Published online Nov 16, 2025. doi: 10.4253/wjge.v17.i11.110207
Revised: June 12, 2025
Accepted: September 23, 2025
Published online: November 16, 2025
Processing time: 167 Days and 0.9 Hours
Endoscopic interventions play a vital role in diagnosing and managing gastroin
To evaluate the role of the gut microbiome in influencing clinical outcomes after endoscopic interventions, focusing on microbial diversity, specific taxa, metabolic functions, and emerging predictive models.
A systematic literature search was conducted in PubMed, EMBASE, and Cochrane databases up to May 2025, selecting human studies that analyzed gut microbiome composition or function in relation to endoscopic interventions and clinical outcomes. Microbiome analysis techniques included 16S rRNA gene sequencing, metagenomics, and metabolomics.
Forty-two studies met the inclusion criteria. Our review identifies key beneficial microbes, such as Faecalibacterium prausnitzii and Bacteroides spp., which support mucosal healing. In contrast, dysbiosis (e.g., an increased abundance of Proteo
The gut microbiome plays a significant role in recovery after endoscopy. Integrating microbiome analysis into clinical decision-making could improve outcomes through personalized predictions and targeted therapies. Future research should focus on standardizing microbiome assessment protocols and validating predictive models to optimize patient care.
Core Tip: The gut microbiome critically influences patient outcomes after endoscopic procedures by modulating mucosal healing and inflammation. Disruptions from bowel preparation and the procedures affect microbial balance, with targeted probiotic and microbiome-based therapies offering promising recovery improvements. Integration of multi-omics and machine learning enables personalized prediction and management of complications, marking a shift toward precision medicine in gastrointestinal endoscopy.
- Citation: Agrawal H, Agarwal N, Gupta N. Impact of gut microbiome on outcomes following endoscopic interventions in gastrointestinal disease. World J Gastrointest Endosc 2025; 17(11): 110207
- URL: https://www.wjgnet.com/1948-5190/full/v17/i11/110207.htm
- DOI: https://dx.doi.org/10.4253/wjge.v17.i11.110207
Endoscopic interventions serve key roles in diagnosing and managing gastrointestinal diseases, including screening colonoscopy and advanced therapies such as endoscopic mucosal resection (EMR) and endoscopic submucosal dissection (ESD)[1] Despite technical progress, predicting patient outcomes remains difficult. Complications like bleeding, infection, delayed healing, and stricture formation can worsen prognosis and increase healthcare costs[2].
The human gastrointestinal tract contains about 1014 bacteria, forming a complex ecosystem that influences host physiology, immune responses, and disease risk. This microbiome regulates immune function, supports epithelial barrier integrity, and affects inflammation and tissue repair[3]. Dysbiosis, an imbalance in microbial composition, associates with several gastrointestinal disorders including inflammatory bowel disease (IBD), colorectal cancer, and functional conditions[4].
Recent evidence indicates the gut microbiome may affect recovery and complications after endoscopic interventions. Variations in microbial diversity and the presence of specific bacterial taxa correlate with clinical outcomes[5]. Microbial metabolites such as short-chain fatty acids contribute to epithelial regeneration and immune regulation essential for mucosal healing[6]. Additionally, bowel preparation before endoscopy temporarily alters the microbiome, which could impact recovery after the procedure[7].
This manuscript reviews the role of the gut microbiome in outcomes after endoscopic interventions. It examines how changes in microbial diversity, specific bacteria, and metabolites influence healing, complications, and disease recurrence. It describes the effects of endoscopy and bowel preparation on gut microbes. The paper presents microbiome-based prediction models and therapies that aim to improve patient care. It highlights the use of multi-omics and machine learning (ML) methods to support personalized treatment and better prognosis in gastrointestinal endoscopy.
A systematic literature search was performed following PRISMA guidelines. We queried PubMed, EMBASE, and Cochrane Library databases from inception until May 2025. Search terms combined keywords and Medical Subject Heading related to gut microbiome (“gut microbiome”, “intestinal microbiome”), endoscopic interventions (“endoscopy”, “colonoscopy”, “endoscopic mucosal resection”), and outcomes (“complications”, “healing”, “prediction”, “response”).
We included original research studies that evaluated the gut microbiome in human patients undergoing any endoscopic procedure, reported clinical outcomes related to procedure success, complications such as bleeding, infection, or strictures, or healing, and used microbiome analysis techniques like 16S rRNA gene sequencing, shotgun metagenomics, or metabolomics. We excluded animal and in vitro studies.
Two independent authors screened titles and abstracts for relevance. Full texts of eligible studies were assessed against inclusion criteria, and disagreements were resolved by consensus. From included studies, we extracted data on study design, population size, patient demographics, type of endoscopic procedure, microbiome sampling method (stool or mucosal biopsy), sequencing technique, and timing relative to the procedure. We collected data on microbial diversity metrics, including alpha and beta diversity, specific taxa associated with outcomes, functional analyses such as metabolic pathways and metabolite levels, and clinical outcomes like complication rates, healing times, and therapeutic response. Study quality was assessed using the Newcastle-Ottawa Scale for observational studies and the Cochrane Risk of Bias tools for interventional trials.
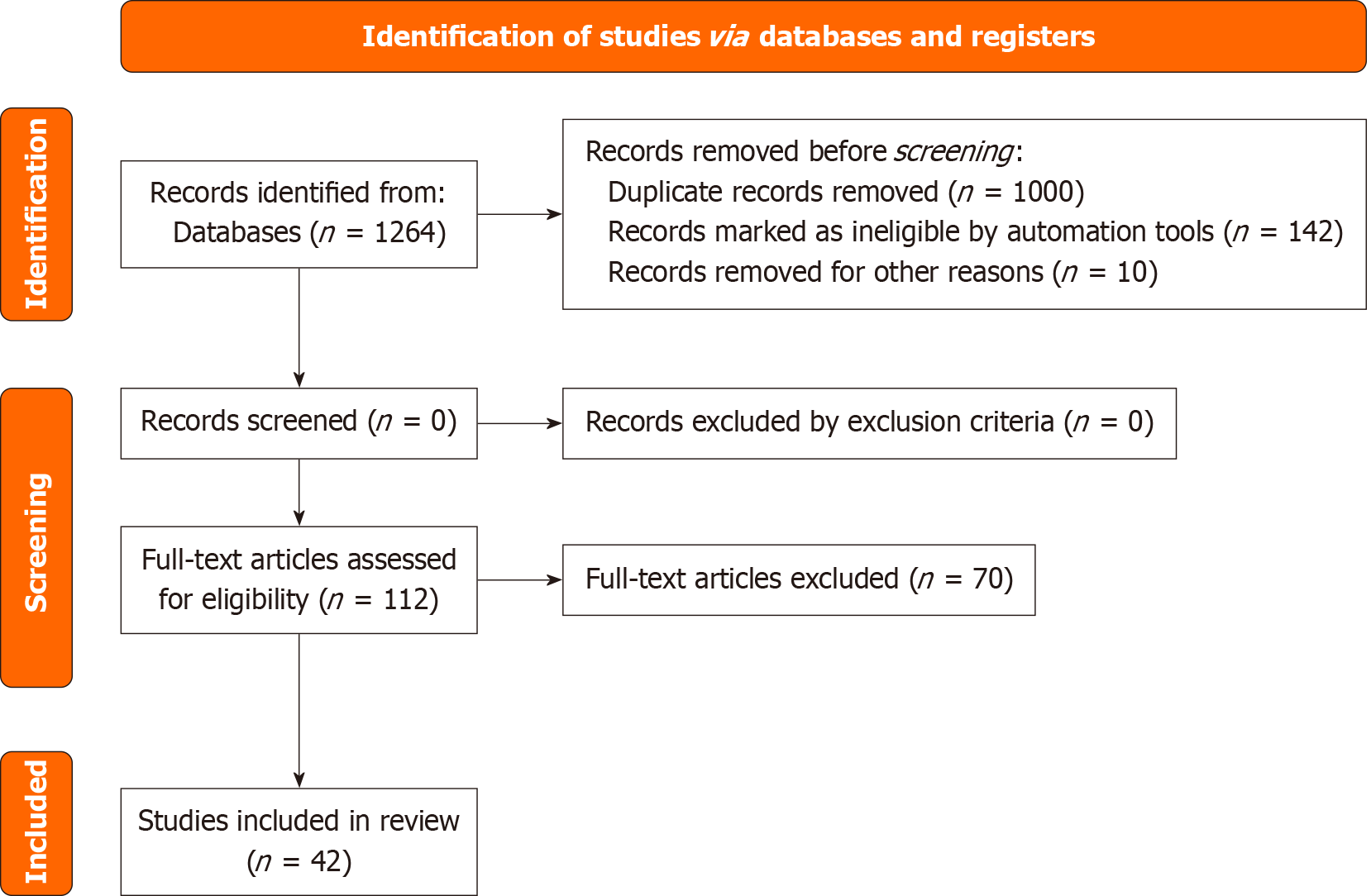
The initial search yielded 1264 records. After removing duplicates and screening abstracts, 112 full-text articles were reviewed. Forty-two studies met inclusion criteria (Figure 1). These studies collectively demonstrate that gut microbiome composition and diversity significantly correlate with mucosal healing, complication rates, and disease recurrence after endoscopic interventions. Interventions such as Helicobacter pylori (H. pylori) eradication, probiotics, and microbiome-targeted therapies showed promising clinical benefits in improving recovery and reducing adverse outcomes. ML models integrating microbiome profiles provide accurate predictions of recurrence risk, supporting personalized post-endoscopy surveillance.
