©The Author(s) 2025.
World J Gastrointest Oncol. Sep 15, 2025; 17(9): 109777
Published online Sep 15, 2025. doi: 10.4251/wjgo.v17.i9.109777
Published online Sep 15, 2025. doi: 10.4251/wjgo.v17.i9.109777
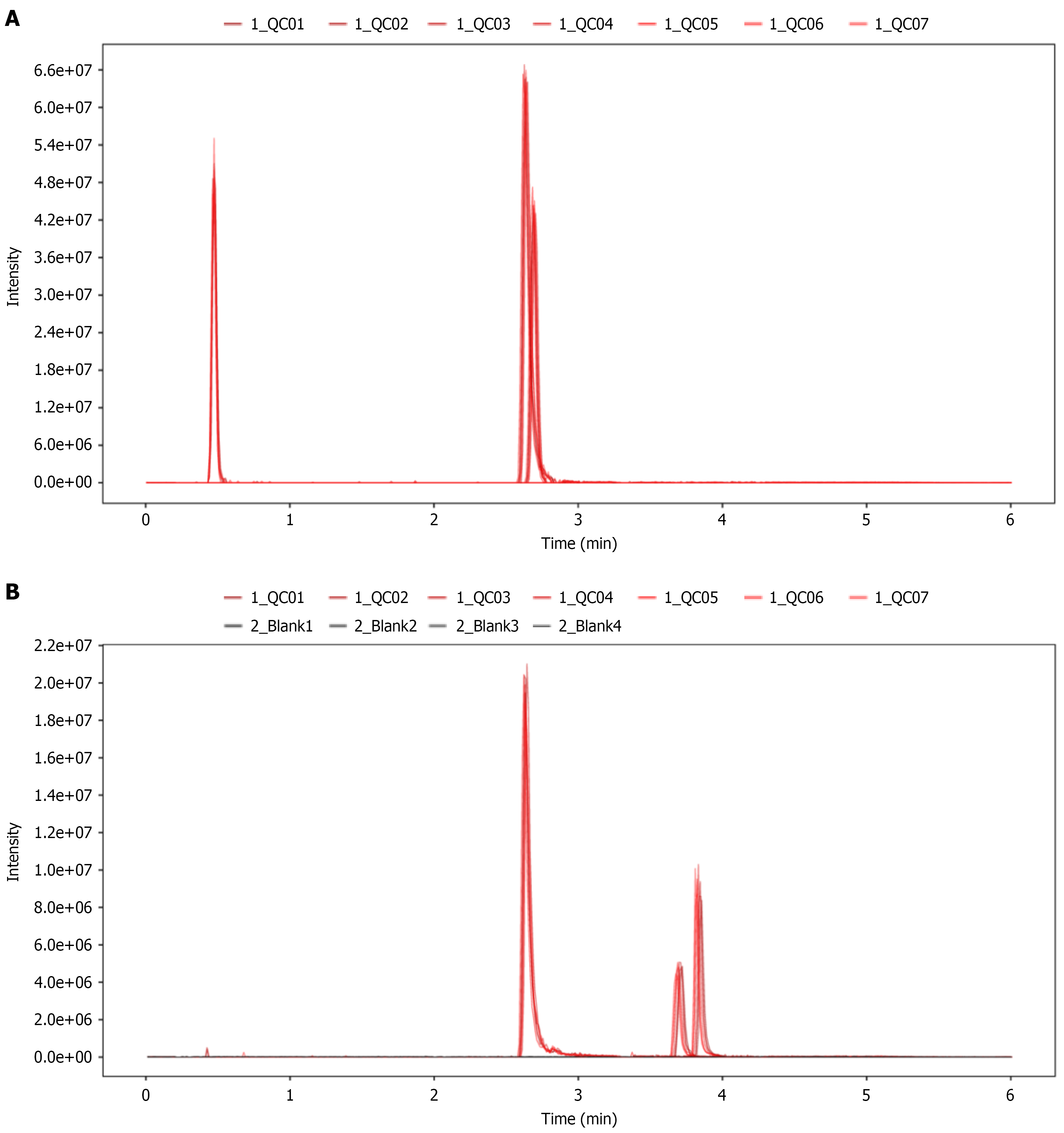
Figure 1 Extracted ion chromatograms of internal standards in all quality control samples.
A: Extracted ion chromatogram (EIC) plot of internal standards in positive for all quality control (QC) samples; B: EIC plot of internal standards in negative ions for all QC samples.
