Published online Nov 15, 2020. doi: 10.4251/wjgo.v12.i11.1272
Peer-review started: June 1, 2020
First decision: September 11, 2020
Revised: September 24, 2020
Accepted: October 12, 2020
Article in press: October 12, 2020
Published online: November 15, 2020
Processing time: 163 Days and 15.1 Hours
Recent studies have proved the important role of many oncogenic long non-coding RNAs (lncRNAs) in the progression of pancreatic cancer, but little is known about the mechanisms of tumor suppression in pancreatic cancer.
To evaluate the function of tumor suppressor lncRNA C9orf139 in pancreatic cancer progression and to study the underlying mechanism.
We assigned 54 patients with pancreatic ductal adenocarcinoma treated at our hospital to the patient group and 30 normal subjects undergoing physical examination to the control group. RT-qPCR was used to measure the relative expression of C9orf139 in the tissue and serum of patients, in an attempt to investigate the prognostic value of C9orf139 in pancreatic cancer patients. The luciferase reporter gene assay was performed to determine the interaction between C9orf139 and miR-663a. The biological function of C9orf139 was assessed by in vitro assays and in vivo subcutaneous tumor formation tests in animal models. To figure out the molecular mechanism of C9orf139 to act on miR-663a/Sox12, RNA pull-down, Western blot assay, RNA immunoprecipitation assay, and co-immunoprecipitation assay were performed.
C9orf139 level significantly increased in the tissue and serum of patients, which had clinical diagnostic value for pancreatic cancer. Patients with high C9orf139 expression had a higher risk of progressing to stage III + IV, lymph node metastasis, and poor differentiation. Cox regression analysis suggested that C9orf139, tumor-node-metastasis stage, and lymph node metastasis were independent prognostic factors in patients. The underlying mechanism of C9orf139 was that it promoted the growth of pancreatic cancer cells by modulating the miR-663a/Sox12 axis.
C9orf139 is highly expressed in pancreatic cancer, qualified to be used as a potential diagnostic and prognostic marker for pancreatic cancer. Its promotion of pancreatic cancer cell growth is achieved by mediating the miR-663a/Sox12 axis.
Core Tip: C9orf139 is highly expressed in pancreatic cancer, qualified to be used as a potential diagnostic and prognostic marker for pancreatic cancer. Its promotion of pancreatic cancer cell growth is achieved by mediating the miR-663a/Sox12 axis.
- Citation: Ge JN, Yan D, Ge CL, Wei MJ. LncRNA C9orf139 can regulate the growth of pancreatic cancer by mediating the miR-663a/Sox12 axis. World J Gastrointest Oncol 2020; 12(11): 1272-1287
- URL: https://www.wjgnet.com/1948-5204/full/v12/i11/1272.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v12.i11.1272
Pancreatic cancer is the most lethal malignant gastrointestinal tumor in the clinic and is reported to be the fourth leading cause of death from cancer worldwide[1]. According to the latest global tumor statistics[2], new cases and death cases of pancreatic cancer surpassed 400000 last year, and its increasing incidence among younger people has a serious impact on people's life. The best existing treatment for pancreatic cancer is surgical resection. But most patients are already at the middle and advanced tumor stage and are with metastasis at the time of diagnosis, no longer suitable for surgery[3,4]. Radiotherapy is a choice for those unsuitable for surgery, but its treatment outcome is unsatisfactory, which is one of the main reasons for the poor 5-year survival of pancreatic cancer no more than 10%[5,6]. Therefore, it is urgent to figure out the molecular mechanism of pancreatic cancer progression and explore more therapeutic regiments for pancreatic cancer.
The study on non-coding RNAs has been popular in various fields in recent years. The well-known mainstream non-coding RNAs are long non-coding RNAs (lncRNAs), microRNAs, and circular RNAs (circRNAs)[7-9]. MicroRNAs have been frequently studied recently, and they are involved in the development and progression of pancreatic cancer, as well as in tumor resistance and autophagy[10-12]. LncRNAs are a group of non-coding RNAs with more than 200 nt, which were first discovered as "metabolism waste" produced during transcription[13]. Scientific advancement revealed differential expression of lncRNAs and their function of gene regulation in various tumors such as lung cancer[14] , liver cancer[15], breast cancer[16], gastric cancer[17], and pancreatic cancer[18]. More and more studies in recent years have found that lncRNAs can compete with sponge microRNAs as competing endogenous RNAs (ceRNAs) to regulate mRNA[19]. C9orf139 is an important member of the lncRNA family. Song et al[20] found that C9orf139 was differentially expressed in various tumors and was highly expressed in pancreatic cancer, which is expected to be a potential target for pancreatic cancer. But whether it can interact with sponge microRNA to regulate tumor growth is unclear. The result of bioinformatics analysis implied a targeting site between C9orf139 and miR-663a. Previous studies have found that miR-663a is lowly expressed in pancreatic cancer, and the up-regulation of miR-663a can inhibit tumor growth[21].
This study analyzed the possibility of C9orf139 as a target to inhibit tumor growth and provide a potential target for clinical treatment of pancreatic cancer by acting as a ceRNA to sponge miR-663a.
We recruited 54 patients with pancreatic ductal adenocarcinoma admitted to our hospital from May 2013 to May 2014 into this study and assigned them to the patient group. This study has obtained ethical approval from the Ethics Committee of the First Affiliated Hospital of China Medical University. Cancer tissue samples and tumor-adjacent tissue samples were collected from the 54 patients during the operation, transported by liquid nitrogen to the laboratory for testing, and then stored at -80 °C. The peripheral blood of patients from the patient group was also collected before treatment. We also recruited 30 normal people undergoing physical examination during the same period to the control group and collected their peripheral blood. All peripheral blood samples were separated to obtain the serum and then sent to the laboratory for testing immediately. The inclusion criteria were: Patients diagnosed with pancreatic ductal adenocarcinoma by pathological examination; patients in line with the tumor-node-metastasis (TNM) staging criteria issued in 2009; patients with no previous anti-tumor treatment before the study. All patients provided informed consent. Normal people from the control group were not affected by diseases involving the pancreatic system according to imaging test results. The exclusion criteria were: Patients with other tumors; patients not capable of participating in the follow-up; patients with an expected survival of less than 1 mo. The tumor tissue was collected during the surgery. This study followed the Declaration of Helsinki[22]. Patients from the patient group were comparable to people from the control group in age and sex.
The human pancreatic cancer cell lines AsPC-1, BxPC-3, PANC-1, PaCa-2, and SW1990, and the human pancreatic ductal epithelial cell line HPDE6-C7 were from ATCC (Rockville, MD, United States). The cells were cultured in RPMI-1640 or DMEM (Gibco, Waltham, MA, United States) containing 10% fetal bovine serum (FBS). Overexpression plasmid containing C9orf139 (sh-C9orf139), plasmids for knockdown (si-C9orf139-#1, si-C9orf139-#2, and si-C9orf139-#3), and Sox12 expression plasmid (pcDNA3.1-Sox12) were constructed by Shanghai GenePharma Co., Ltd. Sox12 shRNA, Sox12 siRNA, control siRNA, miR-663a mimic, miR-NC, miR-663a inhibitor, and control inhibitor were obtained from RiboBio (Guangzhou, China). Cell transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions.
