Published online Sep 27, 2025. doi: 10.4254/wjh.v17.i9.109429
Revised: June 6, 2025
Accepted: July 31, 2025
Published online: September 27, 2025
Processing time: 138 Days and 3.6 Hours
In this letter, we discuss the recently published study by Pan et al, which investigated the relationships between interleukin-36 (IL-36) subfamily cytokines and the gut microbiota in patients diagnosed with liver cirrhosis. This observational study revealed that the serum levels of IL-36α, IL-36γ, and IL-38 were significantly elevated in liver cirrhosis patients, accompanied by a distinct gut microbiota profile. These findings provide novel insights into the role of inflammatory cytokines in the imbalance of the gut-liver axis. Meanwhile, in our studies, it was found that IL-36γ is considerably increased in a mouse model of metabolic dys
Core Tip: This letter discusses the important role of the interleukin-36 (IL-36) subfamily in the pathogenesis of liver diseases, particularly in modulating immune responses and promoting inflammation. Although considerable knowledge gaps remain regarding the precise mechanisms through which IL-36 cytokines contribute to liver damage, emerging evidence suggests that the targeting of IL-36 signaling may present novel therapeutic avenues for the management of chronic liver diseases. Further research is essential to investigate the potential of IL-36 as both a biomarker and a therapeutic target within clinical contexts.
- Citation: Xiong ZK, Gu SM, Zheng YY. Interleukin-36 subfamily cytokines in liver diseases. World J Hepatol 2025; 17(9): 109429
- URL: https://www.wjgnet.com/1948-5182/full/v17/i9/109429.htm
- DOI: https://dx.doi.org/10.4254/wjh.v17.i9.109429
The recent study by Pan et al[1] caught our attention as it examined the link between interleukin-36 (IL-36) cytokines and the gut microbiota in liver cirrhosis (LC) patients. This investigation sheds light on the association between these cytokines and the gut microbiota, thereby offering a new insight into the gut-liver axis. This study reveals an exciting avenue for exploration in the field of hepatology; however, there are several additional considerations that warrant further discussion.
This observational study revealed that the serum levels of IL-36α, IL-36γ, and IL-38 are significantly elevated in LC patients and correlate positively with the Child-Pugh score, thus suggesting a relationship between these cytokines and LC progression. Their data further revealed a distinct gut microbiota profile characterized by increased abundances of pathogenic bacteria, such as Veillonella (3.2% vs 0.8% in healthy control, P < 0.05), Bacteroides (15.6% vs 9.1%), and Fusobacterium (2.1% vs 0.4%), and decreased abundances of beneficial bacteria, including Lactobacillus (0.5% vs 2.3%) and Bifidobacterium (0.3% vs 1.5%). This observation reinforces the possible involvement of IL-36 in managing the gut microbiota among patients with LC. By exploring the connection between the IL-36 subfamily and gut microbiota in LC, this study addresses a considerable gap in the literature. This area remains relatively underexplored, and the findings provide novel insights into the involvement of inflammatory cytokines in imbalances in the gut-liver axis, which may ultimately lead to the identification of innovative therapeutic targets for the management of LC.
While this study pinpointed IL-36 cytokines, including IL-36α, IL-36γ, IL-36 receptor antagonist (IL-36Ra), and IL-38, as significant factors in LC-related inflammation, several limitations warrant careful consideration. First, although the sample size of the study (61 LC patients and 29 healthy controls) is deemed acceptable, it is relatively limited owing to the significant heterogeneity inherent in microbial communities identified through clinical samples, which constrains the generalizability of the findings[2]. Expanding this study to include a larger cohort would increase the statistical power of the findings and allow for more robust conclusions. Moreover, the cross-sectional nature of this study provides only a snapshot of the IL-36 subfamily and microbiota at a single point in time. Longitudinal studies that include monitoring of fluctuations in IL-36 subfamily levels and the composite of the microbiota over extended periods would yield more valuable insights into factors underlying disease occurrence and progression.
Moreover, the primary shortcoming of this study is the lack of mechanistic investigation, such as a lack of receptor expression data. While the serum levels of IL-36 subfamily cytokines were measured in this study, data specific to liver tissue would have offered deeper insights into localized inflammation. Notably, IL-36 cytokine concentrations are frequently lower in the liver, complicating direct investigations into their roles in hepatic inflammation. This gap in understanding could be addressed by examining intrahepatic cytokine levels and their specific interactions with liver-resident cells such as hepatocytes and macrophages.
Interestingly, our studies revealed that although the levels of IL-36α, IL-36β, IL-36Ra, and IL-38 are exceedingly low in both healthy and morbid mouse liver tissues, IL-36γ expression is significantly elevated in a metabolic dysfunction-associated liver disease mouse model. These findings suggest that IL-36γ may be a key cytokine in the progression of chronic liver inflammation, thereby supporting the hypothesis that IL-36 could play an underappreciated role in liver diseases.
On the basis of our observations, the elevation of IL-36γ levels in chronic liver inflammation may be linked to the activation of CD4+ T cells and macrophages. Specifically, IL-36γ may promote T helper type 17 differentiation through dendritic cell activation and IL-23 secretion, while enhancing M1 macrophage polarization via nuclear factor-κB-dependent cytokine production (e.g., IL-1β and tumor necrosis factor-α), and ultimately, this cascade could amplify hepatic inflammation and fibrosis[3]. Consequently, further research into the downstream signaling pathways associated with IL-36γ may facilitate the development of targeted therapeutic strategies aimed at more effectively managing liver diseases.