The gastrointestinal microbiome varies along the digestive tract, reflecting different local environments. Microbial composition shifts significantly from the stomach to the colon. Each section hosts communities adapted to its conditions[7]. The upper gastrointestinal tract, including the duodenum and jejunum, absorbs fats, simple carbohydrates, and proteins. The distal small intestine absorbs bile acids and vitamin B12. The colon receives few dietary carbohydrates and nutrients but supports the highest microbial diversity due to bacterial fermentation[8].
Culturomics and next-generation sequencing show that the colon has greater microbial diversity than upper regions, with distinct bacterial groups in each area[9]. The stomach, previously considered mostly sterile due to acid, contains microbes important for gastric health and disease[10]. Detailed studies link stomach microbiome profiles to conditions like chronic gastritis, peptic ulcers, and gastric cancer risk[11]. These findings challenge earlier views on microbial distribution and highlight the need to account for local differences in microbiome diagnostics and treatments.
Certain bacterial genera influence health outcomes. Faecalibacterium prausnitzii, a key butyrate producer, reduces inflammation and promotes healing by blocking nuclear factor kappa B pathways and activating regulatory T cells. Low levels are linked to delayed healing after endoscopic injury[12,13]. Bacteroides species aid immune tolerance and mucosal repair by breaking down polysaccharides, but their loss during bowel preparation or flare-ups impairs recovery. Proteobacteria, including Escherichia coli and Klebsiella pneumoniae, release lipopolysaccharides during inflammation, triggering cytokine release and worsening mucosal damage. Lactobacillus and Bifidobacterium may help maintain epithelial barriers post-endoscopy, though the benefits of probiotics are mixed. Fusobacterium nucleatum is associated with colorectal cancer and poor healing after polyp resection by promoting inflammation and inhibiting cell death[8,14,15] (Table 1).
| Microbial genera | Function | Impact on healing and complications |
| Faecalibacterium | Butyrate production, anti-inflammatory effects | Promotes mucosal healing, reduces inflammation |
| Bacteroides | Polysaccharide breakdown, immune regulation | Supports immune tolerance, aids mucosal repair |
| Proteobacteria | Pathogenicity, LPS production | Increases inflammation, delays healing |
| Fusobacterium | Inflammation promotion, cancer association | Impairs healing, linked to cancer recurrence |
The gastrointestinal microbiome closely interacts with host physiology, influencing both disease and treatment outcomes. Endomicroscopy reveals how colorectal mucosal traits affect bacterial composition. Inflammation shifts microbial communities, favoring facultative anaerobes. Goblet cells shape bacterial colonization, and the microbiome rapidly responds to host changes, thereby affecting disease progression and treatment. Microbial metabolism, including short-chain fatty acids such as butyrate, supports mucosal integrity and immune regulation[16]. Low butyrate levels or its loss are linked to poor healing and increased risk of complications. Microbial bile acid conversion influences epithelial growth, while disrupted metabolism contributes to stricture formation. Tryptophan metabolism and the aryl hydro
| Metabolite | Source microbes | Role in healing |
| Butyrate | Faecalibacterium prausnitzii, Bacteroides | Fuels colonocytes, regulates immune cells, maintains epithelial integrity |
| SCFAs (acetate, propionate) | Fermentative bacteria (e.g., Bacteroides) | Supports tight junctions, immune modulation |
| LPS | Proteobacteria, pathogenic strains | Triggers cytokine release, exacerbates inflammation |
The gut microbiome influences innate and adaptive immune responses essential for mucosal healing. Commensal microbes stimulate regulatory T cells to produce interleukin-10 and transforming growth factor-beta[23]. These cytokines reduce inflammation and support tissue repair. Losing these microbes shifts the immune system toward inflammation. Microbial signals control neutrophil and macrophage recruitment and activation[24]. Excess neutrophils produce reactive oxygen species that harm the mucosa. A balanced microbiome limits this response and helps resolve inflammation. Microbial metabolites strengthen epithelial tight junctions and increase mucus layer thickness[7]. Dysbiosis weakens these defences, allowing antigens and pathogens to cross, causing chronic inflammation[25].
Bowel preparation for colonoscopy causes lasting changes in the gut microbiome. Longitudinal studies show mucosal and luminal microbes shift significantly after standard prep with bisacodyl and polyethylene glycol[26]. Bacteroidetes and species like Faecalibacterium prausnitzii consistently increase. These changes reorganize microbial communities and affect disease detection and treatment[27].
Effects differ between healthy individuals and those with IBD. In IBD patients, bowel prep reduces differences between mucosal and luminal microbiomes, increasing shared microbial groups in biopsies and stool. This convergence may reduce diagnostic accuracy by masking disease-related markers[28].
Cheon et al[29] found that H. pylori eradication therapy speeds mucosal healing and reduces ulcer size after EMR for gastric ulcers. Shen et al[30] showed that enhanced recovery after surgery-based nursing care improves management of postoperative complications and prognosis in patients with early gastrointestinal tumors treated by endoscopic resection. Shi et al[31] developed a ML model using gut microbiome and clinical data to predict colorectal polyp recurrence one year after EMR (Table 3) (Figure 2).
| Ref. | Title | Study group | Intervention | Inference |
| Cheon et al[29] | H. pylori eradication therapy may facilitate gastric ulcer healing after endoscopic mucosal resection | Patients undergoing endoscopic mucosal resection for gastric ulcers | H. pylori eradication therapy | Eradication therapy accelerates mucosal healing and reduces ulcer size post-endoscopy |
| Shen et al[30] | The impact of ERAS-based nursing interventions on postoperative complication management and prognosis in early gastrointestinal tumor endoscopic resection | Early GI tumor patients undergoing endoscopic resection | ERAS nursing care protocol | ERAS nursing interventions improve complication management and prognosis post-endoscopic resection |
| Shi et al[31] | Construction and validation of machine learning-based predictive model for colorectal polyp recurrence one year after endoscopic mucosal resection | Patients post-endoscopic mucosal resection for colorectal polyps | ML model using microbiome + clinical data | Microbiome-informed ML model predicts colorectal polyp recurrence with good accuracy |
Colonoscopy alters gut microbial composition. Probiotic supplementation helps restore balance and improves patient outcomes. Reviews indicate probiotics support mucosal healing by modulating immune response and repairing the epithelial barrier[32]. Follow-up over three to six months shows some microbes return to baseline, while others remain altered, indicating lasting microbiome effects after endoscopy[33]. Endoscopic interventions impact the microbiome beyond bowel prep. Sampling with ingestible devices shows traditional endoscopy, involving fasting and sedation, may not capture normal microbiome states[34]. Upper endoscopy risks contamination from oral, oesophageal, or gastric microbes, complicating analysis. The invasive nature alters the gut environment, creating artificial conditions that misrepresent microbial communities[35].
Magnetic controlled capsule endoscopy offers a less invasive alternative that preserves natural microbiome conditions while allowing thorough examination[36]. Combined with metabolomic analysis, this method supports noninvasive disease detection and maintains microbiome integrity. Future technologies should minimize disruption to native microbial ecosystems to improve patient outcomes[37].
