Published online Feb 15, 2023. doi: 10.4251/wjgo.v15.i2.276
Peer-review started: November 18, 2022
First decision: January 3, 2023
Revised: January 11, 2023
Accepted: February 2, 2023
Article in press: February 2, 2023
Published online: February 15, 2023
Processing time: 88 Days and 22.4 Hours
Genetic variations are associated with individual susceptibility to gastric cancer. Recently, polygenic risk score (PRS) models have been established based on genetic variants to predict the risk of gastric cancer. To assess the accuracy of current PRS models in the risk prediction, a systematic review was conducted. A total of eight eligible studies consisted of 544842 participants were included for evaluation of the performance of PRS models. The overall accuracy was moderate with Area under the curve values ranging from 0.5600 to 0.7823. Incorporation of epidemiological factors or Helicobacter pylori (H. pylori) status increased the accuracy for risk prediction, while selection of single nucleotide polymorphism (SNP) and number of SNPs appeared to have little impact on the model performance. To further improve the accuracy of PRS models for risk prediction of gastric cancer, we summarized the association between gastric cancer risk and H. pylori genomic variations, cancer associated bacteria members in the gastric microbiome, discussed the potentials for performance improvement of PRS models with these microbial factors. Future studies on comprehensive PRS models established with human SNPs, epidemiological factors and microbial factors are indicated.
Core Tip: A systematic review was conducted to evaluate current polygenic risk score (PRS) models in gastric cancer risk prediction. Our study showed that PRS models had the potential to predict the risk of gastric cancer with a moderate accuracy. The prediction models’ performance could be improved after incorporating epidemiological factors or Helicobacter pylori (H. pylori) status. The potential of H. pylori genomic variations and members of the gastric microbiome were discussed as candidates for gastric cancer prediction models.
- Citation: Wang XY, Wang LL, Xu L, Liang SZ, Yu MC, Zhang QY, Dong QJ. Evaluation of polygenic risk score for risk prediction of gastric cancer. World J Gastrointest Oncol 2023; 15(2): 276-285
- URL: https://www.wjgnet.com/1948-5204/full/v15/i2/276.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v15.i2.276
Gastric cancer (GC) is the fourth most commonly diagnosed type of cancer worldwide and the second leading cause of cancer-related death[1]. According to the latest cancer statistics, there were approximately 26380 new gastric cancer cases and 11090 deaths in the United States in 2022[2]. The occurrence of gastric cancer results from a combination of risk factors, including host genetic factors and Helicobacter pylori (H. pylori) infection[3,4]. Genetic variations play an important role in the occurrence and progression of gastric cancer[5,6]. Genome-wide association studies have identified many single nucleotide polymorphisms (SNPs) in the human genome that are involved in the development of gastric cancer. H. pylori infection affects approximately half of the world’s population. The pathogen is considered a definite carcinogen of gastric cancer[7]. Epidemiological studies have revealed that age, sex, alcohol consumption and smoking are risk factors for gastric cancer[8].
To prevent the development of gastric cancer, it is important to identify individuals at high risk for cancer and apply intervention measures to impede the progression of the disease. Many studies have been conducted to explore the performance of biomarkers or models established with risk factors for predicting gastric cancer risk. Models based on epidemiological factors, including age, sex and H. pylori infection, have adequate performance in the prediction of gastric cancer risk[9,10]. Genetic variations of H. pylori show great potential for use in the prediction of gastric cancer risk[11]. Cancer-associated SNPs have been reported to be valuable in stratifying gastric cancer risk based on the genetic background[12,13].
Polygenic risk score (PRS) models are established with a number of SNPs or genetic variants to explore the combined effect of multiple genetic variations in the risk prediction of disease[14]. They show improved performance in the prediction of the risk of breast, prostate, and colorectal cancer and diseases involving multiple genetic factors[15-17]. Calculations of PRS vary among different models. The simplest way to calculate the PRS is summing the number of all risk alleles[18]. Considering variations in the cancer risk associated with different SNPs, each risk allele is weighted by its odds ratio (OR) value for cancer. PRS is then calculated as a sum of weighted risk alleles[14]. To date, studies have been conducted employing PRS models to predict gastric cancer risk. Different sets of cancer-associated SNPs have been used in the PRS models. In certain studies, epidemiological factors have been included in the establishment of the models. To assess the performance of PRS models in the prediction of gastric cancer risk, this article aimed to comprehensively analyze the accuracy of PRS models for risk prediction through a systematic review of related studies and discuss potentials in the performance improvement of PRS models with the inclusion of H. pylori genetic variations and bacterial members of the gastric microbiome for use in the future.