Cells transfected with plasmids were used to prepare a cell suspension at a density of 4 × 104 cells/mL, which was then seeded in 96-well plates, at 0.1 mL (4 × 103) per well, and incubated at 37 °C under 5% CO2. Then, 10 μL of CCK-8 reagent (Beyotime Biotechnology, Shanghai, CHN) was added to the plates at the 24th, 48th, 72th, and 96th hours of the incubation and cultured for 4 h. The optical density was measured with a microplate reader (MultiskanTM FC Microplate Photometer, CA, Thermo Scientific) at 450 nm.
Cells transfected with plasmids were used to prepare a cell suspension at a density of 4 × 104 cells/mL and suspended in a serum-free medium containing 1 μg/mL mitomycin C to inhibit cell proliferation. Then, the cell suspension was inoculated into the upper chamber of transwell, while 10% FBS was added to the lower chamber and cultured at 37 °C for 24 h. The matrix and cells remaining in the upper chamber that did not migrate through the membrane were wiped off. Then the system was washed three times with PBS and fixed with paraformaldehyde for 10 min, followed by three times of washing with double distilled water. After drying, cells were stained with 0.5% crystal violet and placed under a microscope to observe the cell migration. For wound healing assay, cells were seeded in 6-well plates and scratched artificially with a 200 μL pipette. Wound closure was observed after 24 h and imaged under a microscope.
Cells transfected with plasmids were digested with 0.25% trypsin (Gibco) and washed twice with PBS. Next, cells were added to 100 μL of binding buffer to prepare a suspension at a density of 1 × 106 cells/mL, and the suspension was inoculated with Annexin V-FITC (Yeasen Biotech Co., Ltd.) and PI at room temperature in the dark for 5 min. Finally, cell apoptosis was measured using the FC500MCL flow cytometry system. The assay was repeated three times to obtain the mean value.
Total RNA was extracted from tissues and sera using a TRIzol kit (Invitrogen, United States). The purity, concentration, and integrity of total RNA were measured by ultraviolet spectrophotometry and agarose gel electrophoresis. Reverse transcription was subsequently performed using the TaqManTM Reverse Reverse Transcription Kit (Invitrogen) according to the kit manual, and cDNA was then synthesized for subsequent studies. PCR amplification was performed using the PrimeScript RT Master Mix kit (Takara Bio, Japan). The amplification system was 20 μL in total, containing 10 μL of SYBR qPCR Mix, 0.8 μL of upstream primer, 0.8 μL of downstream primer, 2 μL of cDNA product, 0.4 μL of 50 × ROX reference dye, and RNase-free water to adjust the volume. PCR conditions were pre-denaturation at 95 °C for 60 s and 40 cycles of denaturation at 95 °C for 30 s, and annealing and extension at 60 °C for 40 s. We designed three parallel replicate wells and all specimens were tested three times. U6 was used as the internal reference of microRNAs, and GADPH was used as the internal reference of other genes. The data were analyzed using 2-ΔΔct method[23]. The PCR reaction was performed using the 7500 PCR instrument from ABI.
The cultured cells were lysed using RIPA buffer (Thermo Scientific, Inc., United States) and the protein concentration was measured using a BCA kit (Thermo Scientific, Inc.). The protein was adjusted to a density of 4 μg/μL and separated by 12% SDS-PAGE electrophoresis. The protein was transferred to a 0.22 μm PVDF membrane and blocked with 5% skim milk for 2 h, followed by overnight incubation at 4 °C with Sox12 antibody (dilution 1:1000; Abcam, United States). The primary antibody was washed and the horseradish peroxidase-labeled goat anti-rabbit secondary antibody (dilution 1:5000; Abcam) was added and incubated at 37 °C for 1 h. The membrane was rinsed three times with PBS, 5 min each time. The excess liquid on the membrane was dried with a filter paper, and the ECL reagent was used for color development. The protein bands were scanned and the gray values were analyzed using Quantity One software. GAPDH was used as the internal reference.
RNA immunoprecipitation (RIP) assay was conducted in line with the manual of the EZMagna RIP kit (Millipore, Billerica, MA, United States) to investigate the possibility that C9orf139 and miR-663a interact or bind to the potential binding protein Ago2 in LoVo and HCT116 cells. PaCa-2 cells were lysed and incubated with protein A magbeads that were conjugated with the antibody at 4 °C. Six hours later, the magbeads were washed with washing buffer and then incubated with 0.1% SDS/0.5 mg/mL proteinase K for 30 min at 55 °C to detach proteins. Finally, qRT-PCR analysis of the immunoprecipitated RNA was performed to demonstrate the presence of C9orf139 and miR-663a using specific primers.
PaCa-2 cells were transfected with biotinylated miR-663a-wt, miR-663a-mut, and a negative control (GenePharma, Shanghai, China), respectively. After 48 h, the cell lysate was incubated with M-280 Streptomyces magbeads (Invitrogen) according to the manufacturer's instructions. The level of LINC00152 in the RNA complex bound to the beads was determined using the qRT-PCR method.
A cDNA fragment containing the wild type (C9orf139-WT) or mutant type (C9orf139-mut) fragment was subcloned to the downstream of the luciferase gene of psi-CHECK2 luciferase reporter vector. The 3’ untranslated region (UTR) of Sox12-wt and the corresponding mutant (Sox12-mut) were subcloned to downstream of the luciferase gene of the psi-check2 luciferase reporter vector. The miR-663a mimic or inhibitor was co-transfected with the C9orf139-WT or C9orf139-mut reporter vector (Invitrogen, United States). After 48 h of transfection, the luciferase activity of firefly and renin in the cell lysates was continuously determined using a dual-luciferase reporter kit (Promega, United States). Similarly, Sox12-WT or Sox12-mut was co-transfected with miR-663a mimic.
This animal experiment was approved by the ethics committee of our hospital. Nude mice (4-5 wk old, male, n = 5 per group) were from Charles River Laboratories (China). Subcutaneous injection of 200 mL of transfected PaCa-2 cells (2 × 106) was performed on the left side of the back of each nude mouse. Tumor size was measured periodically using the formula: 0.52 × L × W2 (L refers to the tumor length, W refers to tumor width). Mice were euthanized on the 30th day after the injection, and the tumor was collected.
The survival of patients was followed for 5 years via the telephone or the records of reexamination on the 1st, 3rd, 6th, 9th, and 12th months of each year.
Data visualization and data analyses were performed using GraphPad 7 software package. The analysis of independent prognostic factors was conducted using SPSS20.0 software. The distribution of the measurement data was analyzed by the K-S test. Data with a normal distribution are expressed as the mean ± standard deviation (means ± SD) and were compared between the two groups by the independent sample t-test. Count data are expressed as percentages (%) and were compared between the two groups by the chi-square test. The comparison between multiple groups was performed by one-way ANOVA. The LSD-t test was used for post-hoc pairwise comparison. The comparison between multiple time points was performed by repeated measure ANOVA. The Bonferroni test was used for post-hoc test. Receiver operating characteristic (ROC) curve analysis was employed to assess the diagnostic value of C9orf139 in pancreatic cancer. Pearson correlation coefficient was used to analyze the correlation between genes. Kaplan-Meier survival curve was plotted to display the overall survival that was analyzed by the Log-rank test. Multivariate Cox regression was conducted to predict the prognosis of patients. P < 0.05 indicated a statistical difference.