Microbiome patterns relate to outcomes in endoscopic therapies[38]. Barrett’s esophagus patients with microbial profiles rich in anti-inflammatory taxa show higher mucosal remission rates after treatment. Dysbiosis associates with persistent metaplasia and recurrence[39]. In IBD, higher microbial diversity and more butyrate producers link to better mucosal healing after medical and endoscopic treatments[40]. For colorectal cancer prevention, microbial signatures assess adenoma recurrence risk. High levels of pro-inflammatory bacteria like Fusobacterium nucleatum indicate greater risk and slower healing[41].
Microbiome-based prediction models aid colorectal cancer and polyp detection[42]. ML methods like Naive Bayes and Neural Networks reduce false negatives to about 5% by analyzing stool samples for microbial signatures linked to colorectal neoplasia. This approach may reduce colonoscopy demand and improve early detection[43].
After adenoma resection, taxa such as Butyricimonas, Faecalitalea, and Catenibacterium remain elevated, suggesting use in surveillance[44]. These bacteria correlate with adenoma-related amino acid profiles, linking microbiome changes to tumor development[45]. Bioinformatic analyses separate high- and low-risk colorectal adenocarcinoma patients by microbial patterns related to immune cell infiltration and survival. Reviews identify many bacteria with predictive value, but models require validation in prospective studies before clinical use[46]. Studies find 39 bacterial species differing between colorectal cancer patients with and without lympho-vascular invasion[47]. Random Forest and XGBoost models accurately predict clinical outcomes.
Ben Izhak et al[48] found gut microbiome changes linked to weight loss and metabolic improvement after bariatric surgery, suggesting microbiome monitoring can predict surgical outcomes. Carlini et al[49] showed preoperative gut microbiome interventions reduced anastomotic leak rates in laparoscopic colorectal cancer surgery. Jin et al[50] demonstrated that gut microbiome from patients without anastomotic complications promoted collagen synthesis and healing in colon anastomoses, indicating microbiome manipulation aids recovery. Liu et al[51] reported that Bifidobacterium animalis supplementation after colorectal polyp resection improved immune response and recovery vs placebo. Park et al[52] found probiotics after anterior colon resection reduced postoperative symptoms and complications. Martínez-Sánchez et al[53] and Veziant et al[54] are studying dietary and clinical factors combined with microbiome profiling to predict surgical and cancer outcomes in colorectal patients. These studies highlight the gut microbiome as a modifiable factor to improve colorectal surgery results and patient prognosis (Table 4).
| Ref. | Title | Study group | Intervention | Inference |
| Ben Izhak et al[48] | Projection of gut microbiome pre-and post-bariatric surgery to predict surgery outcome | Patients undergoing bariatric surgery | Bariatric surgery with perioperative microbiome profiling | Microbial shifts post-surgery correlate with weight loss success and metabolic improvements |
| Carlini et al[49] | Implementation of the gut microbiota prevents anastomotic leaks in laparoscopic colorectal surgery for cancer | Colorectal cancer patients undergoing laparoscopic surgery | Preoperative gut microbiota modulation | Microbiota-targeted intervention reduces anastomotic leak rates |
| Jin et al[50] | Gut microbiota from non-complicated anastomotic leak patients promotes colon anastomotic healing | Patients with and without anastomotic leak | Microbiota transfer from non-leak patients to epithelial cells | Microbiota from healthy patients induces collagen synthesis, promoting healing |
| Liu et al[51] | Improvement effect of Bifidobacterium animalis subsp. lactis MH-02 in patients receiving resection of colorectal polyps | Patients undergoing colorectal polyp resection | Bifidobacterium animalis supplementation | Probiotic improves postoperative immune parameters and recovery |
| Park et al[52] | Effects of probiotics on the symptoms and surgical outcomes after anterior resection of colon cancer | Colon cancer patients post anterior resection | Probiotic supplementation | Probiotics reduce postoperative symptoms and complications |
| Martínez-Sánchez et al[53] | Gut microbiome modification through dietary intervention in patients with colorectal cancer (protocol) | Colorectal cancer patients planned for surgery | Dietary intervention to modify microbiome | Protocol to assess dietary impact on microbiome and surgical outcomes |
| Veziant et al[54] | Prognostic value of combination of gut microbiota, sarcopenia, obesity, metabolic syndrome to predict surgical/oncologic outcomes in colorectal cancer | Colorectal cancer surgery patients | Observational with microbiota and clinical/metabolic factor assessment | Protocol to evaluate gut microbiota with clinical/metabolic factors for outcome prediction |
Gastric microbiome analysis aids disease prediction and risk assessment, especially for chronic gastritis and gastric cancer[55]. Oral bacterial profiles, including Prevotella, Haemophilus, Fusobacterium, Neisseria, and Streptococcus, show distinct patterns in patients undergoing upper endoscopy[56]. While alpha diversity does not strongly correlate with findings, the presence and abundance of specific bacteria provide useful diagnostic data alongside endoscopy[57].
Tests for gastric microbiota dysbiosis show promise in cancer risk assessment for atrophic gastritis patients, with specificity near 89% in identifying high-grade dysplasia or cancer[58]. These models analyze antrum and corpus biopsy samples to predict cancer risk and may serve as clinical biomarkers. Low levels of healthy gastric bacteria also signal gastrointestinal disease risk and offer opportunities for early detection[59].
Functional studies using genomic DNA sequencing of gastric biopsies enhance understanding of host-microbe interactions and improve risk prediction[60]. Combining microbiome data with biomarkers like pepsinogen ratios, H. pylori antibodies, and gastrin levels improves screening accuracy over endoscopy alone[61].
Cheon et al[29] conducted a randomized controlled trial on H. pylori eradication after EMR in gastric ulcer patients. The therapy accelerated mucosal healing and reduced ulcer size, supporting its use as standard postoperative care. Shen et al[30] tested enhanced recovery after surgery-based nursing interventions in patients undergoing endoscopic resection for early gastrointestinal tumours. Their trial showed better management of postoperative complications and improved prognosis, emphasizing microbiota-related supportive care. Shi et al[31] created a ML model using gut microbiome and clinical data to predict colorectal polyp recurrence one year after EMR. This approach highlights microbiome profiling for personalized surveillance. Studies show colonoscopy causes significant shifts in gut microbiome. Probiotic supplementation restores balance and improves outcomes. Reviews on probiotics in mucosal healing support their role in restoring the epithelial barrier and modulating immune response after endoscopic interventions. These methods aid personalized detection and risk assessment.
Surgical site infections and anastomotic complications link closely to gut microbiome composition, especially after colorectal surgery. Reviews show strong connections between microbiome profiles and issues like anastomotic leaks, infections, and postoperative ileus. Certain bacteria influence wound healing, immune response, and tissue repair[1].
Despite improved surgical standards and care in high-volume centres, complication rates remain significant: 10%-30% for ileus, 6.5%-20% for infections, and 2.7%-20% for leaks. Preoperative microbiome analysis can predict these risks, opening paths for preventive measures. Surgery also alters gut microbiome, with changes varying by bacterial type and affecting recovery positively or negatively.
Distinct microbial patterns appear in patients who develop complications, highlighting the role of microbiome in outcomes. Studies such as Carlini et al[49] have shown that targeted modulation of gut microbiome prior to colorectal surgery significantly reduces anastomotic leak incidence, underscoring the microbiome’s role in surgical healing. Similarly, Jin et al[50] demonstrated that microbiome from patients without anastomotic complications promotes epithelial collagen synthesis and supports colon anastomosis healing.
Cheon et al[29] found that H. pylori eradication therapy speeds mucosal healing after gastric mucosal resection. Bowel preparation and the procedure can disrupt the microbiome, causing prolonged imbalance and slower recovery. Probiotic supplementation after surgery reduces infection rates and improves immune function[52,62]. Persistent dysbiosis raises risks for complications and disease recurrence, highlighting the need for microbiome-focused perioperative care[63]. Preoperative microbiome optimization through probiotics and diet aims to increase beneficial bacteria before surgery. Microbiome assessment may become part of risk evaluation, and targeted interventions could improve outcomes[64].