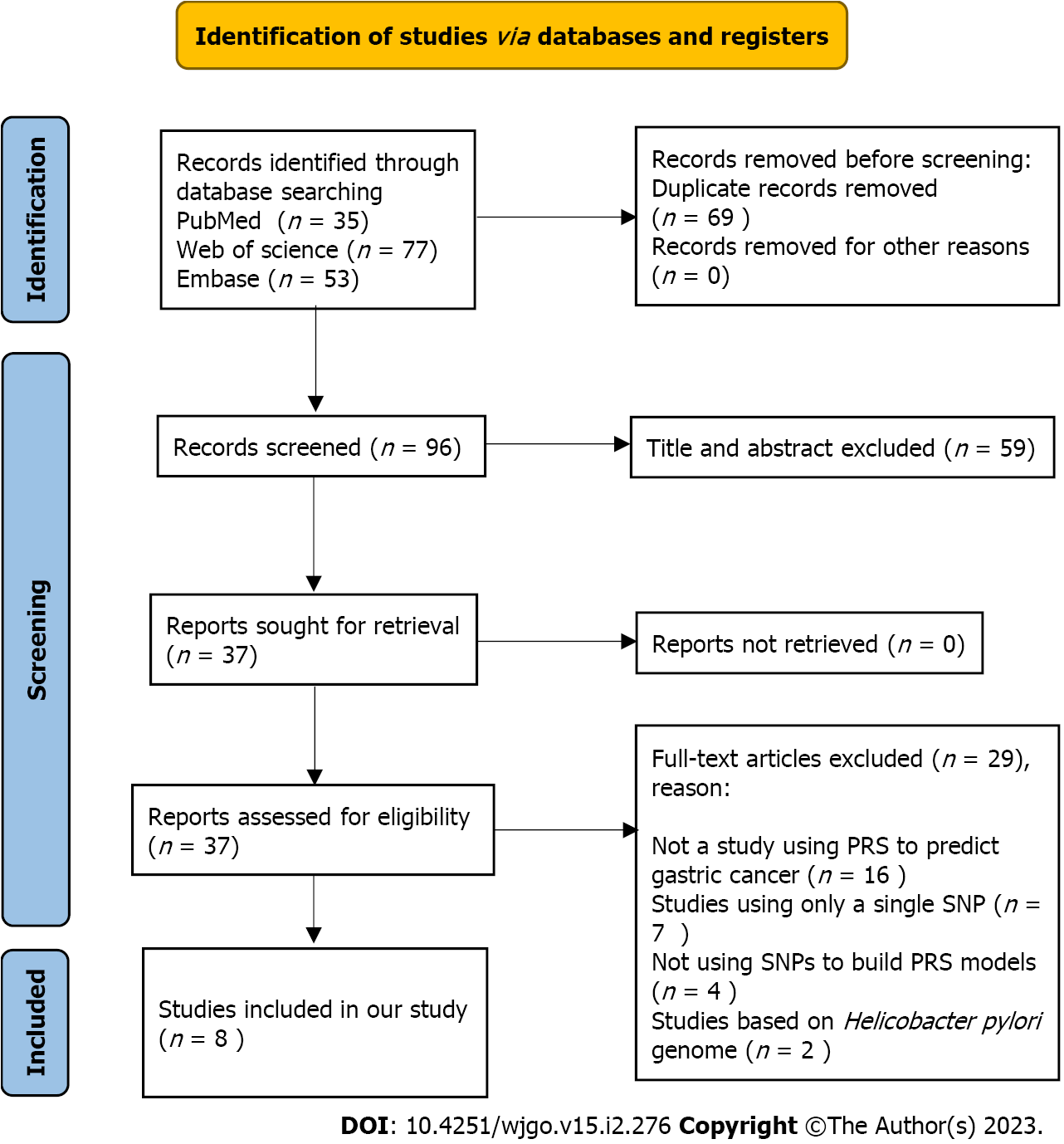
To assess the performance of PRS models for the prediction of gastric cancer risk, a systematic review was conducted. The details of the methods for the systematic review can be found in Supplementary material. The PRISMA flow chart (Figure 1) shows the article retrieval and filtering process. A total of 165 articles were retrieved from PubMed, Web of Science, and Embase electronic database using the search strategies in the Methods (Supplementary material). After removing duplicates, a total of 96 articles were further screened. According to the topics and abstracts, 37 articles were selected and analysed for eligibility. Of them, 8 studies were eligible and included in our systematic review in accordance with the inclusion and exclusion criteria[19-26]. The reasons for exclusion are shown in the PRISMA flow diagram.
The characteristics of the included studies are shown in Table 1. All of the studies were published in the last five years. Six of them were case-control studies, and two were cohort studies. The study areas included China (5), Korea (1), Japan (1) and Europe (1). The sample sizes in the included studies ranged from 1088 to 400807. A total of 544842 participants were included. All the studies established a PRS model to predict the risk of gastric cancer.
| Ref. | Population | Design | Group | Sample size | Sex, (%) | Age, (%) | SNPs (RA, OR) |
| Mao et al[19], 2017 | Chinese | Case-control | GC, HC | 2631, 4373 | Male: 5100 (72.8%); female: 1904 (27.2%) | < 60 yr 3299 (52.9%); ≥ 60 yr 3705 (47.1%) | rs1514175 (A, 1.01), rs2815752 (A, 1.07), rs574367 (T, 1.11), rs12463617 (C, 1.05), rs1861411 (A, 1.02), rs6545814 (G, 1.05), rs10513801 (T, 1.17), rs2535633 (G, 0.98), rs16858082 (T, 0.96), rs261967 (C, 1.02), rs888789 (A, 0.99), rs6890814 (C, 0.99), rs4713766 (A, 1.05), rs9356744 (T, 1.03), rs9473924 (T, 0.98), rs17150703 (G, 1), rs4735692 (A, 1.02), rs11142387 (C, 1.06), rs1211166 (A, 1.02), rs11191580 (C, 0.92), rs10160804 (A, 0.99), rs11030104 (A, 1.02), rs11604680 (G, 0.97), rs2237892 (T, 1), rs671 (G, 1.12), rs897057 (C, 1.04), rs7989336 (A, 1.03), rs9568867 (A, 1.03), rs4776970 (A, 1.05), rs1558902 (A, 1.04), rs2531995 (T, 1.05), rs4788102 (A, 1.08), rs7503807 (A, 1), rs9299 (T, 0.9), rs591166 (A, 1.08), rs11671664 (G, 0.97), rs3810291 (A, 1.02) |
| Choi et al[20], 2020 | European | Cohort | GC | 272 cases in 400807 individuals | Male: 186372 (46.5%); female: 214435 (53.5%) | NR | rs2990223 (G, 1.27), rs10036575 (T, 1.23), rs2294008 (T, 1.21) |
| Jin et al[21], 2020 | Chinese | Cohort | GC | Training set: 10254 cases and 10914 controls; validation set: 692 cases in 100220 individuals | Training set: NR; validation set: Male: 42862 (42.8%); female: 57358 (57.2%) | Training set: NR; validation set: < 60 yr 69805 (69.7%); ≥ 60 yr 30415 (30.3%); mean in case: 60.82 ± 9.33; mean in controls: 53.64 ± 11.00 | NR |
| Qiu et al[22], 2020 | Chinese | Case-control | GC, HC | 1115, 1172 | Male: 1615 (70.6%); female: 672 (29.4%) | < 60 yr 1162 (50.8%); ≥ 60 yr 1125 (49.2%) | rs13361707 (C, 1.47), rs2294008 (T, 1.19), rs4072037 (T, 1.38), rs3762272 (T, 1.21), rs2274223 (G, 1.35), rs80142782 (T, 1.36) |
| Wang et al[23], 2020 | Chinese | Case-control | GC, HC | 2631, 4373 | Male: 5100 (72.8%); female: 1904 (27.2%) | < 60 yr 3299 (52.9%); ≥ 60 yr 3705 (47.1%) | rs1801133 (A, 1.02), rs2275565 (G, 1.01), rs4660306 (T, 1), rs1047891 (A, 1), rs9369898 (A, 1), rs548987 (C, 0.98), rs42648 (G, 1.01), rs1801222 (A, 0.99), rs12780845 (A, 1.01), rs7130284 (C, 1.01), rs2251468 (C, 1.03), rs154657 (A, 1.01), rs12921383 (C, 1.01), rs838133 (A, 1.02), rs234709 (C, 0.99) |
| Duan et al[24], 2021 | Chinese | Case-control | GC, HC | 544, 544 | Male: 825 (75.8%); female: 263 (24.2%) | Mean in case: 57.80 ± 12.06; mean in controls: 57.02 ± 11.97 | rs1859168 (C, 1.09), rs3815254 (A, 0.98), rs4784659 (C, 0.55), rs579501 (A, 0.71), rs77628730 (A, 1.26), rs6989575 (C, 1.03), rs7816475 (A, 1.19), rs6470502 (T, 0.51), rs1518338 (C, 1.08), rs2867837 (G, 0.95), rs12494960 (A, 2.62), rs74798803 (T, 0.97), rs7818137 (T, 1.2), rs550894 (T, 1.13), rs3825071 (A, 1.48), rs580933 (G, 0.98), rs7943779 (A, 1.54), rs911157 (T, 1.74), rs16981280 (C, 0.76), rs2273534 (C, 0.92), rs957313 (T, 1.04) |
| Ishikura et al[25], 2021 | Japanese | Case-control | GC, HC | Training set: 696 cases and 1392 controls; validation set: 795 cases and 795 controls | Training set: Male: 1560 (74.7%); female: 528 (25.3%); validation set: Male: 1180 (74.2%); female: 410 (25.8%) | Training set: < 60 yr 1034 (49.5%); ≥ 60 yr 1054 (50.5%); validation set: < 60 yr 621 (39.1%); ≥ 60 yr 969 (60.9%) | rs4072037 (G, 1.35), rs2294008 (T, 0.62), rs7849280 (G, 0.24) |