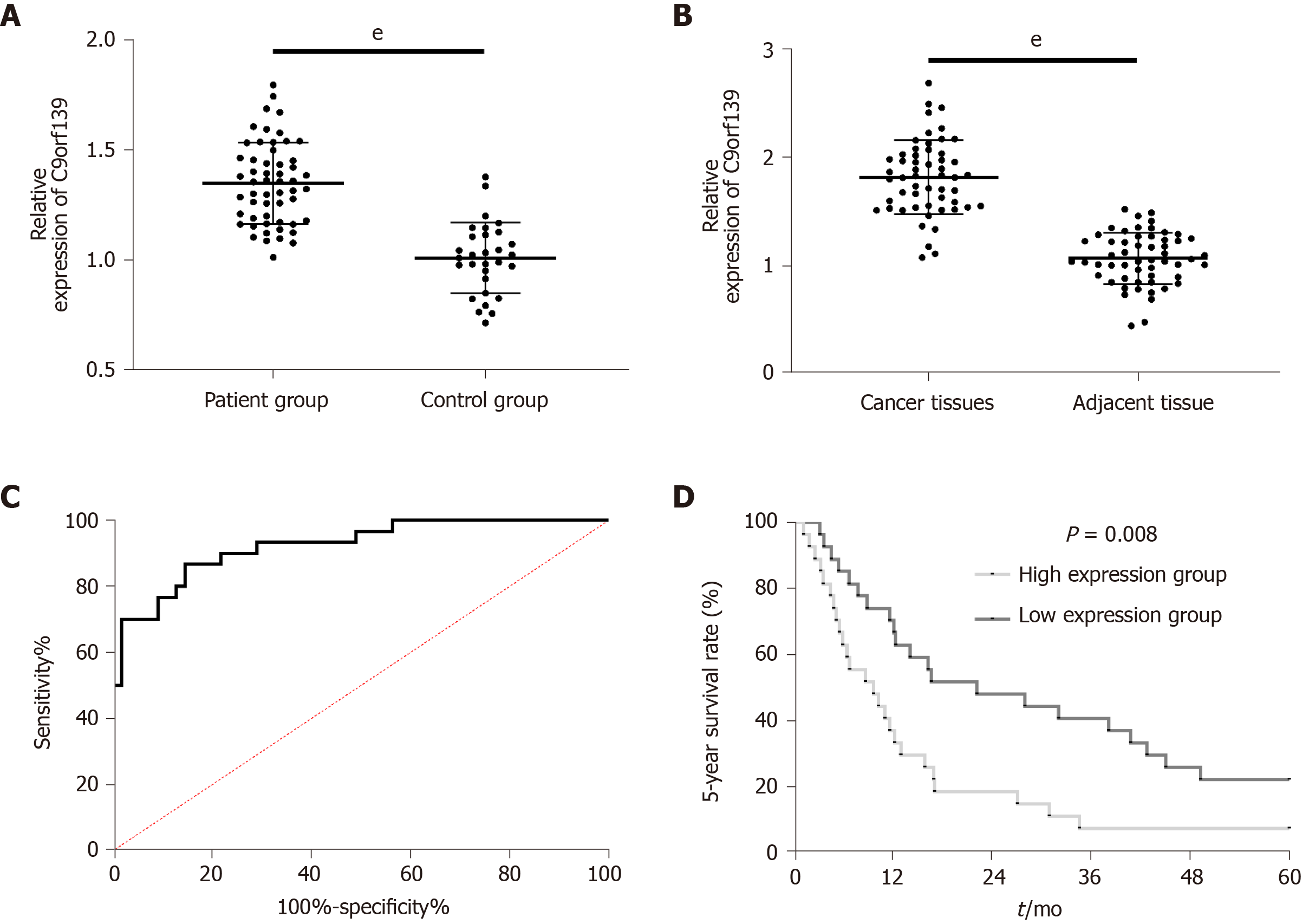
Clinical detection revealed a marked increase in C9orf139 expression in patients' tissues or sera. The area under the ROC curve was 0.923. To investigate the relationship between C9orf139 and pathological data of patients with pancreatic cancer, we divided patients into the high expression group and low expression group according to the median value of C9orf139 level. Patients with high expression had markedly higher risks of progressing to stage III + IV, lymph node metastasis, and poor differentiation. The 5-year survival of patients in the high expression group was significantly reduced. Cox regression analysis demonstrated that C9orf139, TNM stage, and lymph node metastasis were independent prognostic factors for pancreatic cancer patients. More details are shown in Figures 1 and 2 and Tables 1 and 2.
| Variable | LncRNA C9orf139 | χ2 | P value | ||
| High expression (n = 27) | Low expression (n = 27) | ||||
| Age | 0.260 | 1.271 | |||
| ≥ 60 yr (n = 20) | 12 (44.44) | 8 (29.63) | |||
| < 60 yr (n = 34) | 15 (55.56) | 19 (70.37) | |||
| Sex | 0.307 | 0.580 | |||
| Male (n = 32) | 15 (55.56) | 17 (62.96) | |||
| Female (n = 22) | 12 (44.44) | 10 (37.04) | |||
| Tumor size | 0.333 | 0.564 | |||
| ≥ 3 cm (n = 18) | 10 (37.04) | 8 (29.63) | |||
| < 3 cm (n = 36) | 17 (62.96) | 19 (70.37) | |||
| TNM stage | 9.826 | 0.002 | |||
| I-II (n = 35) | 12 (44.44) | 23 (85.19) | |||
| III-IV (n = 19) | 15 (55.56) | 4 (14.81) | |||
| Degree of differentiation | 4.207 | 0.040 | |||
| Poor (n = 17) | 12 (44.44) | 5 (18.52) | |||
| Moderate and high (n = 37) | 15 (55.56) | 22 (81.48) | |||
| Lymph node metastasis | 5.206 | 0.004 | |||
| Yes (n = 13) | 11 (40.74) | 2 (7.41) | |||
| No (n = 41) | 16 (59.26) | 25 (92.59) | |||
| Variable | Univariate Cox regression analysis | Multivariate Cox regression analysis | ||||
| P value | HR | 95CI% | P value | HR | 95CI% | |
| Age (≥ 60 yr vs < 60 yr) | 0.637 | 1.158 | 0.631-2.125 | |||
| Sex (male vs female) | 0.885 | 0.957 | 0.528-1.734 | |||
| Tumor size (≥ 3 cm vs < 3 cm) | 0.160 | 0.648 | 0.354-1.187 | |||
| TNM stage (I + II vs III + IV) | 0.035 | 1.932 | 1.047-3.564 | 0.109 | 1.724 | 0.886-3.353 |
| Degree of differentiation (poor vs moderate and high) | 0.019 | 0.477 | 0.257-0.886 | 0.045 | 0.526 | 0.281-0.987 |