Gut microbiome data is complex and high-dimensional, posing challenges and opportunities for clinical research, especially in predicting postoperative outcomes and disease recurrence[65]. ML has enabled integration of large microbiome datasets with clinical variables to create predictive models that support personalized medicine and postoperative care[66]. Shi et al[31] developed a ML model combining microbial profiles and clinical data to predict colorectal polyp recurrence within one year after EMR. The model showed strong accuracy, supporting its use in personalized surveillance and early intervention. Yu et al[67] used bioinformatics and ML to analyze microbiome and transcriptome data for colon adenocarcinoma risk prediction. This multi-omics approach enhances risk stratification by integrating host gene expression with microbial patterns. Liu et al[58] created gut microbiome-based algorithms to predict metachronous adenoma after colorectal cancer surgery, identifying high-risk patients for improved follow-up. Wang et al[2] reviewed computational methods, including ML, to study microbial interactions linked to colorectal anastomotic leaks, aiding microbiome-targeted prevention. Yu et al[68] applied computational analysis to track microbiome changes after colorectal adenoma resection, providing insights on healing and recurrence risk. Challenges remain in handling data heterogeneity, high dimensionality, and model interpretability. Advances in algorithms, validation, and multi-omics integration will improve clinical application of ML in microbiome analysis[69]. ML on gut microbiome data enables personalized risk assessment and targeted care, advancing precision management in surgery and gastrointestinal diseases.
Integrating gut microbiome profiles with clinical data and biomarkers improves prediction of postoperative outcomes. Combining microbial signatures with patient demographics, metabolic status, inflammatory markers, and imaging provides a clearer view of risks and recovery[70]. Veziant et al[54] designed the METABIOTE cohort to assess gut microbiome, sarcopenia, obesity, and metabolic syndrome in predicting surgical and oncologic outcomes after colorectal cancer surgery. This approach enhances risk assessment beyond clinical data by including microbial and metabolic factors. Liu et al[62] developed algorithms combining microbiome and clinical data to identify patients at risk for metachronous adenoma recurrence, improving surveillance. Caenepeel et al[71] found microbiome composition correlates with disease activity and treatment response in IBD. Combining microbiome analysis with biomarkers like C-reactive protein, fecal calprotectin, and endoscopy refines monitoring and treatment. Yu et al[67] integrated transcriptomic, metabolomic, and microbiome data to capture host-microbe interactions related to healing and inflammation. These multi-omics models, often with ML, identify composite biomarkers that enhance prognosis. Challenges include data standardization, integration, and the need for large cohorts. Gut microbiome analysis offers value in postoperative surveillance and disease monitoring. Tracking microbial composition and function over time gives early insight into recovery, complication risks, and disease recurrence[72]. Shi et al[31] showed combining microbiome data with clinical factors predicts colorectal polyp recurrence after endoscopic resection, supporting early surveillance and intervention. Liu et al[73] developed gut microbiome-based algorithms to forecast metachronous adenoma risk after colorectal cancer surgery, enabling personalized follow-up. In IBD, mucosal and fecal microbiome profiles act as biomarkers for disease activity and relapse[74]. Caenepeel et al[71] and Wang et al[75] identified microbiome signatures linked to endoscopic healing and clinical remission, guiding treatment and reducing invasive interventions. Continuous microbiome monitoring helps detect early postoperative complications like anastomotic leaks and infections. Carlini et al[49] and Wang et al[75] highlight microbiome markers’ role in predicting and preventing these events. Though standardizing sampling and data analysis remains difficult, gut microbiome surveillance provides a non-invasive, dynamic tool to improve patient management, outcomes, and reduce healthcare costs through early detection and personalized care.
Modulating the gut microbiome offers therapeutic strategies to improve outcomes after gastrointestinal surgery and endoscopy. Randomized trials by Liu et al[51] and Park et al[52] show probiotic supplementation enhances immune function, speeds recovery, and reduces postoperative complications. Carlini et al[49] found preoperative microbiome modulation lowers anastomotic leak rates after colorectal surgery, highlighting clinical benefits of microbiome optimization. Dietary interventions are being studied for their role in shaping the microbiome to support surgical recovery[53]. Fecal microbiome transplantation (FMT) also shows potential in restoring microbial balance and promoting mucosal healing, though further research is needed. Advanced microbiome-based predictive models, such as those by Shi et al[31], support personalized postoperative care by identifying high-risk patients for targeted treatment. Microbiome-targeted therapies represent a move toward precision medicine to enhance recovery, reduce complications, and improve outcomes. Clinical trials indicate probiotics aid recovery after colorectal polyp resection. One study found Bifidobacterium animalis subsp. lactis microbiota homeostasis-02, given daily for seven days, lowered postoperative symptoms and improved bowel function vs placebo[76].
Probiotics support mucosal barriers, regulate immunity, and restore microbial balance disrupted by interventions. Early use and strain selection, especially Bifidobacterium species, increase benefits[76].
These findings support including probiotics in post-procedural care. Probiotics are simple, low-cost, safe, and have no major side effects. Future studies should determine optimal probiotic combinations, treatment duration, and patient groups to maximize benefits and minimize risks.
Microbiome research faces technical challenges before routine clinical use. Variations in sample collection, processing, and analysis affect measurement accuracy and interpretation. Bowel preparation for endoscopy alters microbial communities, complicating results. Upper gastrointestinal sampling risks contamination, requiring standardized pro
Advances in sequencing enable detailed identification of species, strains, resistance genes, virulence factors, and metabolic pathways through shotgun metagenomics. Combining microbiome data with metabolomics, proteomics, and transcriptomics creates comprehensive profiles of host-microbe interactions. Multi-omics approaches offer more insight than microbiome analysis alone[75]. Real-time sequencing and point-of-care testing could allow rapid analysis during endoscopy for immediate clinical decisions. Artificial intelligence and ML, especially deep learning, analyze complex multi-omics data and detect clinical patterns[86]. Federated learning supports large-scale research while protecting patient privacy. Cloud platforms increase access to advanced analytics without local infrastructure[87]. Future research should focus on large, longitudinal studies tracking microbiome changes linked to clinical outcomes. Integrating multi-omics will improve understanding of host-microbe interactions. ML can enhance prediction of complications and treatment responses. Clinical trials must test interventions like probiotics, prebiotics, dietary changes, and fecal microbiome transplants[88]. Personalized endoscopy could use microbiome profiles for risk assessment, bowel preparation optimization, and tailored post-procedure care[89]. Targeted microbiome modulation offers new options to improve post-endoscopic outcomes. Precision probiotics designed for individual microbiomes may enhance therapy and reduce side effects. Synthetic microbial communities could deliver consistent, targeted effects beyond traditional probiotics[90]. FMT effectively treats recurrent Clostridioides difficile infection and is under study for other dysbiosis-related conditions[91]. Standardized FMT protocols for endoscopic patients could provide strong treatment options[92]. Selective depletion using targeted antimicrobials or bacteriophages can remove harmful bacteria while preserving beneficial ones. New microbiome-based drugs, such as prebiotics, postbiotics, and microbiome-derived therapeutics, offer precise control over microbiome modification. Integrating microbiome monitoring with treatment could enable adaptive protocols that maximize benefits and minimize risks in real time[62,73,93,94].