| Park et al[26], 2021 | Korean | Case-control | GC, HC | 450, 1136 | Male: 836 (52.7%); female: 750 (47.3%) | Mean in case: 55.4 ± 10.7; mean in control: 52.1 ± 8.5 | rs2294008 (T, 1.2), rs6656150 (C, 0.8), rs8280142782 (C, 0.6), rs760077 (A, 0.8), rs140081212 (A, 0.8), rs4460629 (T, 0.8) |
Details regarding the establishment and evaluation of PRS models in these studies are described in Table 2. The accuracy of the PRS models was assessed with the Area under the curve (AUC) in five included studies. The performance of the models was moderate, with AUC values ranging from 0.5600 to 0.7823. The highest performance has been reported by Ishikura et al[25] from Japan, with an AUC of 0.7677 in the training set and 0.7823 in the validation set. Two studies from China have used ORs to evaluate the performance of PRS models[19,23]. OR values for the highest quartile with respect to the lowest are 1.14 and 1.19, indicating that the performance of the models was unsatisfactory. The hazard ratio (HR) was used in the remaining study with a value of 2.08 for the highest quantile with respect to the lowest. Overall, the performance of the current PRS models varies considerably among the included studies and appears to have a moderate predictive power for gastric cancer. Factors affecting the performance are indicated.
| Ref. | No. of SNPs included | SNP selection | PRS and related methods used to calculate it | AUC or OR (95%CI) of model with PRS | AUC or OR (95%CI) of model with PRS and Clinical risk factors | Difference | Clinical risk factors included |
| Mao et al[19], 2017 | 37 | Significance level and linkage disequilibrium | Weighted PRS using weights derived from the same study | OR for the highest quartile respect to the lower quartile: 1.14 (1.01-1.29) | - | - | - |
| Choi et al[20], 2020 | 3 | Significance level | Weighted PRS using weights derived from literature | AUC: 0.56 (0.53- 0.60); HR for the highest quintiles respect to the lower quintiles: 1.75 (1.18-2.59) | - | - | - |
| Jin et al[21], 2020 | 112 | Significance level | Weighted PRS using weights derived from the same study | HR for the highest quintiles respect to the lower quintiles: 2.08 (1.61-2.69) | HR for participants with a high genetic risk and an unfavorable lifestyle respect to those with a low genetic risk and a favorable lifestyle 5.14 (2.04–12.93) | - | Smoking, alcohol consumption, consumption of preserved foods, intake of fresh fruit and vegetables |
| Qiu et al[22], 2020 | 6 | Significance level and validated to be associated with gastric cancer risk | Weighted PRS using weights derived from the same study | AUC 0.653 | AUC 0.684 | 0.031 | BMI |
| Wang et al[23], 2020 | 15 | Significance level | Weighted PRS using weights derived from literature | OR for the highest quartile respect to the lower quartile: 1.19 (1.04–1.37) | - | - | - |
| Duan et al[24], 2021 | 21 | Prediction functions through bioinformatics tools | Weighted PRS using weights derived from the same study | AUC 0.737 (0.71-0.76); OR for the highest 10% respect to the 40-60%: 5.75 (3.09-10.70) | AUC for PRS + Hp infection: 0.752 (0.690-0.814); AUC for PRS + family history of tumor: 0.773 (0.702-0.843) | PRS + Hp infection: 0.014; PRS + family history of tumor: 0.036 | Hp infection, family history, smoking, alcohol consumption |
| Ishikura et al[25], 2021 | 3 | Significance level | Weighted PRS using weights derived from the same study | AUC for training set: 0.6287 (0.6039–0.6530); AUC for validation set: 0.5673 (0.5391–0.5960) | AUC for training set: 0.7677 (0.7465–0.7890); AUC for validation set: 0.7823 (0.7694–0.8140) | Training set: 0.139; validation set: 0.215 | Smoking, alcohol consumption, fruit and vegetable intake, and ABCD classification |
| Park et al[26], 2021 | 6 | Significance level | Weighted PRS using weights derived from literature | AUC: 0.565 (0.535–0.596); OR for the highest tertile respect to the lower tertile: 2.03 (1.51–2.72) | AUC: 0.607 (0.576–0.638); OR for the highest tertile respect to the lower tertile: 2.53 (1.92–3.34) | 0.042 | A sex-specific prediction model |
Pearson correlation analysis of sample sizes and AUC values demonstrated that there was no significant correlation between them (r = -0.51, P = 0.380). This result suggested that the variations in the sample size of the included studies had minimal influence on the predictive power of the PRS model. The number of genetic variants in the models ranged from 3 to 112 SNPs. Pearson correlation analyses were performed to explore whether the number of SNPs was related to the predictive power of the model. The results showed that there was no significant correlation between the number of SNPs and AUC (r = 0.85, P = 0.067). Our results are consistent with previous systematic reviews of breast cancer[27]. This suggests that the inclusion of more SNPs in the PRS model would not improve the performance in the prediction of gastric cancer risk.