| Lymph node metastasis (yes vs no) | 0.019 | 0.456 | 0.237-0.877 | 0.167 | 0.622 | 0.317-1.219 |
| C9orf139 (low expression vs high expression) | 0.010 | 0.455 | 0.251-0.825 | 0.020 | 0.488 | 0.267-0.893 |
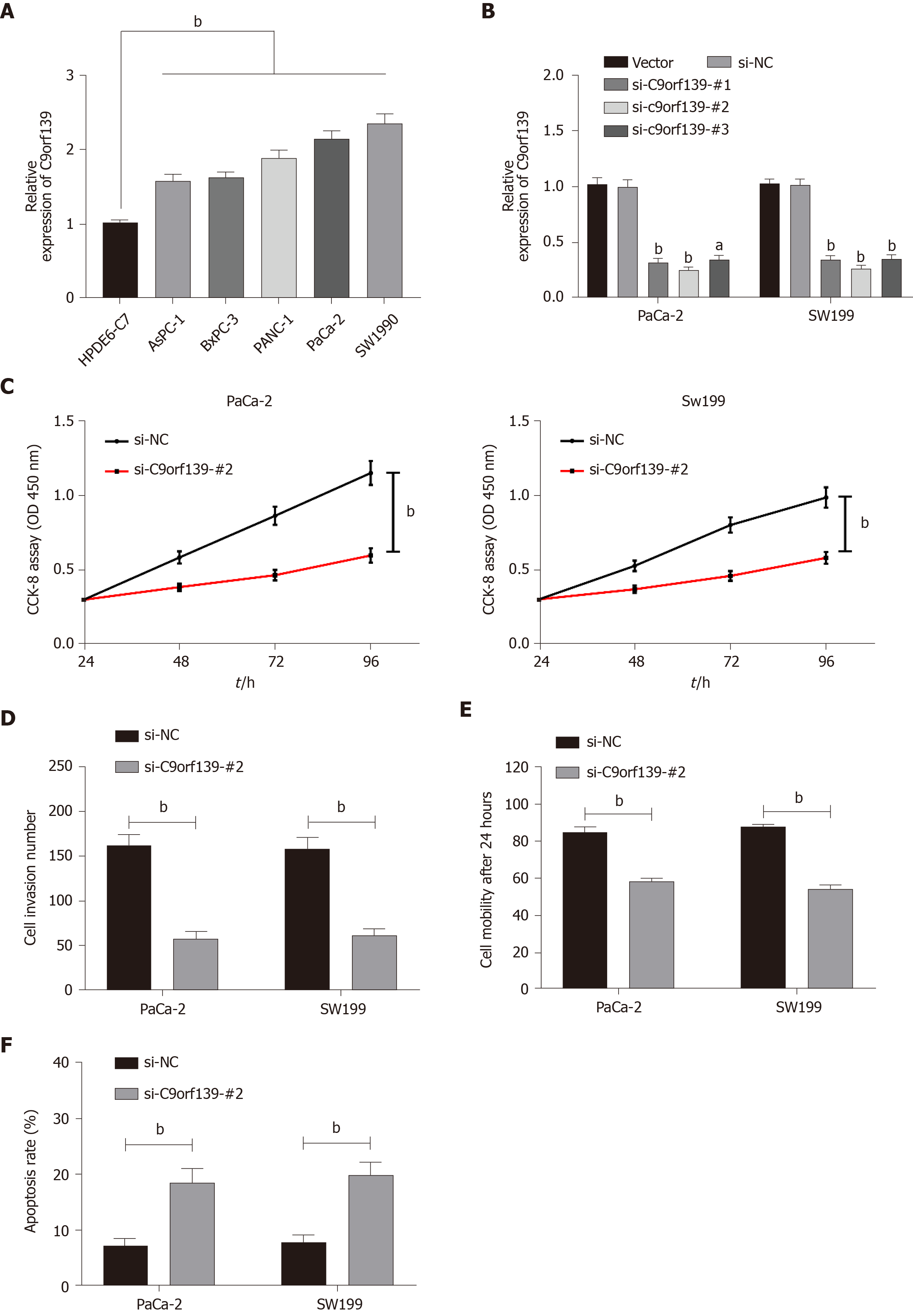
Clinical detection revealed high expression of C9orf139 in pancreatic cancer cells. For further verification, we tested C9orf139 expression in each group of cells and confirmed that C9orf139 was highly expressed in all pancreatic cancer cells. We selected PaCa-2 and SW1990 cells in which C9orf139 expression was especially high for the following experiments. To investigate the effect of C9orf139 on pancreatic cancer cells, we established three C9orf139 knockout expression vectors (si-C9orf139#1-#3) and transfected them into PaCa-2 and SW199 cells. We detected that si-C9orf139 #2 had the lowest relative expression of C9orf139, so we selected si-C9orf139#2 for subsequent experiments. We conducted relevant experiments to test the biological condition of the transfected cells. The results of CCK-8 assay revealed great inhibition of cell growth in pancreatic cancer cells transfected with si-C9orf139#2, and Transwell assay and wound-healing assay demonstrated a marked decrease in the number of cells migrating through the membrane and the migration rate in cells transfected with si-C9orf139#2. Flow cytometry showed a marked increase in the apoptosis of cells transfected with si-C9orf139#2. Such results suggested that knockdown of lncRNA C9orf139 inhibits the growth of pancreatic cancer cells. More details are shown in Figure 2.
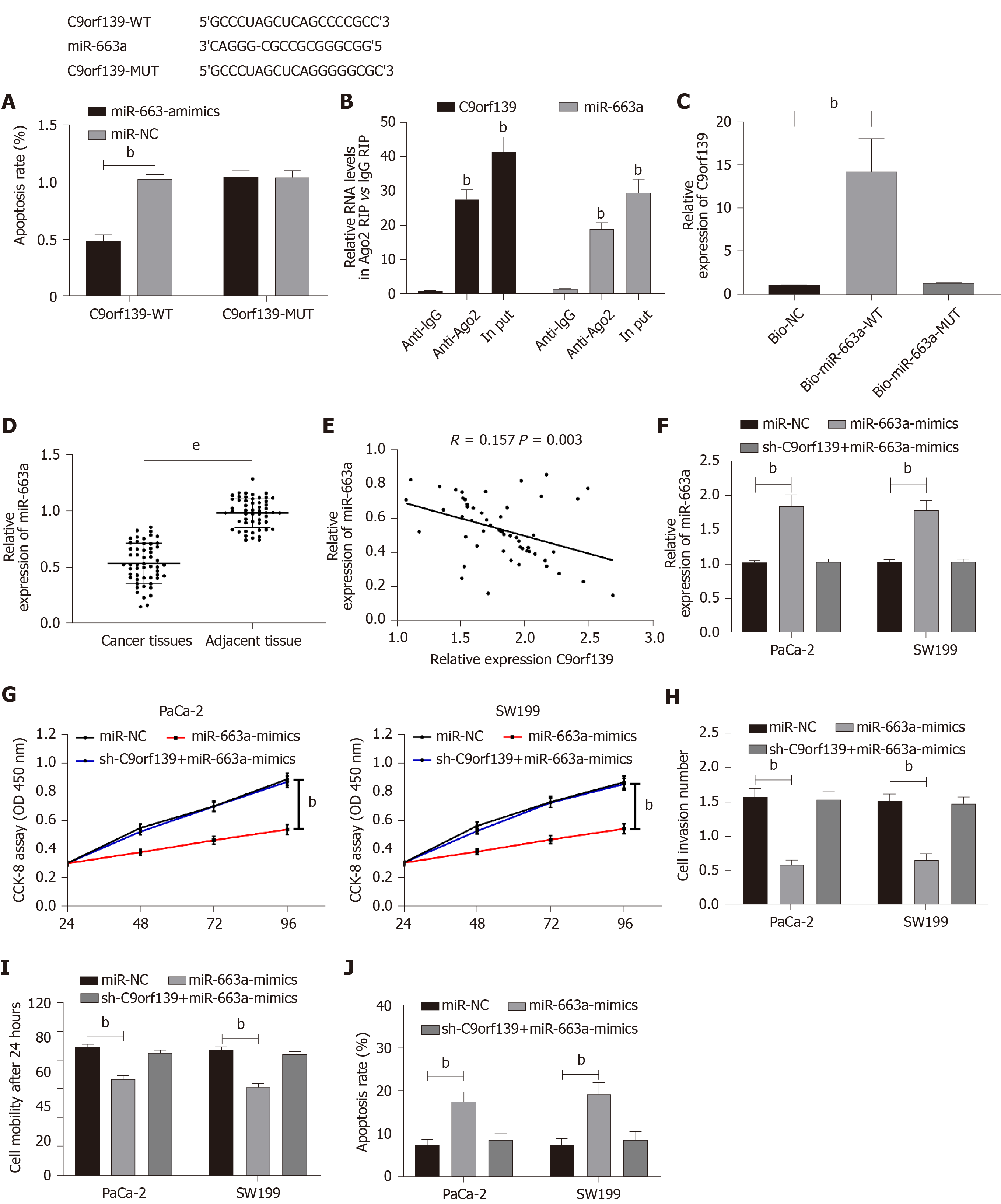
Online miR target prediction of C9orf139 (http://starbase.sysu.edu.cn/) found a potential binding target between C9orf139 and miR-663a. We conducted dual-luciferase reporter assay, RIP assay, and RNA pull-down assay to confirm this. The dual-luciferase report assay showed that the fluorescence activity of C9orf139-WT was significantly inhibited by miR-663a-mimic. RIP assay showed that the levels of C9orf139 and miR-663a precipitated by Ago2 antibody were significantly higher than those precipitated by IgG. RNA pull-down assay revealed that C9orf139 could be pulled down by biotin-labeled miR-663a-WT, not