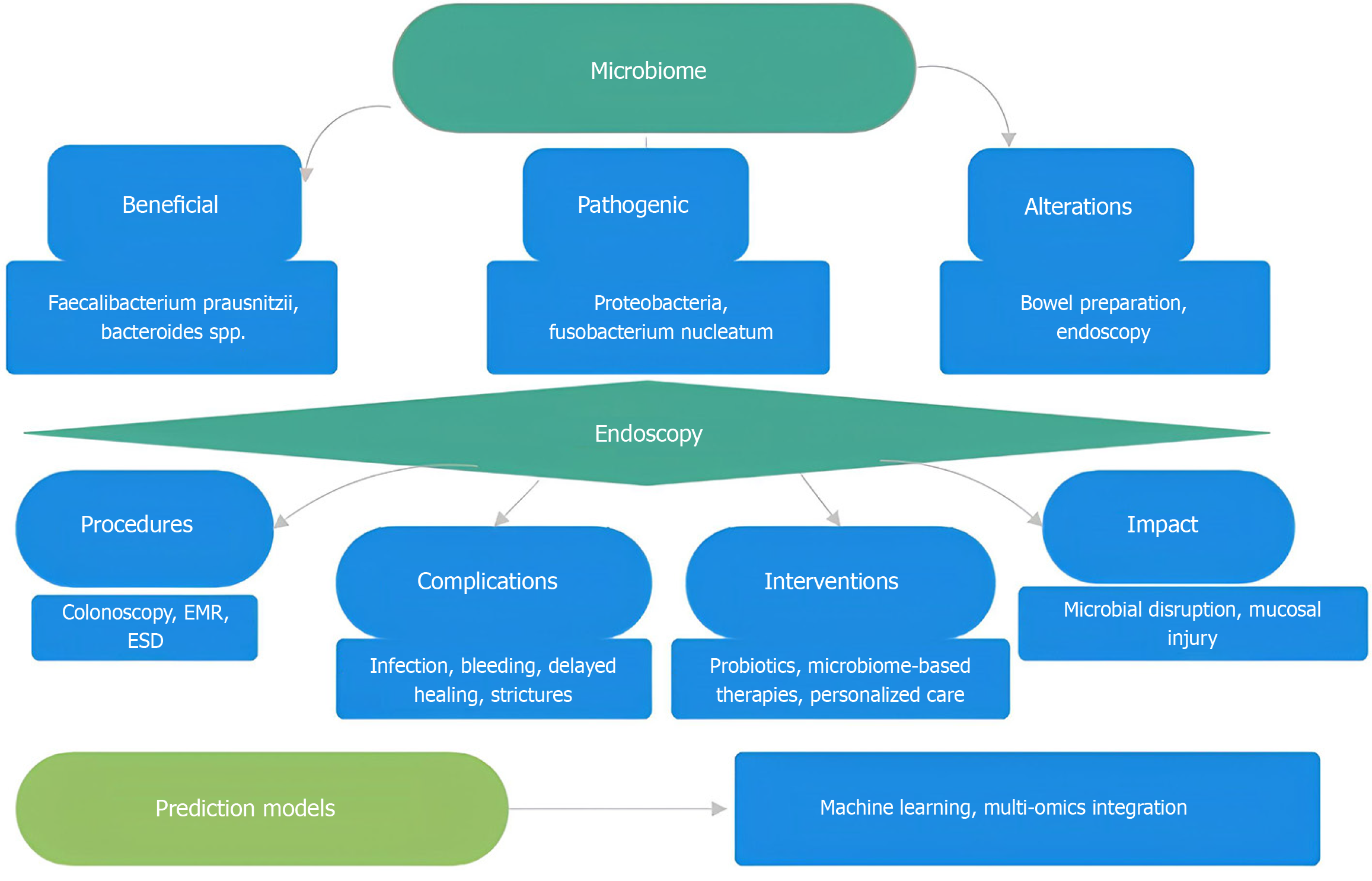
The gastrointestinal microbiome influences outcomes after endoscopic interventions. It aids disease detection, supports recovery, and improves patient management. Combining advanced sequencing with ML produces models that identify high-risk patients, guide treatments, and enhance surveillance. Spatial and temporal microbial variations provide information beyond traditional clinical measures, enabling personalized care. Endoscopic interventions change microbiome composition, making microbial factors relevant in clinical decisions. Microbiome-targeted therapies show potential to improve outcomes. Despite technical and regulatory challenges, research and technology will transform gastroenterology. Standardized protocols, validation methods, and user-friendly tools are needed for routine care and resource optimization (Figure 2). Future efforts should emphasize prospective validation, multi-omics integration, and targeted therapies using microbiome data. Microbiome analysis offers opportunities for early detection, treatment prediction, and personalized strategies. This shift toward precision medicine could improve outcomes after endoscopic interventions.
| 1. | Agnes A, Puccioni C, D'Ugo D, Gasbarrini A, Biondi A, Persiani R. The gut microbiota and colorectal surgery outcomes: facts or hype? A narrative review. BMC Surg. 2021;21:83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 2. | Wang AH, Li M, Li CQ, Kou GJ, Zuo XL, Li YQ. Human colorectal mucosal microbiota correlates with its host niche physiology revealed by endomicroscopy. Sci Rep. 2016;6:21952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 3. | Alam A, Neish A. Role of gut microbiota in intestinal wound healing and barrier function. Tissue Barriers. 2018;6:1539595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 110] [Article Influence: 13.8] [Reference Citation Analysis (1)] |
| 4. | Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, Mele MC. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms. 2019;7:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 969] [Cited by in RCA: 2283] [Article Influence: 326.1] [Reference Citation Analysis (4)] |
| 5. | Allegretti JR, Khanna S, Mullish BH, Feuerstadt P. The Progression of Microbiome Therapeutics for the Management of Gastrointestinal Diseases and Beyond. Gastroenterology. 2024;167:885-902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 18] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 6. | Amoroso C, Perillo F, Strati F, Fantini MC, Caprioli F, Facciotti F. The Role of Gut Microbiota Biomodulators on Mucosal Immunity and Intestinal Inflammation. Cells. 2020;9:1234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 185] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 7. | Gorkiewicz G, Moschen A. Gut microbiome: a new player in gastrointestinal disease. Virchows Arch. 2018;472:159-172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 57] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 8. | Sears CL. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin Microbiol Rev. 2009;22:349-369, Table of Contents. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 337] [Cited by in RCA: 324] [Article Influence: 19.1] [Reference Citation Analysis (1)] |
| 9. | Guyton K, Alverdy JC. The gut microbiota and gastrointestinal surgery. Nat Rev Gastroenterol Hepatol. 2017;14:43-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 165] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 10. | Shi Y, Cui H, Wang F, Zhang Y, Xu Q, Liu D, Wang K, Hou S. Role of gut microbiota in postoperative complications and prognosis of gastrointestinal surgery: A narrative review. Medicine (Baltimore). 2022;101:e29826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 11. | Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM, Thomas LV, Zoetendal EG, Hart A. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65:330-339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1341] [Cited by in RCA: 1639] [Article Influence: 163.9] [Reference Citation Analysis (0)] |
| 12. | Al Radi ZMA, Prins FM, Collij V, Vich Vila A, Festen EAM, Dijkstra G, Weersma RK, Klaassen MAY, Gacesa R. Exploring the Predictive Value of Gut Microbiome Signatures for Therapy Intensification in Patients With Inflammatory Bowel Disease: A 10-Year Follow-up Study. Inflamm Bowel Dis. 2024;30:1642-1653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 13. | Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1199] [Cited by in RCA: 1552] [Article Influence: 103.5] [Reference Citation Analysis (0)] |
| 14. | Arfi S, Sharma P, Das K, Bhaskar Y, Goel I, Singh H, Das R. Low abundance of healthy bacteria in the gastric microbiota can be a potential biomarker for gastrointestinal diseases: A pilot study. Indian J Med Res. 2024;159:254-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 15. | O'Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol. 2016;13:691-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 521] [Cited by in RCA: 815] [Article Influence: 81.5] [Reference Citation Analysis (0)] |
| 16. | David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559-563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8117] [Cited by in RCA: 7170] [Article Influence: 597.5] [Reference Citation Analysis (1)] |
| 17. | Gan L, Wang Y, Huang S, Zheng L, Feng Q, Liu H, Liu P, Zhang K, Chen T, Fang N. Therapeutic Evaluation of Bifidobacterium animalis subsp. lactis MH-02 as an Adjunctive Treatment in Patients with Reflux Esophagitis: A Randomized, Double-Blind, Placebo-Controlled Trial. Nutrients. 2024;16:342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 18. | Bosch S, Acharjee A, Quraishi MN, Rojas P, Bakkali A, Jansen EE, Brizzio Brentar M, Kuijvenhoven J, Stokkers P, Struys E, Beggs AD, Gkoutos GV, de Meij TG, de Boer NK. The potential of fecal microbiota and amino acids to detect and monitor patients with adenoma. Gut Microbes. 2022;14:2038863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 19. | Breton Mdcm-Mstr J, Tanes C, Wilson N, Kachelries K, Bittinger K, Mattei P, Baldassano R. P1311 Mucosal Microbiome Signatures Associated with Postoperative Endoscopic Recurrence in Paediatric Crohn’s Disease. J Crohns Colitis. 2025;19:i2362-i2362. [DOI] [Full Text] |
| 20. | Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17:219-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 724] [Cited by in RCA: 1160] [Article Influence: 128.9] [Reference Citation Analysis (0)] |
| 21. | Buffet-Bataillon S, Bouguen G, Fleury F, Cattoir V, Le Cunff Y. Gut microbiota analysis for prediction of clinical relapse in Crohn's disease. Sci Rep. 2022;12:19929. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 22. | De Cruz P, Kang S, Wagner J, Buckley M, Sim WH, Prideaux L, Lockett T, McSweeney C, Morrison M, Kirkwood CD, Kamm MA. Association between specific mucosa-associated microbiota in Crohn's disease at the time of resection and subsequent disease recurrence: a pilot study. J Gastroenterol Hepatol. 2015;30:268-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 126] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 23. | Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, Ter Horst R, Jansen T, Jacobs L, Bonder MJ, Kurilshikov A, Fu J, Joosten LAB, Zhernakova A, Huttenhower C, Wijmenga C, Netea MG, Xavier RJ. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell. 2016;167:1125-1136.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 776] [Article Influence: 86.2] [Reference Citation Analysis (0)] |