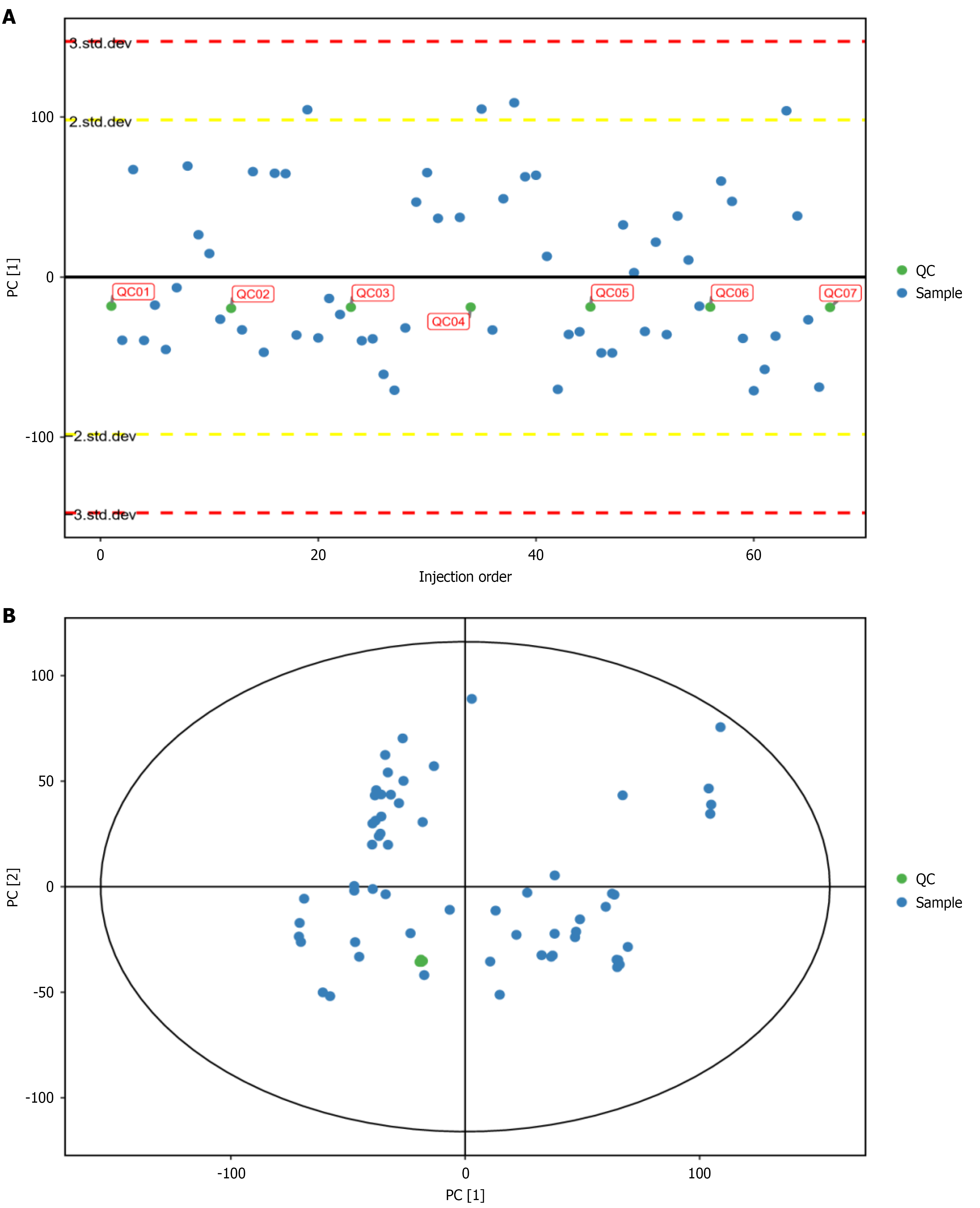
Figure 2 Distribution Plot of principal component analysis-X for quality control Samples.
A: Presentation of quality control (QC) samples in the two-dimensional principal component analysis (PCA) score plot; B: Presentation of QC samples in the one-dimensional PCA-X distribution plot. QC: Quality control.