All of the included studies used the weighted PRS method instead of the simple counting method. The weight of risk alleles is crucial for the performance of the PRS models[14,28]. Of note, the same SNP, such as rs2294008, has been used in different models, but the weights varied greatly (Table 1). Accordingly, the predictive power varied among studies (Table 2). Generally, the weight of a risk allele derives from the OR of risk alleles for the development of gastric cancer. In the included studies, the ORs mostly came directly from the results of case-control comparisons. They have not been, however, confirmed in a validation set. This might account for the different weights that have been used among studies. It appears that a validation of the OR is required to eliminate the bias of weights among studies, improving the consistency in the performance of PRS models.
To improve the accuracy of the PRS model in the prediction of cancer risk, certain epidemiological factors implicated in cancer development have been considered[29,30]. A number of epidemiological factors are associated with the occurrence of gastric cancer. Individuals of male sex and older age are at increased risk for gastric cancer[9,31]. Previous studies have revealed environmental factors in gastric cancer development. A family history has been considered to be significantly associated with the occurrence of gastric cancer, with OR of more than 2[32]. Studies have revealed a close association between alcohol consumption and the risk of gastric cancer. A meta-analysis showed that heavy alcohol consumption increases the risk of gastric cancer[33,34]. Smokers have been reported to be at high risk of gastric cancer[35]. In this study, epidemiological factors were included in five studies in addition to genetic variations (Table 2). After taking into account the epidemiological factors, the AUC achieved using each model increased by 0.014 to 0.215. To explore whether the predictive performance of the PRS model was improved after epidemiological factors were included, the Mann-Whitney test was performed. The results showed that the AUC values were significantly increased from 0.56-0.74 to 0.61-0.78 after epidemiological factors were considered (P = 0.047).
H. pylori is a major cause of gastric cancer. The risk of non-cardia gastric cancer in H. pylori-infected individuals is 6 times higher than that in uninfected individuals[36]. Only one of the included studies took H. pylori into account in the establishment of the PRS model. The performance of the model for the prediction of gastric cancer risk increased with an AUC value increasing from 0.737 to 0.752. H. pylori infection serves as a biomarker for gastric cancer and has been combined with other epidemiological factors to predict gastric cancer risk. Tan et al[37] found that H. pylori infection alone had moderate power for predicting gastric cancer risk with an AUC of 0.66. The accuracy of prediction was improved after other clinical factors were incorporated. A study consisting of 14929 participants demonstrated that H. pylori infection combined with seven epidemiological factors has a high predictive power with an AUC value of 0.76[38]. These findings suggest that incorporating H. pylori status into PRS models boosts the predictive power for gastric cancer risk.
In many types of cancer, PRS models have shown great power for risk prediction[39,40]. Our analyses demonstrated that the performance of current PRS models is promising in predicting the risk of gastric cancer. Nonetheless, the predictive power is not as satisfying as expected. Inclusion of epidemiological factors and H. pylori infections likely enhances the performance of the PRS model for the prediction of gastric cancer risk.
In the reported PRS models, the risk of gastric cancer has been predicted mainly based on the genetic susceptibility resulting from genetic variations and common cancer risk factors, including age, sex, smoking status, and alcohol consumption. Previous studies have reported serum pepsinogen status could reflect the extent of atrophic change in gastric mucosa[41]. The combination of serum pepsinogen status and H. pylori status serves as a valuable marker for stratifying the risk of gastric cancer[42]. Individuals with decrease status of pepsinogen and H. pylori infection had a higher risk of gastric cancer compared with healthy control, with a HR value of 6.0[43]. Furthermore, recent studies have demonstrated that many microbial factors play roles in gastric carcinogenesis. Infection with H. pylori causes gastric cancer in only a minority of individuals[44]. Genetic differences between strains of H. pylori account in part for the differential outcomes of the infection among individuals[45]. The dysbiotic gastric microbiome plays an important role in the development of gastric cancer[46,47]. In addition, studies have shown that other bacteria may play an important role in promoting cancer, following the structural imbalance of the stomach microbiome induced by H. pylori[48,49]. As we mentioned, multiple studies have reported other bacteria that are associated with gastric cancer[50,51]. We believe that the gastric microbiome can be used as a valuable candidate to establish a prediction model for the occurrence of gastric cancer.
The genome of H. pylori is substantially diverse[45]. There is a high level of differences in the gene contents, deletion/insertion, genetic inversion, sequence variations and SNPs[52]. Genetic variations in virulence genes, including cagA, vacA, and babA, are closely associated with gastric cancer risk[53,54]. Genome-wide association studies have identified a number of gastric cancer-associated SNPs in the H. pylori genome[55,56]. These cancer-associated genetic variations of H. pylori can be used in the risk prediction of gastric cancer. During the process of screening the studies (Figure 1), we observed that studies used gastric cancer-associated SNPs of the H. pylori genome to predict gastric cancer risk. Using a model comprising six validated loci in the cag pathogenicity island, a study on 1220 subjects demonstrated a sound predictive power for gastric cancer with an AUC of 0.65[57]. Berthenet et al[55] generated a risk score model with 12 gastric cancer-associated SNPs identified by a GWAS study of H. pylori. The results of this study have shown that the model is capable of predicting gastric cancer risk. A recent report established a PRS model with gastric cancer-associated SNPs selected from previous studies[11]. The model based on H. pylori SNPs achieved good predictive performance. These results convincingly support that the incorporation of H. pylori genomic variations into current PRS models would considerably enhance the accuracy in the prediction of gastric cancer risk.
Dysbiosis of the gastric microbiome promotes the development of gastric cancer[46,47]. Many bacteria in the gastric microbiome possess carcinogenic potential[50,51,58].
An observational study of 1043 patients demonstrated a significant enrichment of Streptococcus anginosus (S. anginosus) and Streptococcus constellatus (S. constellatus) in gastric cancer[58,59]. The abundances of S. anginosus and S. constellatus serve as novel faecal signatures of early gastric cancer. Coker et al demonstrated an association between S. anginosus, Peptostreptococcus stomatis, Parvimonas micra, Slackia exigua, Dialister pneumosintes and gastric cancer[60]. These bacteria could form a synergistic network, leading to additional contributions to the disease. They could be used as potential tissue markers for gastric cancer with AUC values of 0.82 and 0.81 in the discovery and validation cohorts, respectively. Png et al[61] conducted a cohort study involving 43 participants to identify potential carcinogenic bacteria. The study demonstrates that the Moryella genus, Vibro genus, Comamonadaceae family, Paludibacter genus, Agrobacterium genus, and Clostridiales order in the gastric microbiome are associated with gastric cancer. The model containing analyses of these bacteria is capable of predicting early gastric cancer with an AUC of 0.82. It has been shown that a random forest model generated with bacterial members of the gastric microbiome has a high performance in risk prediction[62,63]. These findings collectively support that bacterial members of the gastric microbiome have potential in the risk stratification of gastric cancer. Despite the requirement of further validation, the inclusion of the analysis of these bacteria in PRS models most likely enhances the accuracy in the prediction of gastric cancer.