by miR-663a-mut. The detection of miR-663a expression in the patient tissue showed that miR-663a expression was significantly decreased in the patient tissue, and correlation analysis revealed a negative correlation between miR-663a and C9orf139 in the patient tissue. To further confirm the effects of C9orf139 and miR-663a on pancreatic cancer cells, we transfected pancreatic cancer cells with miR-663a-mimic, sh-C9orf139 + miR-663a-mimic, and miR-NC, respectively. We found that cell proliferation, invasion, and migration were significantly inhibited, and the apoptosis was significantly increased in cells transfected with miR-663a-mimic, while the biological behaviors of cells transfected with sh-C9orf139 + miR-663a-mimic were reversed and contrary to cells transfected with miR-663a-mimic alone. Such results indicated that up-regulated C9orf139 can inhibit the expression of miR-663a and promote tumor cell growth. More details are shown in Figure 3.
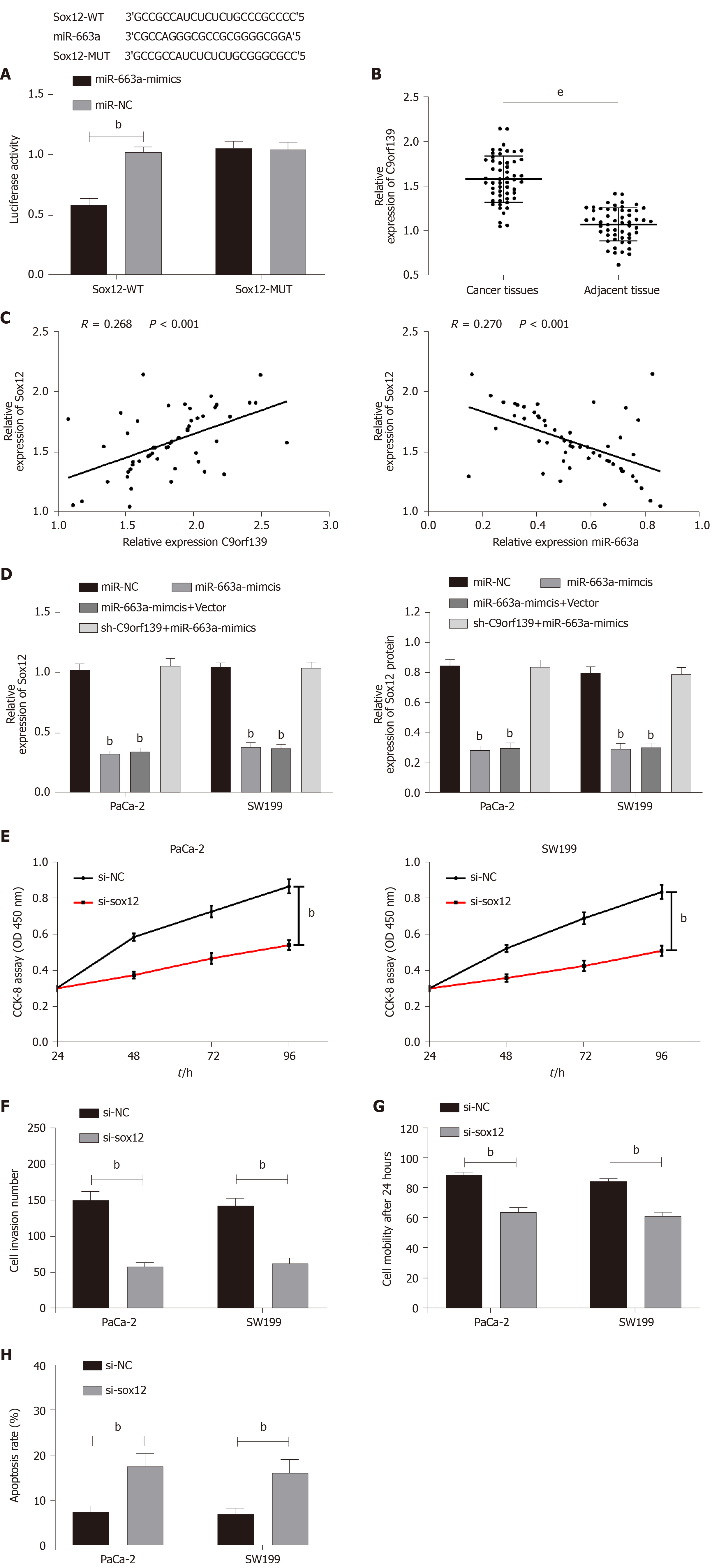
The main regulatory mode of microRNAs is performed through the regulation of transcription of downstream target genes. Bioinformatics analysis revealed a targeted binding site for miR-663a and Sox12. We conducted the dual-luciferase reporter assay and detected Sox12 expression in cells after the transfection and confirmed that there was targeting binding between Sox12 and miR-663a. We transfected cells with miR-NC, miR-663a-mimic, miR-663a-mimic + vector, and sh-C9orf139 + miR-663a-mimic and then detected the expression of Sox12. The results demonstrated that the expression of Sox12 protein and mRNA was suppressed in cells with up-regulated miR-663a, but the expression results were reversed after the co-transfection of sh-C9orf139 and miR-663a-mimic. We detected significantly higher Sox12 expression in the cancer tissues than in the adjacent tissues. Further correlation analysis showed that Sox12 was positively correlated with C9orf139 expression, but negatively correlated with miR-663a. To verify the effect of Sox12 on pancreatic cancer cells, we transfected pancreatic cancer cells with si-Sox12 and si-NC. The results showed that knockdown of Sox12 led to inhibited proliferation, invasion, migration, and increased apoptosis. More details are shown in Figure 4.