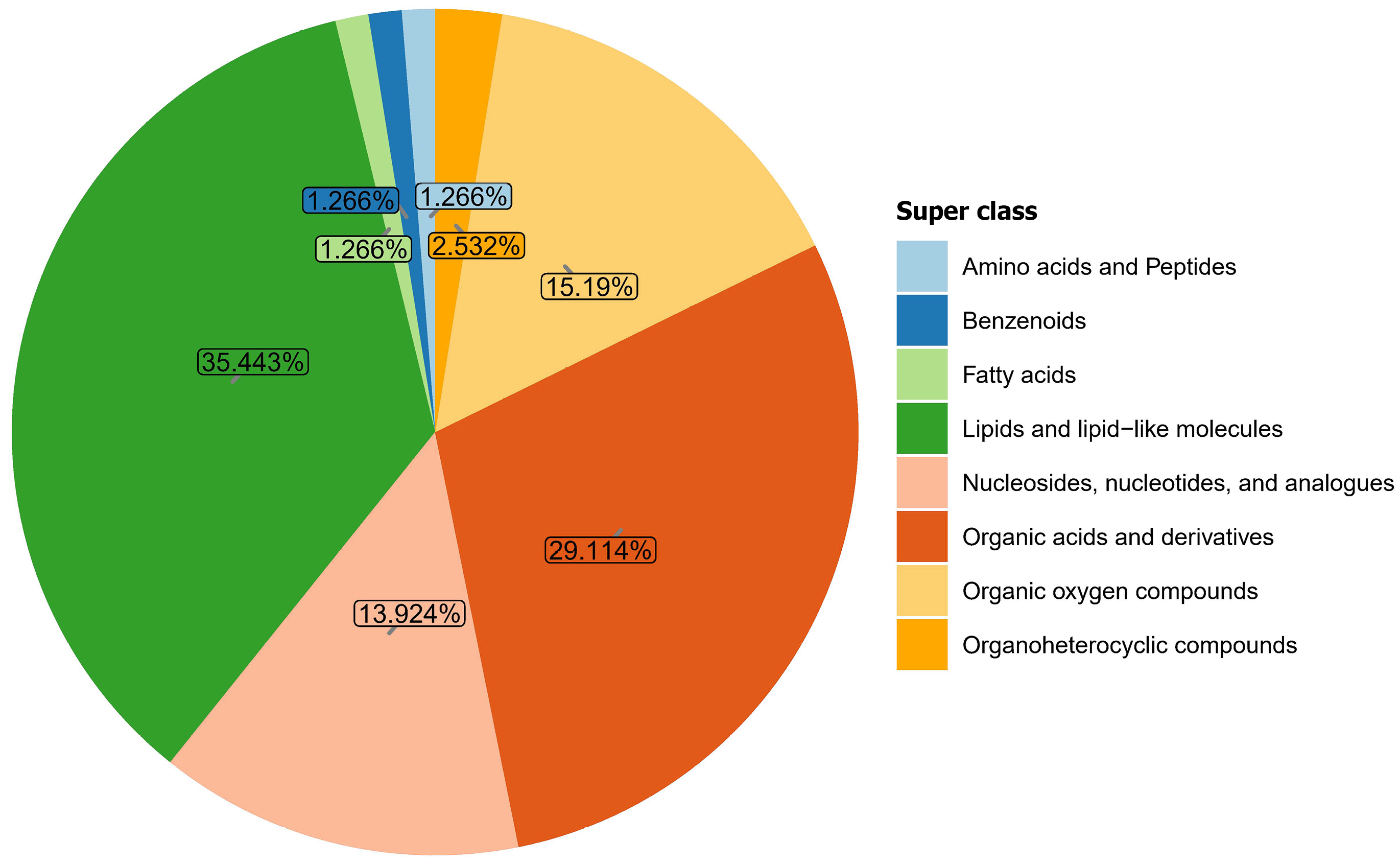
Figure 3 Gut microbial metabolite composition in the gastric cancer and control groups.