Our systematic review showed that PRS models have great potential in the prediction of gastric cancer. Incorporation of other risk factors for gastric cancer could increase the accuracy of the models. To further increase the predictive performance of PRS models for gastric cancer, a comprehensive PRS model generated with the analysis of epidemiological risk factors, genetic variations of H. pylori, and bacterial members of the gastric microbiome in addition to human genetic variations requires further evaluation. PRS models with high accuracy would benefit the development of individual risk scores, facilitating the prevention of gastric cancer.
| 1. | Fuchs CS, Mayer RJ. Gastric carcinoma. N Engl J Med. 1995;333:32-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 489] [Cited by in RCA: 446] [Article Influence: 14.4] [Reference Citation Analysis (1)] |
| 2. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4235] [Cited by in RCA: 11963] [Article Influence: 2990.8] [Reference Citation Analysis (9)] |
| 3. | Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ. Gastric cancer. Lancet. 2009;374:477-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 692] [Cited by in RCA: 798] [Article Influence: 46.9] [Reference Citation Analysis (1)] |
| 4. | McLean MH, El-Omar EM. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol. 2014;11:664-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 296] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 5. | Zhang H, Jin G, Li H, Ren C, Ding Y, Zhang Q, Deng B, Wang J, Hu Z, Xu Y, Shen H. Genetic variants at 1q22 and 10q23 reproducibly associated with gastric cancer susceptibility in a Chinese population. Carcinogenesis. 2011;32:848-852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 67] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 6. | Mocellin S, Verdi D, Pooley KA, Nitti D. Genetic variation and gastric cancer risk: a field synopsis and meta-analysis. Gut. 2015;64:1209-1219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 145] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 7. | Chmiela M, Karwowska Z, Gonciarz W, Allushi B, Stączek P. Host pathogen interactions in Helicobacter pylori related gastric cancer. World J Gastroenterol. 2017;23:1521-1540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 79] [Cited by in RCA: 95] [Article Influence: 10.6] [Reference Citation Analysis (1)] |
| 8. | Machlowska J, Baj J, Sitarz M, Maciejewski R, Sitarz R. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 897] [Cited by in RCA: 970] [Article Influence: 161.7] [Reference Citation Analysis (0)] |
| 9. | Charvat H, Sasazuki S, Inoue M, Iwasaki M, Sawada N, Shimazu T, Yamaji T, Tsugane S; JPHC Study Group. Prediction of the 10-year probability of gastric cancer occurrence in the Japanese population: the JPHC study cohort II. Int J Cancer. 2016;138:320-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 76] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 10. | Iida M, Ikeda F, Hata J, Hirakawa Y, Ohara T, Mukai N, Yoshida D, Yonemoto K, Esaki M, Kitazono T, Kiyohara Y, Ninomiya T. Development and validation of a risk assessment tool for gastric cancer in a general Japanese population. Gastric Cancer. 2018;21:383-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 11. | Wang XY, Wang LL, Liang SZ, Yang C, Xu L, Yu MC, Wang YX, Dong QJ. Prediction of gastric cancer risk by a polygenic risk score of Helicobacter pylori. World J Gastrointest Oncol. 2022;14:1844-1855. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 2] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 12. | Cai M, Dai S, Chen W, Xia C, Lu L, Qi J, Wang M, Zhou L, Lei F, Zuo T, Zeng H, Zhao X. Environmental factors, seven GWAS-identified susceptibility loci, and risk of gastric cancer and its precursors in a Chinese population. Cancer Med. 2017;6:708-720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 13. | Ji X, Yan Y, Ma N, He G, Wang K, Zhang Y, Yin J, Song C, Wang P, Ye H, Dai L, Zhang J. Variant of SNPs at lncRNA NEAT1 contributes to gastric cancer susceptibility in Chinese Han population. Int J Clin Oncol. 2021;26:694-700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Collister JA, Liu X, Clifton L. Calculating Polygenic Risk Scores (PRS) in UK Biobank: A Practical Guide for Epidemiologists. Front Genet. 2022;13:818574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 116] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 15. | Seibert TM, Fan CC, Wang Y, Zuber V, Karunamuni R, Parsons JK, Eeles RA, Easton DF, Kote-Jarai Z, Al Olama AA, Garcia SB, Muir K, Grönberg H, Wiklund F, Aly M, Schleutker J, Sipeky C, Tammela TL, Nordestgaard BG, Nielsen SF, Weischer M, Bisbjerg R, Røder MA, Iversen P, Key TJ, Travis RC, Neal DE, Donovan JL, Hamdy FC, Pharoah P, Pashayan N, Khaw KT, Maier C, Vogel W, Luedeke M, Herkommer K, Kibel AS, Cybulski C, Wokolorczyk D, Kluzniak W, Cannon-Albright L, Brenner H, Cuk K, Saum KU, Park JY, Sellers TA, Slavov C, Kaneva R, Mitev V, Batra J, Clements JA, Spurdle A, Teixeira MR, Paulo P, Maia S, Pandha H, Michael A, Kierzek A, Karow DS, Mills IG, Andreassen OA, Dale AM; PRACTICAL Consortium*. Polygenic hazard score to guide screening for aggressive prostate cancer: development and validation in large scale cohorts. BMJ. 2018;360:j5757. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 142] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 16. | Lecarpentier J, Silvestri V, Kuchenbaecker KB, Barrowdale D, Dennis J, McGuffog L, Soucy P, Leslie G, Rizzolo P, Navazio AS, Valentini V, Zelli V, Lee A, Amin Al Olama A, Tyrer JP, Southey M, John EM, Conner TA, Goldgar DE, Buys SS, Janavicius R, Steele L, Ding YC, Neuhausen SL, Hansen TVO, Osorio A, Weitzel JN, Toss A, Medici V, Cortesi L, Zanna I, Palli D, Radice P, Manoukian S, Peissel B, Azzollini J, Viel A, Cini G, Damante G, Tommasi S, Peterlongo P, Fostira F, Hamann U, Evans DG, Henderson A, Brewer C, Eccles D, Cook J, Ong KR, Walker L, Side LE, Porteous ME, Davidson R, Hodgson S, Frost D, Adlard J, Izatt L, Eeles R, Ellis S, Tischkowitz M; EMBRACE, Godwin AK, Meindl A, Gehrig A, Dworniczak B, Sutter C, Engel C, Niederacher D, Steinemann D, Hahnen E, Hauke J, Rhiem K, Kast K, Arnold N, Ditsch N, Wang-Gohrke S, Wappenschmidt B, Wand D, Lasset C, Stoppa-Lyonnet D, Belotti M, Damiola F, Barjhoux L, Mazoyer S; GEMO Study Collaborators, Van Heetvelde M, Poppe B, De Leeneer K, Claes KBM, de la Hoya M, Garcia-Barberan V, Caldes T, Perez Segura P, Kiiski JI, Aittomäki K, Khan S, Nevanlinna H, van Asperen CJ; HEBON, Vaszko T, Kasler M, Olah E, Balmaña J, Gutiérrez-Enríquez S, Diez O, Teulé A, Izquierdo A, Darder E, Brunet J, Del Valle J, Feliubadalo L, Pujana MA, Lazaro C, Arason A, Agnarsson BA, Johannsson OT, Barkardottir RB, Alducci E, Tognazzo S, Montagna M, Teixeira MR, Pinto P, Spurdle AB, Holland H; KConFab Investigators, Lee JW, Lee MH, Lee J, Kim SW, Kang E, Kim Z, Sharma P, Rebbeck TR, Vijai J, Robson M, Lincoln A, Musinsky J, Gaddam P, Tan YY, Berger A, Singer CF, Loud JT, Greene MH, Mulligan AM, Glendon G, Andrulis IL, Toland AE, Senter L, Bojesen A, Nielsen HR, Skytte AB, Sunde L, Jensen UB, Pedersen IS, Krogh L, Kruse TA, Caligo MA, Yoon SY, Teo SH, von Wachenfeldt A, Huo D, Nielsen SM, Olopade OI, Nathanson KL, Domchek SM, Lorenchick C, Jankowitz RC, Campbell I, James P, Mitchell G, Orr N, Park SK, Thomassen M, Offit K, Couch FJ, Simard J, Easton DF, Chenevix-Trench G, Schmutzler RK, Antoniou AC, Ottini L. Prediction of Breast and Prostate Cancer Risks in Male BRCA1 and BRCA2 Mutation Carriers Using Polygenic Risk Scores. J Clin Oncol. 2017;35:2240-2250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 135] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 17. | Xin J, Du M, Gu D, Ge Y, Li S, Chu H, Meng Y, Shen H, Zhang Z, Wang M. Combinations of single nucleotide polymorphisms identified in genome-wide association studies determine risk for colorectal cancer. Int J Cancer. 2019;145:2661-2669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 18. | Hung CF, Breen G, Czamara D, Corre T, Wolf C, Kloiber S, Bergmann S, Craddock N, Gill M, Holsboer F, Jones L, Jones I, Korszun A, Kutalik Z, Lucae S, Maier W, Mors O, Owen MJ, Rice J, Rietschel M, Uher R, Vollenweider P, Waeber G, Craig IW, Farmer AE, Lewis CM, Müller-Myhsok B, Preisig M, McGuffin P, Rivera M. A genetic risk score combining 32 SNPs is associated with body mass index and improves obesity prediction in people with major depressive disorder. BMC Med. 2015;13:86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 19. | Mao Y, Yan C, Lu Q, Zhu M, Yu F, Wang C, Dai J, Ma H, Hu Z, Shen H, Jin G. Genetically predicted high body mass index is associated with increased gastric cancer risk. Eur J Hum Genet. 2017;25:1061-1066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 20. | Choi J, Jia G, Wen W, Long J, Zheng W. Evaluating polygenic risk scores in assessing risk of nine solid and hematologic cancers in European descendants. Int J Cancer. 2020;147:3416-3423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 21. | Jin G, Lv J, Yang M, Wang M, Zhu M, Wang T, Yan C, Yu C, Ding Y, Li G, Ren C, Ni J, Zhang R, Guo Y, Bian Z, Zheng Y, Zhang N, Jiang Y, Chen J, Wang Y, Xu D, Zheng H, Yang L, Chen Y, Walters R, Millwood IY, Dai J, Ma H, Chen K, Chen Z, Hu Z, Wei Q, Shen H, Li L. Genetic risk, incident gastric cancer, and healthy lifestyle: a meta-analysis of genome-wide association studies and prospective cohort study. Lancet Oncol. 2020;21:1378-1386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 185] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 22. | Qiu L, Qu X, He J, Cheng L, Zhang R, Sun M, Yang Y, Wang J, Wang M, Zhu X, Guo W. Predictive model for risk of gastric cancer using genetic variants from genome-wide association studies and high-evidence meta-analysis. Cancer Med. 2020;9:7310-7316. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 23. | Wang T, Ren C, Ni J, Ding H, Qi Q, Yan C, Deng B, Dai J, Li G, Ding Y, Jin G. Genetic Association of Plasma Homocysteine Levels with Gastric Cancer Risk: A Two-Sample Mendelian Randomization Study. Cancer Epidemiol Biomarkers Prev. 2020;29:487-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 24. | Duan F, Song C, Wang P, Ye H, Dai L, Zhang J, Wang K. Polygenic Risk Scores for Prediction of Gastric Cancer Based on Bioinformatics Screening and Validation of Functional lncRNA SNPs. Clin Transl Gastroenterol. 2021;12:e00430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 25. | Ishikura N, Ito H, Oze I, Koyanagi YN, Kasugai Y, Taniyama Y, Kawakatsu Y, Tanaka T, Ito S, Tajika M, Shimizu Y, Niwa Y, Matsuo K. Risk Prediction for Gastric Cancer Using GWAS-Identifie Polymorphisms, Helicobacter pylori Infection and Lifestyle-Related Risk Factors in a Japanese Population. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 26. | Park B, Yang S, Lee J, Choi IJ, Kim YI, Kim J. Gastric Cancer Risk Prediction Using an Epidemiological Risk Assessment Model and Polygenic Risk Score. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 27. | Fung SM, Wong XY, Lee SX, Miao H, Hartman M, Wee HL. Performance of Single-Nucleotide Polymorphisms in Breast Cancer Risk Prediction Models: A Systematic Review and Meta-analysis. Cancer Epidemiol Biomarkers Prev. 2019;28:506-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 28. | Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 2020;12:44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 630] [Cited by in RCA: 893] [Article Influence: 148.8] [Reference Citation Analysis (0)] |