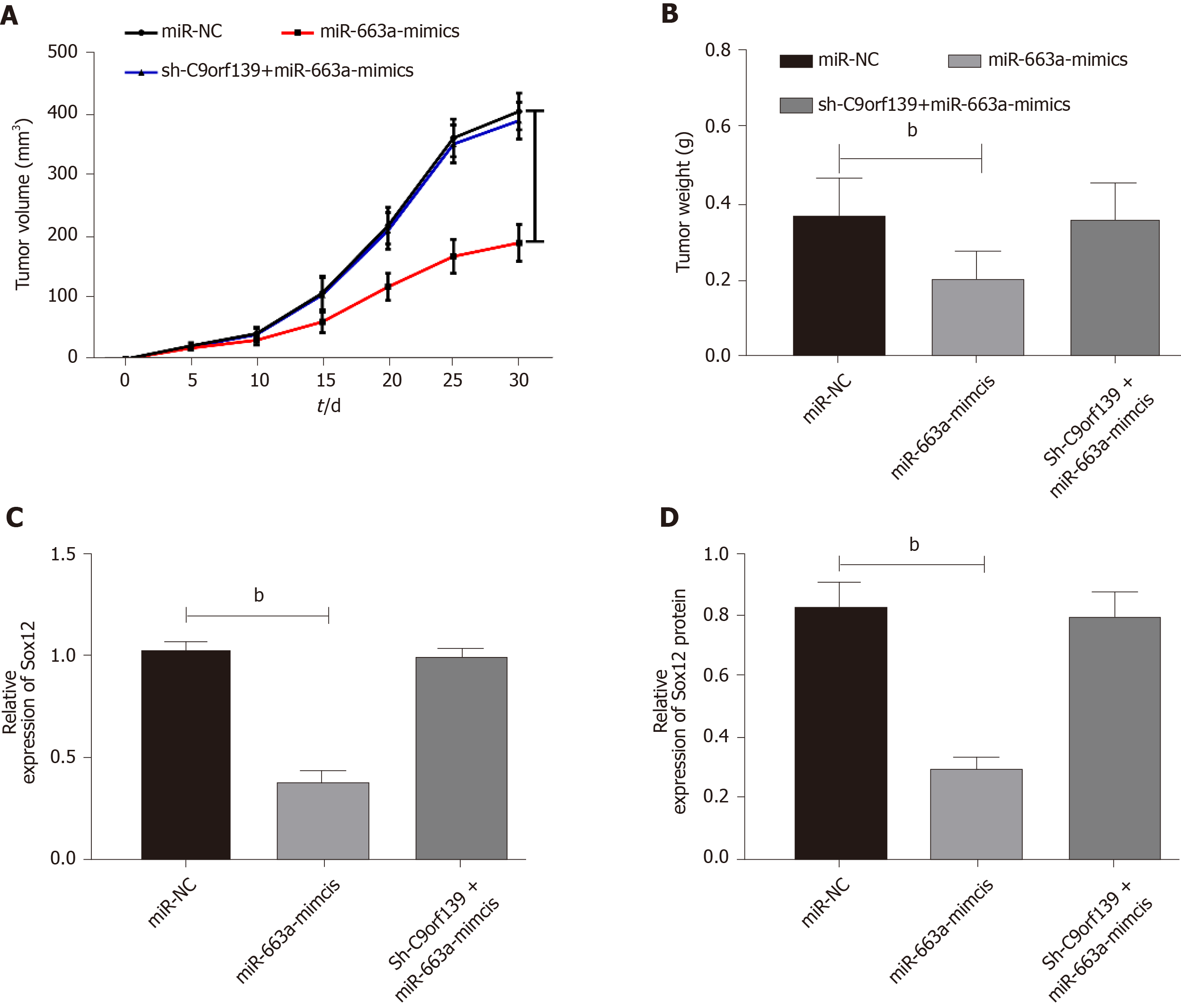
Tumor formation in nude mice was conducted to observe whether lncRNA C9orf139 affects solid tumors through the miR-663a/Sox12 axis. We subcutaneously injected miR-NC, miR-663a-mimic, or sh-C9orf139 + miR-663a-mimic into nude mice, and found that the tumor size and weight were markedly decreased in nude mice injected with miR-663a-mimic, while the tumor size and weight in nude mice injected with sh-C9orf139 + miR-663a-mimic were not significantly different from those in nude mice injected with miR-NC. The expression of Sox12 protein and mRNA in the tumor in nude mice injected with miR-663a-mimic and mice injected with miR-663a-mimic + vector was significantly inhibited, while the inhibition on Sox12 protein and mRNA was reversed in mice injected with C9orf139 + miR-663a-mimic. More details are shown in Figure 5.
In this study, we detected high C9orf139 expression in pancreatic cancer and found that patients with higher C9orf139 expression had markedly decreased 5-year survival and increased risks of progressing to stage III + IV, poor differentiation, and lymph node metastasis. We also found that C9orf139 could promote the growth of pancreatic cancer cells via the miR-663a/Sox12 axis, which indicates the capacity of C9orf139 to be a potential indicator for the treatment of pancreatic cancer.
The5-year survival of pancreatic cancer patients was at the bottom among digestive tract tumors and even among all malignant tumors[24]. The low 5-year survival of pancreatic cancer is mostly attributed to the fact that most patients are already at middle or advanced stages with high risks of lesion metastasis at the time of diagnosis, which disqualifies patients for surgery. Chemoradiotherapy can improve the prognosis of patients, but its treatment efficacy is poor, so the relevant mechanisms of pancreatic cancer should be figured out[25,26].
LncRNAs are non-coding RNAs longer than 200 nt. They have been found to play an important role in epigenetic, transcriptional, and post-transcriptional regulation of gene expression[27]. It is also reported to be closely related to the prognosis of a variety of tumors. Huang et al[28] found that the overexpression of lncRNA PVT1 led to a poor prognosis in patients with pancreatic cancer. A previous study[29] reported that lncRNA-ATB low expression was an independent predictor of poor prognosis in patients with pancreatic cancer. C9orf139, a newly discovered lncRNA located on human 9q34.3 chromosome, was found to be highly expressed in pancreatic cancer and a promising prognostic indicator for pancreatic cancer in the studies by Shi et al[30] and Wei et al[31]. The mechanism of C9orf139 in pancreatic cancer remains unclear. In this study, we found that C9orf139 was highly expressed in tissues and sera of pancreatic cancer patients and in pancreatic cancer cell lines, which was consistent with the results of the above-mentioned studies. Then we further analyzed the relationship between C9orf139 and pathological data and survival prognosis of patients with pancreatic cancer. The results demonstrated that patients with higher expression of C9orf139 were subjected to higher risks of progressing to stage III + IV, poor differentiation, and lymph node metastasis. ROC curve analysis revealed high diagnostic value of C9orf139 for pancreatic cancer and a markedly reduced 5-year survival in patients with high C9orf139 expression. Cox regression analysis found that high C9orf139 expression was an independent predictor of poor prognosis in patients with pancreatic cancer. Such results proved that C9orf139 is close related to the development of pancreatic cancer.