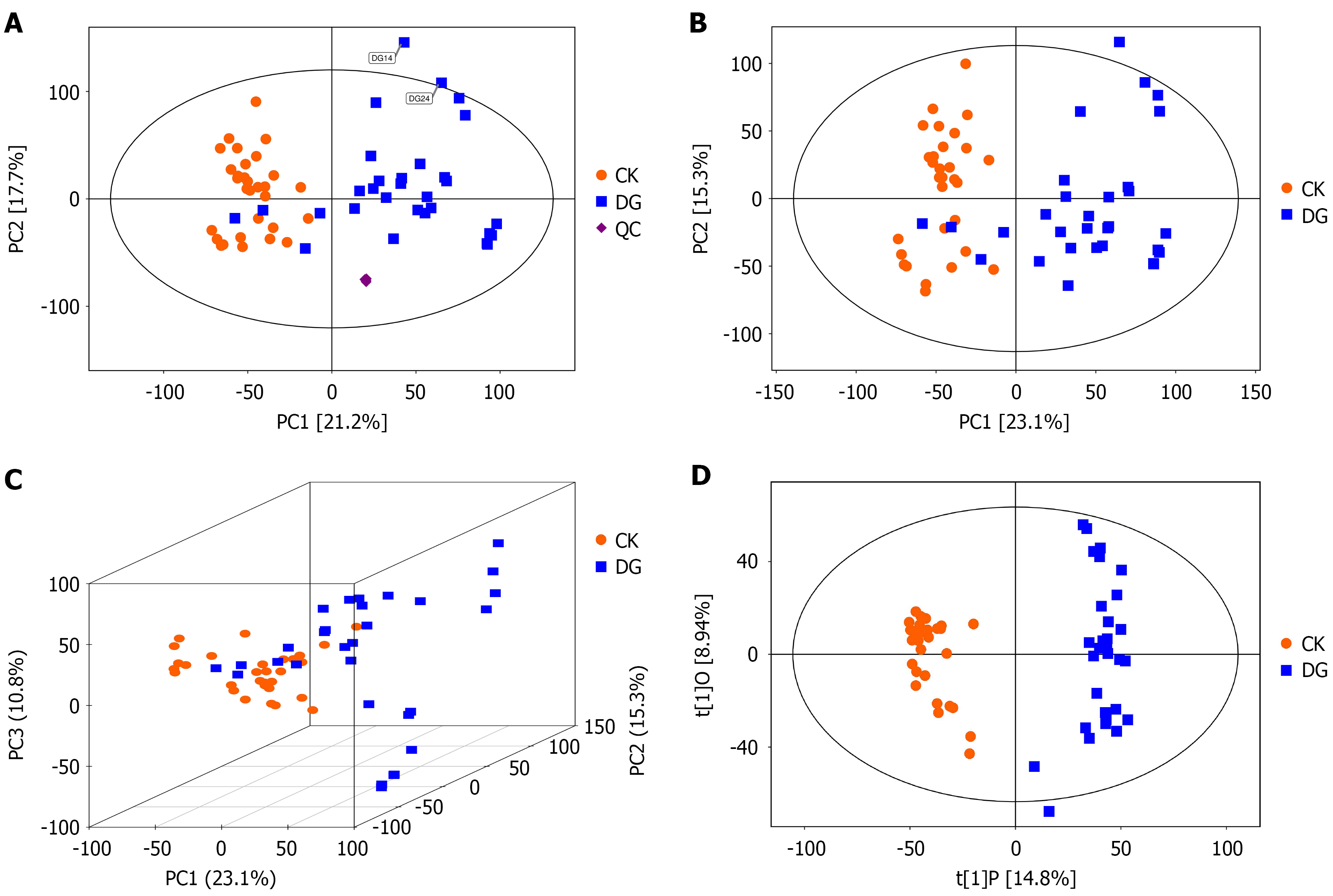
Figure 4 Principal component analysis score scatter plot for all samples (including quality control samples).
A: The principal component analysis (PCA) score scatter plot [score scatter plot for PCA model total with quality control (QC)] of all samples (including QC samples) shows the first principal component (PC1) on the horizontal axis and the second principal component (PC2) on the vertical axis. Each scatter point represents a sample, with color and shape indicating different groups. The closer the sample points are distributed, the more similar the types and concentrations of metabolites in the samples; conversely, the farther apart the samples are, the greater the overall metabolic differences. Most samples fall within the 95% confidence interval (Hotelling’s T-squared ellipse). Observing the PCA score plot of all samples reveals the overall distribution trend of the samples; B: Score scatter plot of PCA model for healthy group (CK) vs gastric cancer group (DG). The results from the PCA score plot indicate that the samples are mostly within the 95% confidence interval (Hotelling’s T-squared ellipse); C: Score scatter plot 3D of PCA model for group CK vs DG; D: Score scatter plot of orthogonal partial least squares discriminant analysis model for group CK vs DG. In the plot, the horizontal axis t[1]P represents the predicted principal component scores of the first principal component, displaying inter-group differences among sample groups. The vertical axis t[1]O represents the orthogonal principal component scores, showing intra-group variations within sample groups. Each scatter point represents an individual sample, with point shapes and colors indicating different experimental groupings. Greater horizontal distance between samples reflects larger inter-group differences, while closer vertical distance indicates better intra-group reproducibility. CK: Healthy group; DG: Gastric cancer group; QC: Quality control.