| 29. | Liyanarachchi S, Gudmundsson J, Ferkingstad E, He H, Jonasson JG, Tragante V, Asselbergs FW, Xu L, Kiemeney LA, Netea-Maier RT, Mayordomo JI, Plantinga TS, Hjartarson H, Hrafnkelsson J, Sturgis EM, Brock P, Nabhan F, Thorleifsson G, Ringel MD, Stefansson K, de la Chapelle A. Assessing thyroid cancer risk using polygenic risk scores. Proc Natl Acad Sci U S A. 2020;117:5997-6002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 30. | Yiangou K, Kyriacou K, Kakouri E, Marcou Y, Panayiotidis MI, Loizidou MA, Hadjisavvas A, Michailidou K. Combination of a 15-SNP Polygenic Risk Score and Classical Risk Factors for the Prediction of Breast Cancer Risk in Cypriot Women. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 31. | Eom BW, Joo J, Kim S, Shin A, Yang HR, Park J, Choi IJ, Kim YW, Kim J, Nam BH. Prediction Model for Gastric Cancer Incidence in Korean Population. PLoS One. 2015;10:e0132613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 32. | Yaghoobi M, Bijarchi R, Narod SA. Family history and the risk of gastric cancer. Br J Cancer. 2010;102:237-242. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 156] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 33. | Rota M, Pelucchi C, Bertuccio P, Matsuo K, Zhang ZF, Ito H, Hu J, Johnson KC, Palli D, Ferraroni M, Yu GP, Muscat J, Lunet N, Peleteiro B, Ye W, Song H, Zaridze D, Maximovitch D, Guevara M, Fernández-Villa T, Vioque J, Navarrete-Muñoz EM, Wolk A, Orsini N, Bellavia A, Håkansson N, Mu L, Persiani R, Kurtz RC, Lagiou A, Lagiou P, Galeone C, Bonzi R, Boffetta P, Boccia S, Negri E, La Vecchia C. Alcohol consumption and gastric cancer risk-A pooled analysis within the StoP project consortium. Int J Cancer. 2017;141:1950-1962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 91] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 34. | Tramacere I, Negri E, Pelucchi C, Bagnardi V, Rota M, Scotti L, Islami F, Corrao G, La Vecchia C, Boffetta P. A meta-analysis on alcohol drinking and gastric cancer risk. Ann Oncol. 2012;23:28-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 221] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 35. | Poorolajal J, Moradi L, Mohammadi Y, Cheraghi Z, Gohari-Ensaf F. Risk factors for stomach cancer: a systematic review and meta-analysis. Epidemiol Health. 2020;42:e2020004. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 186] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 36. | Yang L, Kartsonaki C, Yao P, de Martel C, Plummer M, Chapman D, Guo Y, Clark S, Walters RG, Chen Y, Pei P, Lv J, Yu C, Jeske R, Waterboer T, Clifford GM, Franceschi S, Peto R, Hill M, Li L, Millwood IY, Chen Z; China Kadoorie Biobank Collaborative Group. The relative and attributable risks of cardia and non-cardia gastric cancer associated with Helicobacter pylori infection in China: a case-cohort study. Lancet Public Health. 2021;6:e888-e896. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 152] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 37. | Tan MC, Ho Q, Nguyen TH, Liu Y, El-Serag HB, Thrift AP. Risk Score Using Demographic and Clinical Risk Factors Predicts Gastric Intestinal Metaplasia Risk in a U.S. Population. Dig Dis Sci. 2022;67:4500-4508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 38. | Cai Q, Zhu C, Yuan Y, Feng Q, Feng Y, Hao Y, Li J, Zhang K, Ye G, Ye L, Lv N, Zhang S, Liu C, Li M, Liu Q, Li R, Pan J, Yang X, Zhu X, Li Y, Lao B, Ling A, Chen H, Li X, Xu P, Zhou J, Liu B, Du Z, Du Y, Li Z; Gastrointestinal Early Cancer Prevention & Treatment Alliance of China (GECA). Development and validation of a prediction rule for estimating gastric cancer risk in the Chinese high-risk population: a nationwide multicentre study. Gut. 2019;68:1576-1587. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 161] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 39. | Opstal-van Winden AWJ, de Haan HG, Hauptmann M, Schmidt MK, Broeks A, Russell NS, Janus CPM, Krol ADG, van der Baan FH, De Bruin ML, van Eggermond AM, Dennis J, Anton-Culver H, Haiman CA, Sawyer EJ, Cox A, Devilee P, Hooning MJ, Peto J, Couch FJ, Pharoah P, Orr N, Easton DF, Aleman BMP, Strong LC, Bhatia S, Cooke R, Robison LL, Swerdlow AJ, van Leeuwen FE. Genetic susceptibility to radiation-induced breast cancer after Hodgkin lymphoma. Blood. 2019;133:1130-1139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 40. | Sassano M, Mariani M, Quaranta G, Pastorino R, Boccia S. Polygenic risk prediction models for colorectal cancer: a systematic review. BMC Cancer. 2022;22:65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 41. | Miki K, Ichinose M, Kawamura N, Matsushima M, Ahmad HB, Kimura M, Sano J, Tashiro T, Kakei N, Oka H. The significance of low serum pepsinogen levels to detect stomach cancer associated with extensive chronic gastritis in Japanese subjects. Jpn J Cancer Res. 1989;80:111-114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 86] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 42. | Yamaji Y, Mitsushima T, Ikuma H, Okamoto M, Yoshida H, Kawabe T, Shiratori Y, Saito K, Yokouchi K, Omata M. Inverse background of Helicobacter pylori antibody and pepsinogen in reflux oesophagitis compared with gastric cancer: analysis of 5732 Japanese subjects. Gut. 2001;49:335-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 79] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 43. | Watabe H, Mitsushima T, Yamaji Y, Okamoto M, Wada R, Kokubo T, Doi H, Yoshida H, Kawabe T, Omata M. Predicting the development of gastric cancer from combining Helicobacter pylori antibodies and serum pepsinogen status: a prospective endoscopic cohort study. Gut. 2005;54:764-768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 315] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 44. | Kumar S, Metz DC, Ellenberg S, Kaplan DE, Goldberg DS. Risk Factors and Incidence of Gastric Cancer After Detection of Helicobacter pylori Infection: A Large Cohort Study. Gastroenterology. 2020;158:527-536.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 203] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 45. | Dong QJ, Zhan SH, Wang LL, Xin YN, Jiang M, Xuan SY. Relatedness of Helicobacter pylori populations to gastric carcinogenesis. World J Gastroenterol. 2012;18:6571-6576. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 10] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 46. | Castaño-Rodríguez N, Goh KL, Fock KM, Mitchell HM, Kaakoush NO. Dysbiosis of the microbiome in gastric carcinogenesis. Sci Rep. 2017;7:15957. