in postmitotic mammalian tissues. Nat Commun. 2021;12:3100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 145] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 26. | Sun HL, Men JR, Liu HY, Liu MY, Zhang HS. FOXM1 facilitates breast cancer cell stemness and migration in YAP1-dependent manner. Arch Biochem Biophys. 2020;685:108349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 27. | He Q, Lin Z, Wang Z, Huang W, Tian D, Liu M, Xia L. SIX4 promotes hepatocellular carcinoma metastasis through upregulating YAP1 and c-MET. Oncogene. 2020;39:7279-7295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 28. | Chen Z, Wang HW, Wang S, Fan L, Feng S, Cai X, Peng C, Wu X, Lu J, Chen D, Chen Y, Wu W, Lu D, Liu N, You Y, Wang H. USP9X deubiquitinates ALDH1A3 and maintains mesenchymal identity in glioblastoma stem cells. J Clin Invest. 2019;129:2043-2055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 76] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 29. | Inoue K. Feedback enrichment analysis for transcription factor-target genes in signaling pathways. Biosystems. 2020;198:104262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 30. | Turner D, Kim R, Guo JT. TFinDit: transcription factor-DNA interaction data depository. BMC Bioinformatics. 2012;13:220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 31. | Sun J, Li H, Huo Q, Cui M, Ge C, Zhao F, Tian H, Chen T, Yao M, Li J. The transcription factor FOXN3 inhibits cell proliferation by downregulating E2F5 expression in hepatocellular carcinoma cells. Oncotarget. 2016;7:43534-43545. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 32. | Chen C, Ge C, Liu Z, Li L, Zhao F, Tian H, Chen T, Li H, Yao M, Li J. ATF3 inhibits the tumorigenesis and progression of hepatocellular carcinoma cells via upregulation of CYR61 expression. J Exp Clin Cancer Res. 2018;37:263. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 33. | An T, Dong T, Zhou H, Chen Y, Zhang J, Zhang Y, Li Z, Yang X. The transcription factor Krüppel-like factor 5 promotes cell growth and metastasis via activating PI3K/AKT/Snail signaling in hepatocellular carcinoma. Biochem Biophys Res Commun. 2019;508:159-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 34. | Kaul S, Blackford JA Jr, Chen J, Ogryzko VV, Simons SS Jr. Properties of the glucocorticoid modulatory element binding proteins GMEB-1 and -2: potential new modifiers of glucocorticoid receptor transactivation and members of the family of KDWK proteins. Mol Endocrinol. 2000;14:1010-1027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 35. | Oshima H, Szapary D, Simons SS Jr. The factor binding to the glucocorticoid modulatory element of the tyrosine aminotransferase gene is a novel and ubiquitous heteromeric complex. J Biol Chem. 1995;270:21893-21901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 36. | Pan Y, Tong JHM, Lung RWM, Kang W, Kwan JSH, Chak WP, Tin KY, Chung LY, Wu F, Ng SSM, Mak TWC, Yu J, Lo KW, Chan AWH, To KF. RASAL2 promotes tumor progression through LATS2/YAP1 axis of hippo signaling pathway in colorectal cancer. Mol Cancer. 2018;17:102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 37. | Deng J, Lei W, Xiang X, Zhang L, Yu F, Chen J, Feng M, Xiong J. MicroRNA-506 inhibits gastric cancer proliferation and invasion by directly targeting Yap1. Tumour Biol. 2015;36:6823-6831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 38. | Yu SJ, Hu JY, Kuang XY, Luo JM, Hou YF, Di GH, Wu J, Shen ZZ, Song HY, Shao ZM. MicroRNA-200a promotes anoikis resistance and metastasis by targeting YAP1 in human breast cancer. Clin Cancer Res. 2013;19:1389-1399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 130] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 39. | Zhu X, Bu F, Tan T, Luo Q, Zhu J, Lin K, Huang J, Luo C, Zhu Z. Long noncoding RNA RP11-757G1.5 sponges miR-139-5p and upregulates YAP1 thereby promoting the proliferation and liver, spleen metastasis of colorectal cancer. J Exp Clin Cancer Res. 2020;39:207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 40. | Miao W, Li N, Gu B, Yi G, Su Z, Cheng H. MiR-27b-3p suppresses glioma development via targeting YAP1. Biochem Cell Biol. 2020;98:466-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 41. | Zhang P, Liu L, Zhang L, He X, Xu X, Lu Y, Li F. Runx2 is required for activity of CD44(+)/CD24(-/Low) breast cancer stem cell in breast cancer development. Am J Transl Res. 2020;12:2305-2318. [PubMed] |
| 42. | Wu S, Wang H, Li Y, Xie Y, Huang C, Zhao H, Miyagishi M, Kasim V. Transcription Factor YY1 Promotes Cell Proliferation by Directly Activating the Pentose Phosphate Pathway. Cancer Res. 2018;78:4549-4562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 122] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 43. | Xu M, Gu M, Zhou J, Da J, Wang Z. Interaction of YAP1 and mTOR promotes bladder cancer progression. Int J Oncol. 2020;56:232-242. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 44. | Guo L, Chen Y, Luo J, Zheng J, Shao G. YAP1 overexpression is associated with poor prognosis of breast cancer patients and induces breast cancer cell growth by inhibiting PTEN. FEBS Open Bio. 2019;9:437-445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 45. | Guo L, Zheng J, Luo J, Zhang Z, Shao G. Targeting Yes1 Associated Transcriptional Regulator Inhibits Hepatocellular Carcinoma Progression and Improves Sensitivity to Sorafenib: An in vitro and in vivo Study. Onco Targets Ther. 2020;13:11071-11087. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 46. | Roßner F, Sinn BV, Horst D. Pathology of Combined Hepatocellular Carcinoma-Cholangiocarcinoma: An Update. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 47. | Liu B, Nie J, Liang H, Liang Z, Huang J, Yu W, Wen S. Pharmacological inhibition of SETD7 by PFI-2 attenuates renal fibrosis following folic acid and obstruction injury. Eur J Pharmacol. 2021;901:174097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 48. | Ueda H, Neyama H, Sasaki K, Miyama C, Iwamoto R. Lysophosphatidic acid LPA(1) and LPA(3) receptors play roles in the maintenance of late tissue plasminogen activator-induced central poststroke pain in mice. Neurobiol Pain. 2019;5:100020. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Elshimi E, Egypt; He D, China S-Editor: Gong ZM L-Editor: A P-Editor: Zhang YL