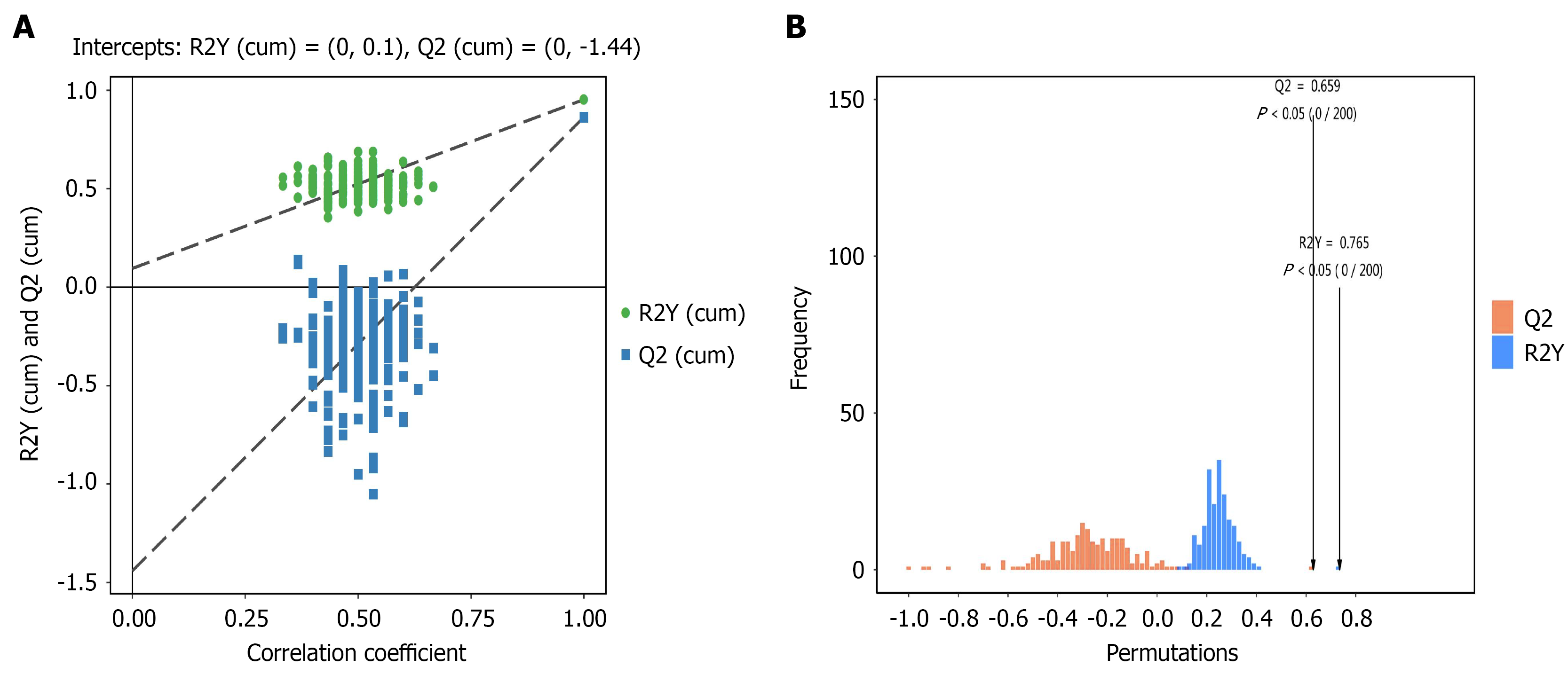
Figure 5 Fecal metabolomics analysis of orthogonal partial least squares discriminant analysis between gastric cancer group and healthy group.
A: Permutation plot test results of the orthogonal partial least squares discriminant analysis (OPLS-DA) model comparison between healthy group (CK) and gastric cancer group (DG) groups (permutation plot test of OPLS-DA model for group CK vs DG). The X-axis shows the permutation retention (proportion consistent with the original model's Y variable order, where a point at 1 represents the original model's R2Y and Q2 values), while the Y-axis displays the values of R2Y or Q2. Green dots indicate the permutation test-derived R2Y values, blue squares represent the permutation test-derived Q2 values, and two dashed lines show the regression lines for R2Y and Q2; B: Permutation histogram test results for the OPLS-DA model comparison between CK and DG groups (CK vs DG). The X-axis shows the accuracy rate of the randomized model in permutation tests, while the Y-axis indicates the number of random models. Red bars represent the frequency of Q values obtained through permutation tests, and blue bars show the occurrence count of RY values. P < 0.05 indicates optimal model performance.