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 193] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 47. | Liu X, Shao L, Liu X, Ji F, Mei Y, Cheng Y, Liu F, Yan C, Li L, Ling Z. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine. 2019;40:336-348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 222] [Cited by in RCA: 222] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 48. | de Assumpção PP, Araújo TMT, de Assumpção PB, Barra WF, Khayat AS, Assumpção CB, Ishak G, Nunes DN, Dias-Neto E, Coelho LGV. Suicide journey of H. pylori through gastric carcinogenesis: the role of non-H. pylori microbiome and potential consequences for clinical practice. Eur J Clin Microbiol Infect Dis. 2019;38:1591-1597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 49. | Guo Y, Zhang Y, Gerhard M, Gao JJ, Mejias-Luque R, Zhang L, Vieth M, Ma JL, Bajbouj M, Suchanek S, Liu WD, Ulm K, Quante M, Li ZX, Zhou T, Schmid R, Classen M, Li WQ, You WC, Pan KF. Effect of Helicobacter pylori on gastrointestinal microbiota: a population-based study in Linqu, a high-risk area of gastric cancer. Gut. 2020;69:1598-1607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 228] [Article Influence: 38.0] [Reference Citation Analysis (1)] |
| 50. | Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, Mantilla A, Torres J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci Rep. 2014;4:4202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 278] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 51. | Yang I, Woltemate S, Piazuelo MB, Bravo LE, Yepez MC, Romero-Gallo J, Delgado AG, Wilson KT, Peek RM, Correa P, Josenhans C, Fox JG, Suerbaum S. Different gastric microbiota compositions in two human populations with high and low gastric cancer risk in Colombia. Sci Rep. 2016;6:18594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 124] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 52. | Ge Z, Taylor DE. Contributions of genome sequencing to understanding the biology of Helicobacter pylori. Annu Rev Microbiol. 1999;53:353-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 41] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 53. | da Costa DM, Pereira Edos S, Rabenhorst SH. What exists beyond cagA and vacA? World J Gastroenterol. 2015;21:10563-10572. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 36] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 54. | Nejati S, Karkhah A, Darvish H, Validi M, Ebrahimpour S, Nouri HR. Influence of Helicobacter pylori virulence factors CagA and VacA on pathogenesis of gastrointestinal disorders. Microb Pathog. 2018;117:43-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 95] [Article Influence: 11.9] [Reference Citation Analysis (1)] |
| 55. | Berthenet E, Yahara K, Thorell K, Pascoe B, Meric G, Mikhail JM, Engstrand L, Enroth H, Burette A, Megraud F, Varon C, Atherton JC, Smith S, Wilkinson TS, Hitchings MD, Falush D, Sheppard SK. A GWAS on Helicobacter pylori strains points to genetic variants associated with gastric cancer risk. BMC Biol. 2018;16:84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 56. | Tuan VP, Yahara K, Dung HDQ, Binh TT, Huu Tung P, Tri TD, Thuan NPM, Khien VV, Trang TTH, Phuc BH, Tshibangu-Kabamba E, Matsumoto T, Akada J, Suzuki R, Okimoto T, Kodama M, Murakami K, Yano H, Fukuyo M, Takahashi N, Kato M, Nishiumi S, Azuma T, Ogura Y, Hayashi T, Toyoda A, Kobayashi I, Yamaoka Y. Genome-wide association study of gastric cancer- and duodenal ulcer-derived Helicobacter pylori strains reveals discriminatory genetic variations and novel oncoprotein candidates. Microb Genom. 2021;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 57. | Canzian F, Rizzato C, Obazee O, Stein A, Flores-Luna L, Camorlinga-Ponce M, Mendez-Tenorio A, Vivas J, Trujillo E, Jang H, Chen W, Kasamatsu E, Bravo MM, Torres J, Muñoz N, Kato I. Genetic polymorphisms in the cag pathogenicity island of Helicobacter pylori and risk of stomach cancer and high-grade premalignant gastric lesions. Int J Cancer. 2020;147:2437-2445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 58. | Schulz C, Schütte K, Mayerle J, Malfertheiner P. The role of the gastric bacterial microbiome in gastric cancer: Helicobacter pylori and beyond. Therap Adv Gastroenterol. 2019;12:1756284819894062. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 59. | Zhou CB, Pan SY, Jin P, Deng JW, Xue JH, Ma XY, Xie YH, Cao H, Liu Q, Xie WF, Zou XP, Sheng JQ, Wang BM, Wang H, Ren JL, Liu SD, Sun YW, Meng XJ, Zhao G, Chen JX, Cui Y, Wang PQ, Guo HM, Yang L, Chen X, Ding J, Yang XN, Wang XK, Qian AH, Hou LD, Wang Z, Chen YX, Fang JY. Fecal Signatures of Streptococcus anginosus and Streptococcus constellatus for Noninvasive Screening and Early Warning of Gastric Cancer. Gastroenterology. 2022;162:1933-1947.e18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 77] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 60. | Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, Wu WK, Wong SH, Chen Z, Sung JJY, Yu J. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. 2018;67:1024-1032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 304] [Cited by in RCA: 513] [Article Influence: 64.1] [Reference Citation Analysis (0)] |
| 61. | Png CW, Lee WJJ, Chua SJ, Zhu F; Gastric Consortium5, Yeoh KG, Zhang Y. Mucosal microbiome associates with progression to gastric cancer. Theranostics. 2022;12:48-58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 62. | Liu D, Chen S, Gou Y, Yu W, Zhou H, Zhang R, Wang J, Ye F, Liu Y, Sun B, Zhang K. Gastrointestinal Microbiota Changes in Patients With Gastric Precancerous Lesions. Front Cell Infect Microbiol. 2021;11:749207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 63. | Liu D, Zhang R, Chen S, Sun B, Zhang K. Analysis of gastric microbiome reveals three distinctive microbial communities associated with the occurrence of gastric cancer. BMC Microbiol. 2022;22:184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (1)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kawabata H, Japan; Yuan XX, China S-Editor: Chen YL L-Editor: A P-Editor: Chen YL