| 24. | Rosen C, Shezen E, Aronovich A, Klionsky YZ, Yaakov Y, Assayag M, Biton IE, Tal O, Shakhar G, Ben-Hur H, Shneider D, Vaknin Z, Sadan O, Evron S, Freud E, Shoseyov D, Wilschanski M, Berkman N, Fibbe WE, Hagin D, Hillel-Karniel C, Krentsis IM, Bachar-Lustig E, Reisner Y. Preconditioning allows engraftment of mouse and human embryonic lung cells, enabling lung repair in mice. Nat Med. 2015;21:869-879. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 81] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 25. | Ganesh BP, Klopfleisch R, Loh G, Blaut M. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PLoS One. 2013;8:e74963. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 269] [Cited by in RCA: 391] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 26. | Dey N, Soergel DA, Repo S, Brenner SE. Association of gut microbiota with post-operative clinical course in Crohn's disease. BMC Gastroenterol. 2013;13:131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 105] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 27. | Ticinesi A, Milani C, Lauretani F, Nouvenne A, Mancabelli L, Lugli GA, Turroni F, Duranti S, Mangifesta M, Viappiani A, Ferrario C, Maggio M, Ventura M, Meschi T. Gut microbiota composition is associated with polypharmacy in elderly hospitalized patients. Sci Rep. 2017;7:11102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 156] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 28. | Yeh JH, Lin CW, Wang WL, Lee CT, Chen JC, Hsu CC, Wang JY. Positive Fecal Immunochemical Test Strongly Predicts Adenomas in Younger Adults With Fatty Liver and Metabolic Syndrome. Clin Transl Gastroenterol. 2021;12:e00305. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 29. | Cheon JH, Kim JH, Lee SK, Kim TI, Kim WH, Lee YC. Helicobacter pylori eradication therapy may facilitate gastric ulcer healing after endoscopic mucosal resection: a prospective randomized study. Helicobacter. 2008;13:564-571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 30. | Shen Y, Xi Y, Ru LGX, Liu H. The impact of ERAS-based nursing interventions on postoperative complication management and prognosis in early gastrointestinal tumor endoscopic resection: a prospective randomized controlled study. Langenbecks Arch Surg. 2025;410:135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 31. | Shi YH, Liu JL, Cheng CC, Li WL, Sun H, Zhou XL, Wei H, Fei SJ. Construction and validation of machine learning-based predictive model for colorectal polyp recurrence one year after endoscopic mucosal resection. World J Gastroenterol. 2025;31:102387. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (2)] |
| 32. | Jo HH, Lee MY, Ha SE, Yeom DH, Kim YS. Alteration in gut microbiota after colonoscopy: proposed mechanisms and the role of probiotic interventions. Clin Endosc. 2025;58:25-39. [RCA] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 33. | Filidou E, Kolios G. Probiotics in Intestinal Mucosal Healing: A New Therapy or an Old Friend? Pharmaceuticals (Basel). 2021;14:1181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 34. | Liu K, Qin M, Huang J. The prescreening tool for gastric cancer in China. Gut. 2020;69:1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 35. | Palleja A, Mikkelsen KH, Forslund SK, Kashani A, Allin KH, Nielsen T, Hansen TH, Liang S, Feng Q, Zhang C, Pyl PT, Coelho LP, Yang H, Wang J, Typas A, Nielsen MF, Nielsen HB, Bork P, Wang J, Vilsbøll T, Hansen T, Knop FK, Arumugam M, Pedersen O. Recovery of gut microbiota of healthy adults following antibiotic exposure. Nat Microbiol. 2018;3:1255-1265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 560] [Article Influence: 70.0] [Reference Citation Analysis (0)] |
| 36. | Drago L, Toscano M, De Grandi R, Casini V, Pace F. Persisting changes of intestinal microbiota after bowel lavage and colonoscopy. Eur J Gastroenterol Hepatol. 2016;28:532-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 93] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 37. | Barnes EL, Nowell WB, Venkatachalam S, Dobes A, Kappelman MD. Racial and Ethnic Distribution of Inflammatory Bowel Disease in the United States. Inflamm Bowel Dis. 2022;28:983-987. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 38. | Hamilton AL, Kamm MA, De Cruz P, Wright EK, Feng H, Wagner J, Sung JJY, Kirkwood CD, Inouye M, Teo SM. Luminal microbiota related to Crohn's disease recurrence after surgery. Gut Microbes. 2020;11:1713-1728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 39. | Hattori S, Nakamura M, Yamamura T, Maeda K, Sawada T, Mizutani Y, Yamamoto K, Ishikawa T, Furukawa K, Ohno E, Honda T, Kawashima H, Ishigami M, Hirooka Y, Fujishiro M. The microbiome can predict mucosal healing in small intestine in patients with Crohn's disease. J Gastroenterol. 2020;55:1138-1149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 40. | Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790-801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 917] [Cited by in RCA: 1085] [Article Influence: 83.5] [Reference Citation Analysis (0)] |
| 41. | Hernández-Rocha C, Turpin W, Borowski K, Stempak JM, Sabic K, Gettler K, Tastad C, Chasteau C, Korie U, Hanna M, Khan A, Mengesha E, Bitton A, Schwartz MB, Barrie A, Datta LW, Lazarev M, Brant SR, Rioux JD, McGovern DPB, Duerr RH, Schumm LP, Cho JH, Silverberg MS. After Surgically Induced Remission, Ileal and Colonic Mucosa-Associated Microbiota Predicts Crohn's Disease Recurrence. Clin Gastroenterol Hepatol. 2025;23:612-620.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 42. | Kurlinkutė A. Gut Microbiome Impact on Postoperative Morbidity after Major Abdominal Oncological Surgery. M.Sc. Thesis, Vilniaus universitetas. 2024. Available from: https://epublications.vu.lt/object/elaba:210539739/index.html. |
| 43. | Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Jia W, Cai S, Zhao L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 738] [Cited by in RCA: 974] [Article Influence: 64.9] [Reference Citation Analysis (0)] |
| 44. | Lederer AK, Pisarski P, Kousoulas L, Fichtner-Feigl S, Hess C, Huber R. Postoperative changes of the microbiome: are surgical complications related to the gut flora? A systematic review. BMC Surg. 2017;17:125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 45. | Wang R, Taubenberger JK. Methods for molecular surveillance of influenza. Expert Rev Anti Infect Ther. 2010;8:517-527. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 66] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 46. | Ivanov II, Littman DR. Modulation of immune homeostasis by commensal bacteria. Curr Opin Microbiol. 2011;14:106-114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 151] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 47. | Weaver L, Troester A, Jahansouz C. The Impact of Surgical Bowel Preparation on the Microbiome in Colon and Rectal Surgery. Antibiotics (Basel). 2024;13:580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 48. | Ben Izhak M, Eshel A, Cohen R, Madar-Shapiro L, Meiri H, Wachtel C, Leung C, Messick E, Jongkam N, Mavor E, Sapozhnikov S, Maharshak N, Abu-Abeid S, Alis A, Mahler I, Meoded A, Meron Eldar S, Koren O, Louzoun Y. Projection of Gut Microbiome Pre- and Post-Bariatric Surgery To Predict Surgery Outcome. mSystems. 2021;6:e0136720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 49. | Carlini M, Grieco M, Spoletini D, Menditto R, Napoleone V, Brachini G, Mingoli A, Marcellinaro R. Implementation of the gut microbiota prevents anastomotic leaks in laparoscopic colorectal surgery for cancer:the results of the MIRACLe study. Updates Surg. 2022;74:1253-1262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 50. | Jin X, Liu Y, Yan W, Shi S, Liu L, Lin B, Guo X, Cai T, Wei Y. Gut microbiota from nCAL patients promotes colon anastomotic healing by inducing collagen synthesis in epithelial cells. J Gastroenterol Hepatol. 