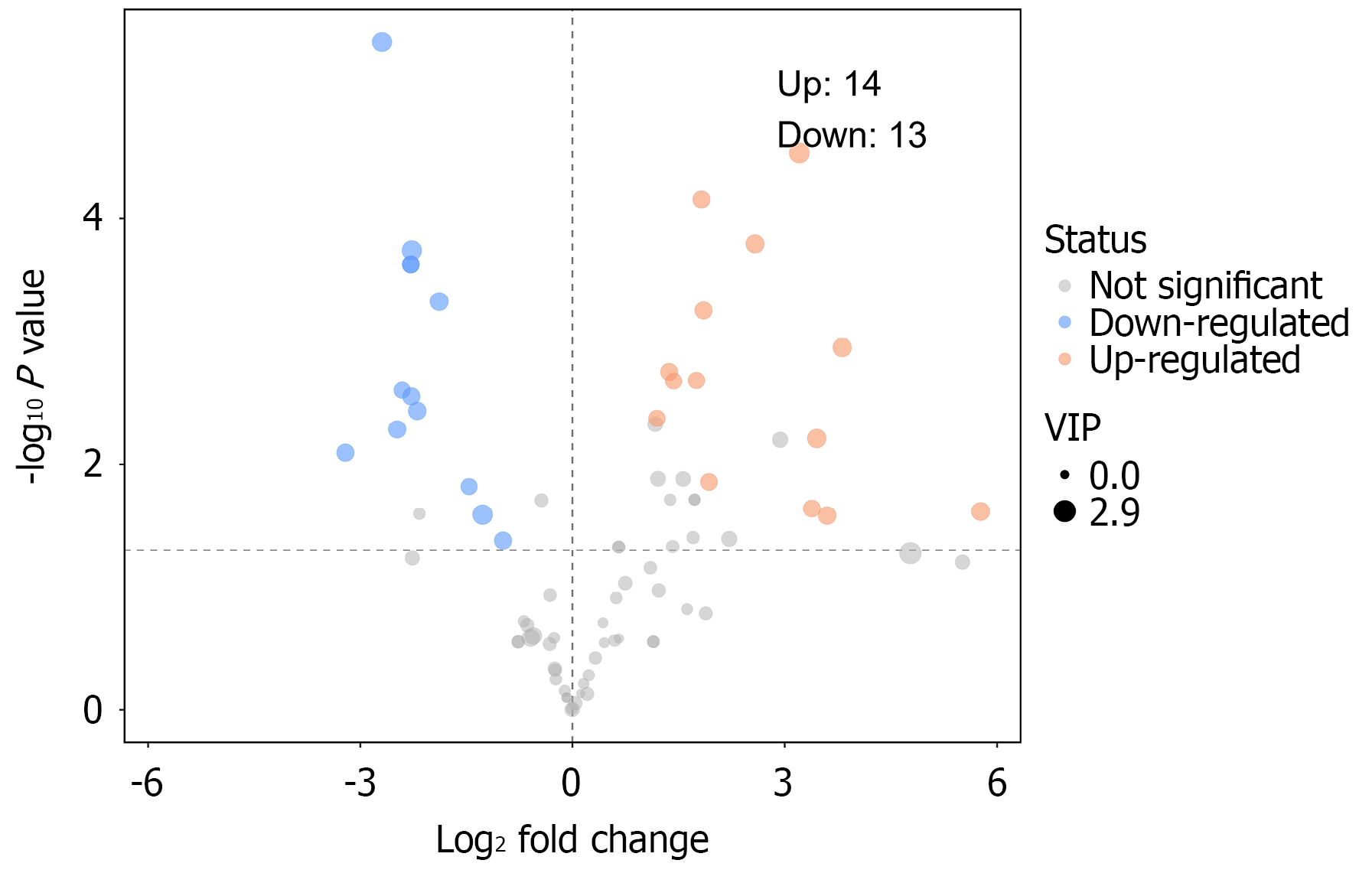
Figure 6 Each point in the volcano plot represents a peak.