The underlying mechanism of action of C9orf139 in pancreatic cancer has not been figured out. The main mechanism of lncRNAs lies in their regulation of gene expression by competing with microRNA elements as ceRNAs[32]. Our bioinformatics prediction suggested binding sites between miR-663a and C9orf139. miR-663a is an important member of the microRNA family. A former study concluded that[33] miR-663a was lowly expressed in the serum of patients with pancreatic cancer and could be used as a potential diagnostic marker for pancreatic cancer, suggesting that miR-663a may also be involved in the progression of pancreatic cancer. We conducted a dual-luciferase reporter assay, RIP assay, and RNA pull-down assay to verify the relationship between the miR-663a and C9orf139. Dual-luciferase reporter assay revealed targeting binding between miR-663a and C9orf139. RIP assay showed that the levels of C9orf139 and miR-663a precipitated with Ago2 antibody were significantly higher than those precipitated with IgG. RNA pull-down assay found that C9orf139 could be pulled down by biotin-labeled miR-663a-wt, not by miR-663a-mut. The above assays suggested that C9orf139 can regulate miR-663a as a ceRNA. To investigate the effects of C9orf139 and miR-663a on the growth of pancreatic cancer cells, we transfected pancreatic cancer cells with si-C9orf139 and miR-663a-mimic and discovered that the up-regulation of miR-663a and knockout of C9orf139 resulted in inhibited cell proliferation, invasion, and migration, as well as increased apoptosis. Such results indicated that the growth of pancreatic cancer cells can be affected by the expression of C9orf139 and miR-663a. To verify this, we co-transfected pancreatic cancer cells with miR-663a-mimic and sh-C9orf139 and found that the inhibited cell proliferation, invasion, and migration, and increased apoptosis caused by the up-regulated miR-663a were completely reversed by the transfection with sh-C9orf139. This demonstrates that the up-regulation of C9orf139 can regulate miR-663a to promote the growth of pancreatic cancer cells.
One of the important mechanisms of microRNAs lies in their regulation of mRNA to affect the biological functions[34]. Our bioinformatics analysis discovered a targeted binding site between miR-663a and Sox12. Sox12 is a member of the SOX transcription factor family. In the study by Wang et al[35], Sox12 was found to be highly expressed in pancreatic cancer and miR-26a could inhibit the proliferation, migration, and invasion of pancreatic cancer cells by regulating Sox12. Dual-luciferase report assay revealed a targeting binding site between miR-663a and Sox12. In this study, knockdown of Sox12 led to inhibited proliferation, invasion, migration, and increased apoptosis. In order to confirm whether C9orf139 can affect pancreatic cancer via the miR-663a/Sox12 axis, we tested the expression of C9orf139, miR-663a, and Sox12 in patient tissues and conducted correlation analysis. It was found that Sox12 was positively correlated with C9orf139 expression and negatively correlated with miR-663a, and miR-663a was negatively correlated with C9orf139. We also found that up-regulated miR-663a led to inhibited Sox12 expression, but the expression of Sox12 mRNA and protein was reversed by the co-transfection with up-regulated C9orf139 plasmid. At the end of this study, we conducted tumor formation in nude mice and confirmed that the C9orf139-mediated miR-663a/Sox12 axis was involved in the growth of pancreatic cancer.
This study was subject to some limitations. First, we did not collect patients with benign pancreatic lesions to measure their C9orf139 expression, so whether C9orf139 can be used as the diagnostic indicator for benign pancreatic lesions and pancreatic cancer has not confirmed. Second, the presence or absence of the connection between C9orf139 and drug resistance, which is the most common condition in patients with pancreatic cancer, is not clear. To perfect our findings, we will collect more samples and research cases with different types and conduct bioinformatics analysis to explore the potential links between C9orf139 and drug resistance in pancreatic cancer.
In conclusion, C9orf139 is highly expressed in pancreatic cancer, qualified to be used as a potential diagnostic and prognostic marker for pancreatic cancer. Its promotion of pancreatic cancer cell growth is achieved via the miR-663a/Sox12 axis.
Pancreatic cancer is one of the tumors with the lowest 5-year survival rate, and its incidence has been surging in recent years. Many studies have confirmed the critical role of long non-coding RNAs (lncRNAs) in the development and progression of pancreatic cancer, but little has been known about C9orf139 in pancreatic cancer.
To identify biomarkers for the diagnosis and treatment of pancreatic cancer.
To explore the mechanism of action of lncRNA-C9orf139 in pancreatic cancer.
The relative expression of C9orf139 in tissues and sera of patients with pancreatic cancer was tested by RT-qPCR. The predictive value of C9orf139 for pancreatic cancer prognosis and the interaction between C9orf139 and miR-663a were assessed. The biological functions of C9orf139 were evaluated by in vitro assays and in vivo subcutaneous tumor formation experiments in animal models. The molecular mechanism of C9orf139 on miR-663a/Sox12 was investigated through assays including RNA pull-down, Western blot, RNA immunoprecipitation, and co-immunoprecipitation.
RT-qPCR results revealed markedly high C9orf139 levels in the serum and tissue of pancreatic cancer patients, which showed clinical diagnostic and prognostic value. Biological and functional analyses suggested that C9orf139 may promote the growth of pancreatic cancer cells by regulating the miR-663a/Sox12 axis.
C9orf139 is highly expressed in pancreatic cancer and may work as a diagnostic and prognostic marker for pancreatic cancer. It promotes pancreatic cancer cell growth via the miR-663a/Sox12 axis.
The role of C9orf139 in other tumors may be uncovered in the future, and its application in anti-cancer therapy will be promoted.