2022;37:1756-1767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 51. | Liu H, Zhang K, Liu P, Xu X, Zhou Y, Gan L, Yao L, Li B, Chen T, Fang N. Improvement Effect of Bifidobacterium animalis subsp. lactis MH-02 in Patients Receiving Resection of Colorectal Polyps: A Randomized, Double-Blind, Placebo-Controlled Trial. Front Immunol. 2022;13:940500. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 52. | Park IJ, Lee JH, Kye BH, Oh HK, Cho YB, Kim YT, Kim JY, Sung NY, Kang SB, Seo JM, Sim JH, Lee JL, Lee IK. Effects of PrObiotics on the Symptoms and Surgical ouTComes after Anterior REsection of Colon Cancer (POSTCARE): A Randomized, Double-Blind, Placebo-Controlled Trial. J Clin Med. 2020;9:2181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 53. | Martínez-Sánchez MA, Núñez-Sánchez MÁ, Balaguer-Román A, Oliva-Bolarín A, Pujante-Gilabert G, Hernández-Agüera Q, Mesa-López MJ, Egea-Valenzuela J, Queipo-Ortuño MI, Ruiz-Alcaraz AJ, Ferrer-Gómez M, Gil-Martínez J, Ramos-Molina B. Gut Microbiome Modification through Dietary Intervention in Patients with Colorectal Cancer: Protocol for a Prospective, Interventional, Controlled, Randomized Clinical Trial in Patients with Scheduled Surgical Intervention for CRC. J Clin Med. 2022;11:3613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 54. | Veziant J, Poirot K, Chevarin C, Cassagnes L, Sauvanet P, Chassaing B, Robin F, Godfraind C, Barnich N, Pezet D, Pereira B, Gagniere J, Bonnet M. Prognostic value of a combination of innovative factors (gut microbiota, sarcopenia, obesity, metabolic syndrome) to predict surgical/oncologic outcomes following surgery for sporadic colorectal cancer: a prospective cohort study protocol (METABIOTE). BMJ Open. 2020;10:e031472. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 55. | Li Y, Tan WH, Wu JC, Huang ZX, Shang YY, Liang B, Chen JH, Pang R, Xie XQ, Zhang JM, Ding Y, Xue L, Chen MT, Wang J, Wu QP. Microbiologic risk factors of recurrent choledocholithiasis post-endoscopic sphincterotomy. World J Gastroenterol. 2022;28:1257-1271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (1)] |
| 56. | Liu H, Cheng G, Xu YL, Fang Q, Ye L, Wang CH, Liu XS. Preoperative Status of Gut Microbiota Predicts Postoperative Delirium in Patients With Gastric Cancer. Front Psychiatry. 2022;13:852269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 57. | Vipperla K, O'Keefe SJ. The microbiota and its metabolites in colonic mucosal health and cancer risk. Nutr Clin Pract. 2012;27:624-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 93] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 58. | Liu Y, Geng R, Liu L, Jin X, Yan W, Zhao F, Wang S, Guo X, Ghimire G, Wei Y. Gut Microbiota-Based Algorithms in the Prediction of Metachronous Adenoma in Colorectal Cancer Patients Following Surgery. Front Microbiol. 2020;11:1106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 59. | Nassani N, Alsheikh M, Carroll B, Nguyen D, Carroll RE. Theranostic Gastrointestinal Endoscopy: Bringing Healing Light to the Lumen. Clin Transl Gastroenterol. 2020;11:e00119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 60. | Machiels K, Pozuelo Del Río M, Martinez-De la Torre A, Xie Z, Pascal Andreu V, Sabino J, Santiago A, Campos D, Wolthuis A, D'Hoore A, De Hertogh G, Ferrante M, Manichanh C, Vermeire S. Early Postoperative Endoscopic Recurrence in Crohn's Disease Is Characterised by Distinct Microbiota Recolonisation. J Crohns Colitis. 2020;14:1535-1546. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 61. | Machiels K, Sabino J, Vandermosten L, Joossens M, Arijs I, de Bruyn M, Eeckhaut V, Van Assche G, Ferrante M, Verhaegen J, Van Steen K, Van Immerseel F, Huys G, Verbeke K, Wolthuis A, de Buck Van Overstraeten A, D'Hoore A, Rutgeerts P, Vermeire S. Specific members of the predominant gut microbiota predict pouchitis following colectomy and IPAA in UC. Gut. 2017;66:79-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 109] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 62. | Liu Y, Méric G, Havulinna AS, Teo SM, Åberg F, Ruuskanen M, Sanders J, Zhu Q, Tripathi A, Verspoor K, Cheng S, Jain M, Jousilahti P, Vázquez-Baeza Y, Loomba R, Lahti L, Niiranen T, Salomaa V, Knight R, Inouye M. Early prediction of incident liver disease using conventional risk factors and gut-microbiome-augmented gradient boosting. Cell Metab. 2022;34:719-730.e4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 57] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 63. | Martins FP, Andrade-Silva J, Teixeira BL, Ferrari A, Christoff AP, Cruz GNF, Paladino FV, de Oliveira LFV, Hernandes C. Oral microbiome test as an alternative diagnostic tool for gastric alterations: A prospective, bicentric cross-sectional study. PLoS One. 2024;19:e0314660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 64. | Mima K, Sakamoto Y, Kosumi K, Ogata Y, Miyake K, Hiyoshi Y, Ishimoto T, Iwatsuki M, Baba Y, Iwagami S, Miyamoto Y, Yoshida N, Ogino S, Baba H. Mucosal cancer-associated microbes and anastomotic leakage after resection of colorectal carcinoma. Surg Oncol. 2020;32:63-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 65. | Mondot S, Lepage P, Seksik P, Allez M, Tréton X, Bouhnik Y, Colombel JF, Leclerc M, Pochart P, Doré J, Marteau P; GETAID. Structural robustness of the gut mucosal microbiota is associated with Crohn's disease remission after surgery. Gut. 2016;65:954-962. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 121] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 66. | Moss A, Bourke MJ, Williams SJ, Hourigan LF, Brown G, Tam W, Singh R, Zanati S, Chen RY, Byth K. Endoscopic mucosal resection outcomes and prediction of submucosal cancer from advanced colonic mucosal neoplasia. Gastroenterology. 2011;140:1909-1918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 479] [Cited by in RCA: 451] [Article Influence: 30.1] [Reference Citation Analysis (1)] |
| 67. | Yu L, Zhao G, Wang L, Zhou X, Sun J, Li X, Zhu Y, He Y, Kofonikolas K, Bogaert D, Dunlop M, Zhu Y, Theodoratou E, Li X. A systematic review of microbial markers for risk prediction of colorectal neoplasia. Br J Cancer. 2022;126:1318-1328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 68. | Yu J, Nong C, Zhao J, Meng L, Song J. An Integrative Bioinformatic Analysis of Microbiome and Transcriptome for Predicting the Risk of Colon Adenocarcinoma. Dis Markers. 2022;2022:7994074. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 69. | Nardone OM, Bazarova A, Bhandari P, Cannatelli R, Daperno M, Ferraz J, Goetz M, Gui X, Hayee B, De Hertogh G, Lazarev M, Li J, Parra-Blanco A, Pastorelli L, Panaccione R, Occhipinti V, Rath T, Smith SCL, Shivaji UN, Tontini GE, Vieth M, Villanacci V, Zardo D, Bisschops R, Kiesslich R, Ghosh S, Iacucci M. PICaSSO virtual electronic chromendoscopy accurately reflects combined endoscopic and histological assessment for prediction of clinical outcomes in ulcerative colitis. United European Gastroenterol J. 2022;10:147-159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 70. | Ó Cuív P, Ljungberg JK, Zhou J, Nissen M, Krueger A, Boyd J, Bongers M, Jeimy JL, Vivian C, Reich E, Rabellino A, Newell R, Fang L, Macdonald S, Pribyl A, Caban S, Mccarthy H, Soh J, Reid L, Wills B, Mchale D, Wood DL, Frazer IH, Angel N, Chipperfield H, Begun J, Keely S, Tyson GW, Hugenholtz P, Munro T, Krause L. A human microbiome-derived therapeutic for ulcerative colitis promotes mucosal healing and immune homeostasis. 2025 Preprint. Available from: bioRxiv: 10.1101/2025.04.18.25325957. [DOI] [Full Text] |