The horizontal axis indicates the fold change (log2-transformed) between groups, and the vertical axis shows the t-test P value (negative log10-transformed). Point size reflects the VIP value of the orthogonal partial least squares discriminant analysis model; larger points indicate higher VIP values. Significantly upregulated metabolites are shown in red, downregulated metabolites in blue, and nonsignificant metabolites in gray.
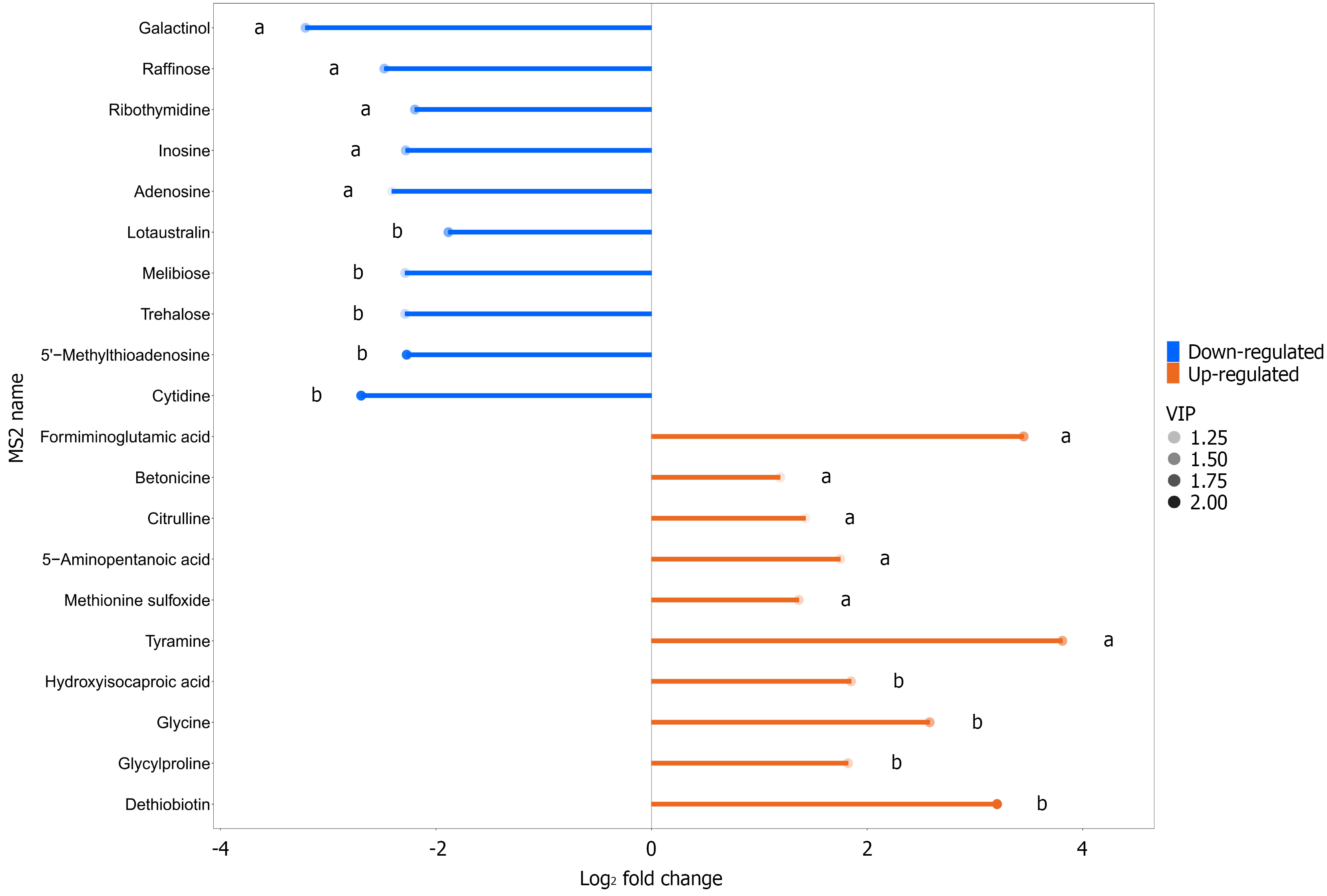
Figure 7 Matchstick analysis for differential metabolites in the control group vs the gastric cancer group.
aP < 0.05; bP < 0.01.