| 1. | Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1397] [Cited by in RCA: 1790] [Article Influence: 179.0] [Reference Citation Analysis (1)] |
| 2. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 56675] [Article Influence: 7084.4] [Reference Citation Analysis (135)] |
| 3. | Suker M, Beumer BR, Sadot E, Marthey L, Faris JE, Mellon EA, El-Rayes BF, Wang-Gillam A, Lacy J, Hosein PJ, Moorcraft SY, Conroy T, Hohla F, Allen P, Taieb J, Hong TS, Shridhar R, Chau I, van Eijck CH, Koerkamp BG. FOLFIRINOX for locally advanced pancreatic cancer: a systematic review and patient-level meta-analysis. Lancet Oncol. 2016;17:801-810. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 738] [Cited by in RCA: 716] [Article Influence: 71.6] [Reference Citation Analysis (0)] |
| 4. | Ferrone CR, Marchegiani G, Hong TS, Ryan DP, Deshpande V, McDonnell EI, Sabbatino F, Santos DD, Allen JN, Blaszkowsky LS, Clark JW, Faris JE, Goyal L, Kwak EL, Murphy JE, Ting DT, Wo JY, Zhu AX, Warshaw AL, Lillemoe KD, Fernández-del Castillo C. Radiological and surgical implications of neoadjuvant treatment with FOLFIRINOX for locally advanced and borderline resectable pancreatic cancer. Ann Surg. 2015;261:12-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 565] [Cited by in RCA: 672] [Article Influence: 61.1] [Reference Citation Analysis (0)] |
| 5. | Zhang L, Sanagapalli S, Stoita A. Challenges in diagnosis of pancreatic cancer. World J Gastroenterol. 2018;24:2047-2060. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 272] [Cited by in RCA: 404] [Article Influence: 50.5] [Reference Citation Analysis (12)] |
| 6. | Khadka R, Tian W, Hao X, Koirala R. Risk factor, early diagnosis and overall survival on outcome of association between pancreatic cancer and diabetes mellitus: Changes and advances, a review. Int J Surg. 2018;52:342-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 7. | Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 422] [Cited by in RCA: 608] [Article Influence: 67.6] [Reference Citation Analysis (0)] |
| 8. | Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661-5667. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 757] [Cited by in RCA: 1301] [Article Influence: 144.6] [Reference Citation Analysis (0)] |
| 9. | Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1024] [Cited by in RCA: 1384] [Article Influence: 125.8] [Reference Citation Analysis (6)] |
| 10. | Lin YH. MicroRNA Networks Modulate Oxidative Stress in Cancer. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 84] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 11. | Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2492] [Cited by in RCA: 3637] [Article Influence: 404.1] [Reference Citation Analysis (0)] |
| 12. | Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer--a brief overview. Adv Biol Regul. 2015;57:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 501] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 13. | Jathar S, Kumar V, Srivastava J, Tripathi V. Technological Developments in lncRNA Biology. Adv Exp Med Biol. 2017;1008:283-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 271] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 14. | Chen R, Li WX, Sun Y, Duan Y, Li Q, Zhang AX, Hu JL, Wang YM, Gao YD. Comprehensive Analysis of lncRNA and mRNA Expression Profiles in Lung Cancer. Clin Lab. 2017;63:313-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 15. | Jiang B, Yang B, Wang Q, Zheng X, Guo Y, Lu W. lncRNA PVT1 promotes hepatitis B virus-positive liver cancer progression by disturbing histone methylation on the c-Myc promoter. Oncol Rep. 2020;43:718-726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 16. | Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, Wang C, Hawke DH, Wang S, Zhang Y, Wei Y, Ma G, Park PK, Zhou J, Zhou Y, Hu Z, Zhou Y, Marks JR, Liang H, Hung MC, Lin C, Yang L. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18:213-224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 330] [Cited by in RCA: 448] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 17. | Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D, Xie M, Xu L, De W, Wang Z, Wang J. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299-6310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 409] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 18. | Wei W, Liu Y, Lu Y, Yang B, Tang L. LncRNA XIST Promotes Pancreatic Cancer Proliferation Through miR-133a/EGFR. J Cell Biochem. 2017;118:3349-3358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 168] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 19. | Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA Interactions. Methods Mol Biol. 2016;1402:271-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 492] [Cited by in RCA: 692] [Article Influence: 69.2] [Reference Citation Analysis (0)] |
| 20. | Song J, Xu Q, Zhang H, Yin X, Zhu C, Zhao K, Zhu J. Five key lncRNAs considered as prognostic targets for predicting pancreatic ductal adenocarcinoma. J Cell Biochem. 2018;119:4559-4569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 21. | Huang B, Wang J, Chen Q, Qu C, Zhang J, Chen E, Zhang Y, Wang Y, Ni L, Liang T. Gemcitabine enhances OSI-027 cytotoxicity by upregulation of miR-663a in pancreatic ductal adenocarcinoma cells. Am J Transl Res. 2019;11:473-485. [PubMed] |
| 22. | Issue Information-Declaration of Helsinki. J Bone Miner Res. 2018;33:BM i-BM ii. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 23. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149116] [Cited by in RCA: 139159] [Article Influence: 5566.4] [Reference Citation Analysis (3)] |
| 24. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13326] [Article Influence: 1332.6] [Reference Citation Analysis (4)] |
| 25. | Qi Q, Geng Y, Sun M, Chen H, Wang P, Chen Z. Hyperfibrinogen Is Associated With the Systemic Inflammatory Response and Predicts Poor Prognosis in Advanced Pancreatic Cancer. Pancreas. 2015;44:977-982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 26. | Luo G, Guo M, Liu Z, Xiao Z, Jin K, Long J, Liu L, Liu C, Xu J, Ni Q, Yu X. Blood neutrophil-lymphocyte ratio predicts survival in patients with advanced pancreatic cancer treated with chemotherapy. Ann Surg Oncol. 2015;22:670-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 132] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 27. | Li J, Meng H, Bai Y, Wang K. Regulation of lncRNA and Its Role in Cancer Metastasis. Oncol Res. 2016;23:205-217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 246] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 28. | Huang C, Yu W, Wang Q, Cui H, Wang Y, Zhang L, Han F, Huang T. Increased expression of the lncRNA PVT1 is associated with poor prognosis in pancreatic cancer patients. Minerva Med. 2015;106:143-149. [PubMed] |
| 29. | Qu S, Yang X, Song W, Sun W, Li X, Wang J, Zhong Y, Shang R, Ruan B, Zhang Z, Zhang X, Li H. Downregulation of lncRNA-ATB correlates with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2016;37:3933-3938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 30. | Shi X, Zhao Y, He R, Zhou M, Pan S, Yu S, Xie Y, Li X, Wang M, Guo X, Qin R. Three-lncRNA signature is a potential prognostic biomarker for pancreatic adenocarcinoma. Oncotarget. 2018;9:24248-24259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 31. | Wei C, Liang Q, Li X, Li H, Liu Y, Huang X, Chen X, Guo Y, Li J. Bioinformatics profiling utilized a nine immune-related long noncoding RNA signature as a prognostic target for pancreatic cancer. J Cell Biochem. 2019;120:14916-14927. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 89] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 32. | Zhou M, Diao Z, Yue X, Chen Y, Zhao H, Cheng L, Sun J. Construction and analysis of dysregulated lncRNA-associated ceRNA network identified novel lncRNA biomarkers for early diagnosis of human pancreatic cancer. Oncotarget. 2016;7:56383-56394. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 121] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 33. | Mody HR, Hung SW, AlSaggar M, Griffin J, Govindarajan R. Inhibition of S-Adenosylmethionine-Dependent Methyltransferase Attenuates TGFβ1-Induced EMT and Metastasis in Pancreatic Cancer: Putative Roles of miR-663a and miR-4787-5p. Mol Cancer Res. 2016;14:1124-1135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 34. | Amirkhah R, Schmitz U, Linnebacher M, Wolkenhauer O, Farazmand A. MicroRNA-mRNA interactions in colorectal cancer and their role in tumor progression. Genes Chromosomes Cancer. 2015;54:129-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 72] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 35. | Wang L, Wang Z, Huang L, Wu C, Zhang B. MiR-29b suppresses proliferation and mobility by targeting SOX12 and DNMT3b in pancreatic cancer. Anticancer Drugs. 2019;30:281-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/Licenses/by-nc/4.0/
Manuscript source: Unsolicited manuscript
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B, B, B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Aykan NF, Casella C, Dinç T, Moschovi MA, Ooi L S-Editor: Huang P L-Editor: Wang TQ P-Editor: Li JH