| 71. | Caenepeel C, Sadat Seyed Tabib N, Vieira-Silva S, Vermeire S. Review article: how the intestinal microbiota may reflect disease activity and influence therapeutic outcome in inflammatory bowel disease. Aliment Pharmacol Ther. 2020;52:1453-1468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 72. | O'Brien CL, Allison GE, Grimpen F, Pavli P. Impact of colonoscopy bowel preparation on intestinal microbiota. PLoS One. 2013;8:e62815. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 77] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 73. | Liu Y, Li B, Wei Y. New understanding of gut microbiota and colorectal anastomosis leak: A collaborative review of the current concepts. Front Cell Infect Microbiol. 2022;12:1022603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 74. | Williamson AJ, Alverdy JC. Influence of the Microbiome on Anastomotic Leak. Clin Colon Rectal Surg. 2021;34:439-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 75. | Wang H, Yan G, Wu Y, Zhuoma D, Liu Z, Gao X, Wang X. Fecal microbiota related to postoperative endoscopic recurrence in patients with Crohn's disease. Gastroenterol Rep (Oxf). 2024;12:goae017. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 76. | Wira CR, Ghosh M, Smith JM, Shen L, Connor RI, Sundstrom P, Frechette GM, Hill ET, Fahey JV. Epithelial cell secretions from the human female reproductive tract inhibit sexually transmitted pathogens and Candida albicans but not Lactobacillus. Mucosal Immunol. 2011;4:335-342. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 55] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 77. | Puca P, Petito V, Laterza L, Lopetuso LR, Neri M, Del Chierico F, Boskoski I, Gasbarrini A, Scaldaferri F. Bariatric procedures and microbiota: patient selection and outcome prediction. Ther Adv Gastrointest Endosc. 2021;14:26317745211014746. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 78. | Reinink AR, Lee TC, Higgins PD. Endoscopic Mucosal Healing Predicts Favorable Clinical Outcomes in Inflammatory Bowel Disease: A Meta-analysis. Inflamm Bowel Dis. 2016;22:1859-1869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 110] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 79. | Seo H, Park JY, You HS, Kim BJ, Kim JG. Comparative Changes in Fecal Microbiome After Endoscopic Resection and Surgical Resection in Gastric Cancer Patients. J Pers Med. 2025;15:144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 80. | Shobar RM, Velineni S, Keshavarzian A, Swanson G, DeMeo MT, Melson JE, Losurdo J, Engen PA, Sun Y, Koenig L, Mutlu EA. The Effects of Bowel Preparation on Microbiota-Related Metrics Differ in Health and in Inflammatory Bowel Disease and for the Mucosal and Luminal Microbiota Compartments. Clin Transl Gastroenterol. 2016;7:e143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 86] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 81. | Sokol H, Brot L, Stefanescu C, Auzolle C, Barnich N, Buisson A, Fumery M, Pariente B, Le Bourhis L, Treton X, Nancey S, Allez M, Seksik P; REMIND Study Group Investigators. Prominence of ileal mucosa-associated microbiota to predict postoperative endoscopic recurrence in Crohn's disease. Gut. 2020;69:462-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 92] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 82. | Stavrou G, Kotzampassi K. Gut microbiome, surgical complications and probiotics. Ann Gastroenterol. 2017;30:45-53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 83. | Ruseckaitė S. The Influence of The Microbiota On Anastomosis Complications After Primary Colorectal Surgery: A Literature Review. M.Sc. Thesis, Lithuanian University Of Health Sciences. 2025. Available from: https://portalcris.lsmuni.lt/server/api/core/bitstreams/57152b8a-fd54-43ce-90dc-082a676422ff/content. |
| 84. | Tsigalou C, Paraschaki A, Bragazzi NL, Aftzoglou K, Bezirtzoglou E, Tsakris Z, Vradelis S, Stavropoulou E. Alterations of gut microbiome following gastrointestinal surgical procedures and their potential complications. Front Cell Infect Microbiol. 2023;13:1191126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 85. | van Praagh JB, de Goffau MC, Bakker IS, Harmsen HJ, Olinga P, Havenga K. Intestinal microbiota and anastomotic leakage of stapled colorectal anastomoses: a pilot study. Surg Endosc. 2016;30:2259-2265. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 86. | Xiao H, Luo H, Qin A, Shu W, Liu X, Xiao F, Liao X, Shi Z, Zou Y, Xu K, Cao S, Li C, Hu Y, Zhang S, Guo J, Wang S, Yan S. Comparing Participation and Interim Effectiveness of Endoscopy and Biomarker-Based Screening for Gastric Cancer: A Cluster Randomized Controlled Trial. J Cancer. 2024;15:6110-6121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 87. | Zaramella A, Arcidiacono D, Duci M, Benna C, Pucciarelli S, Fantin A, Rosato A, De Re V, Cannizzaro R, Fassan M, Realdon S. Predictive Value of a Gastric Microbiota Dysbiosis Test for Stratifying Cancer Risk in Atrophic Gastritis Patients. Nutrients. 2024;17:142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 88. | Zheng J, Sun Q, Zhang J, Ng SC. The role of gut microbiome in inflammatory bowel disease diagnosis and prognosis. United European Gastroenterol J. 2022;10:1091-1102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 89. | Zheng Z, Hu Y, Tang J, Xu W, Zhu W, Zhang W. The implication of gut microbiota in recovery from gastrointestinal surgery. Front Cell Infect Microbiol. 2023;13:1110787. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 34] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 90. | Dadkhah E, Sikaroodi M, Korman L, Hardi R, Baybick J, Hanzel D, Kuehn G, Kuehn T, Gillevet PM. Gut microbiome identifies risk for colorectal polyps. BMJ Open Gastroenterol. 2019;6:e000297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 91. | Ferrie S, Webster A, Wu B, Tan C, Carey S. Gastrointestinal surgery and the gut microbiome: a systematic literature review. Eur J Clin Nutr. 2021;75:12-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 92. | Hajjar R, Gonzalez E, Fragoso G, Oliero M, Alaoui AA, Calvé A, Vennin Rendos H, Djediai S, Cuisiniere T, Laplante P, Gerkins C, Ajayi AS, Diop K, Taleb N, Thérien S, Schampaert F, Alratrout H, Dagbert F, Loungnarath R, Sebajang H, Schwenter F, Wassef R, Ratelle R, Debroux E, Cailhier JF, Routy B, Annabi B, Brereton NJB, Richard C, Santos MM. Gut microbiota influence anastomotic healing in colorectal cancer surgery through modulation of mucosal proinflammatory cytokines. Gut. 2023;72:1143-1154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 77] [Article Influence: 25.7] [Reference Citation Analysis (4)] |
| 93. | Jacob V, Crawford C, Cohen-Mekelburg S, Viladomiu M, Putzel GG, Schneider Y, Chabouni F, OʼNeil S, Bosworth B, Woo V, Ajami NJ, Petrosino JF, Gerardin Y, Kassam Z, Smith M, Iliev ID, Sonnenberg GF, Artis D, Scherl E, Longman RS. Single Delivery of High-Diversity Fecal Microbiota Preparation by Colonoscopy Is Safe and Effective in Increasing Microbial Diversity in Active Ulcerative Colitis. Inflamm Bowel Dis. 2017;23:903-911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 94] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 94. | Koliarakis I, Athanasakis E, Sgantzos M, Mariolis-Sapsakos T, Xynos E, Chrysos E, Souglakos J, Tsiaoussis J. Intestinal Microbiota in Colorectal Cancer Surgery. Cancers (Basel). 2020;12:3011. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/