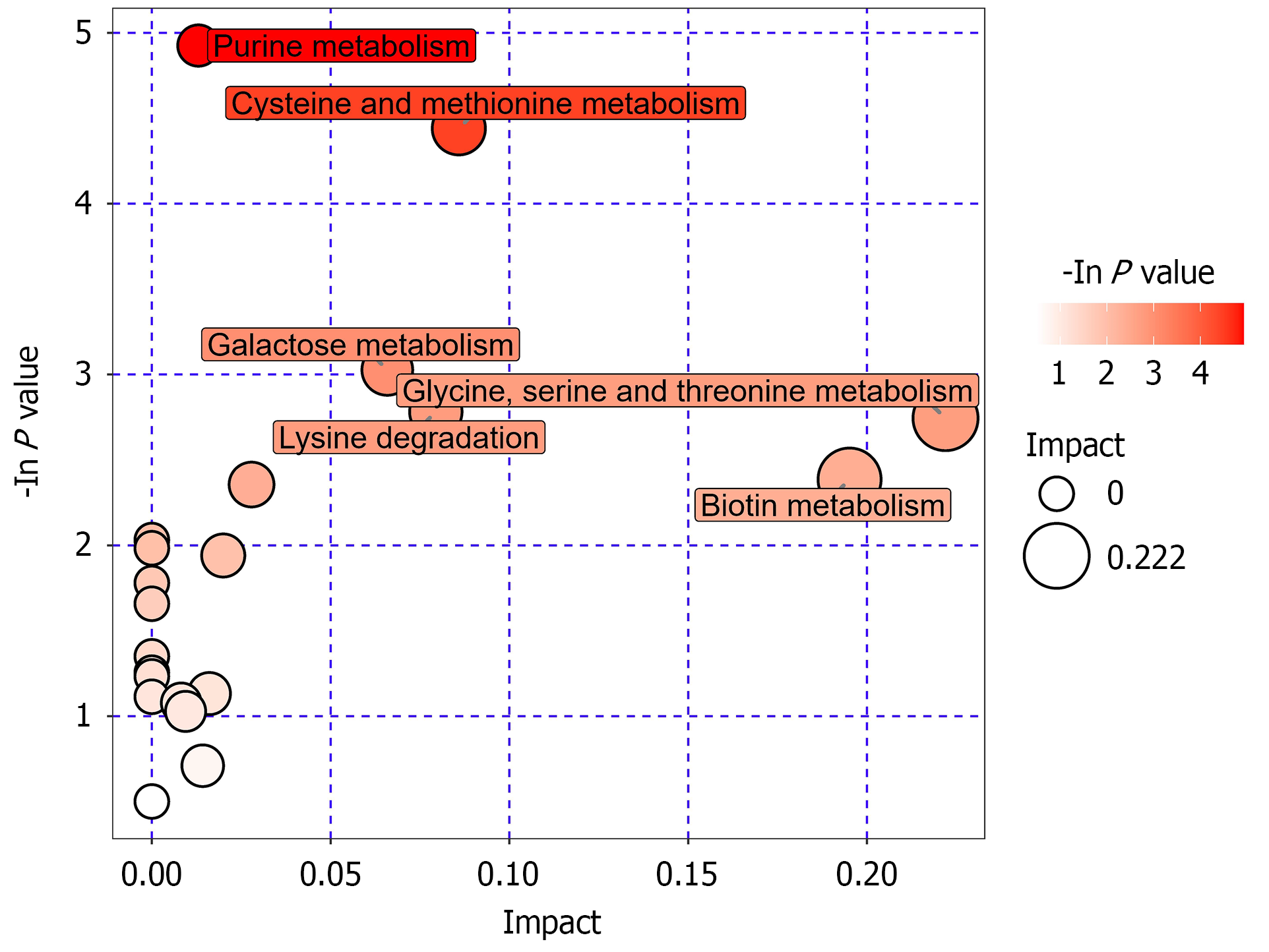
Figure 8 MetaboAnalyst pathway analysis of differential fecal metabolites between gastric cancer and control groups.
- Citation: Zhu LH, Jin ZX, Ma YQ, Feng X, Ci CH, Zhou YS, Gu QL, Lan YM, Zhang ZL. Untargeted metabolomics analysis of metabolite changes in gastric cancer patients from plateau regions. World J Gastrointest Oncol 2025; 17(9): 109777
- URL: https://www.wjgnet.com/1948-5204/full/v17/i9/109777.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i9.109777