IL-36 constitutes a cytokine family within the broader IL-1 superfamily, which encompasses IL-36α, IL-36β, and IL-36γ, as well as the antagonist IL-36Ra and regulatory cytokine IL-38[4]. The biological functions of IL-36 cytokines have primarily been explored in the context of inflammatory and autoimmune diseases, with emerging evidence indicating their significant roles in various liver diseases[5]. For example, IL-36 may contribute to post-transplant immunity or anesthetic effects[6,7].
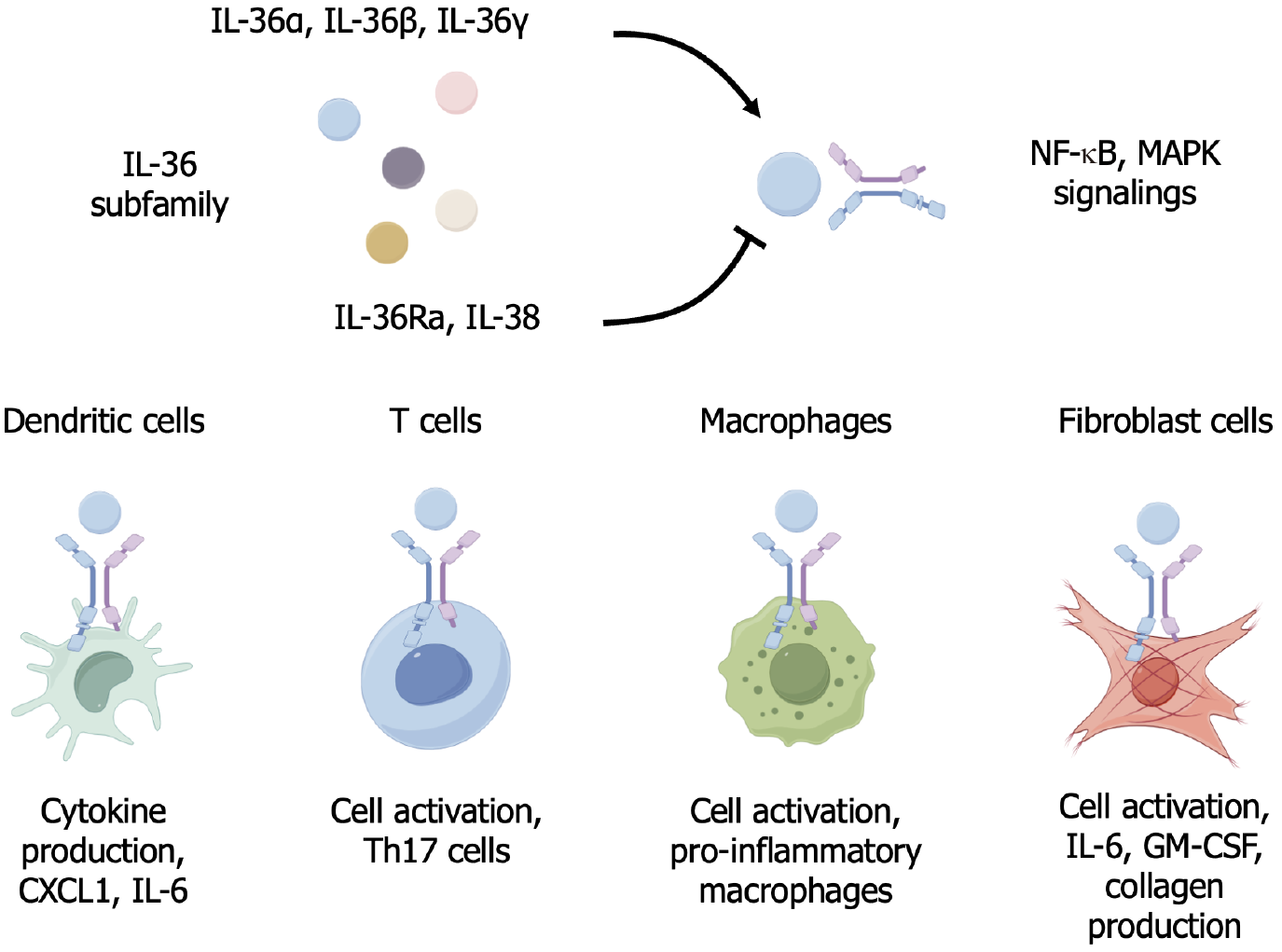
IL-36 cytokines, particularly IL-36α, IL-36β, and IL-36γ, are produced in response to tissue damage or inflammatory stimuli and are implicated in both acute and chronic liver conditions, including hepatitis, fibrosis, and cirrhosis[8,9]. These cytokines influence a variety of immune and nonimmune cells in the liver, including dendritic cells, macrophages, T cells, and fibroblasts (Figure 1).
IL-36 cytokines initiate signaling cascades by binding to IL-36 receptor (IL-36R), which is also referred to as IL-1 receptor-related protein 2, a receptor widely expressed in various immune and parenchymal cells in the liver that is capable of activating the downstream nuclear factor-κB and mitogen-activated protein kinase pathways[10]. Specifically, upon binding to IL-36R, these cytokines recruit the IL-1 receptor accessory protein, thereby forming a signaling complex that triggers the activation of a cascade of intracellular pathways, which are essential for inducing the transcription of proinflammatory genes, including those encoding tumor necrosis factor-α, IL-1β, and IL-6, among others. In the context of various diseases, this signaling can contribute to sustained inflammation, fibrosis, and hepatocyte injury[11-13].
Given the central role of IL-36 in mediating liver inflammation, targeting the IL-36 signaling pathway has emerged as a promising therapeutic strategy. Anti-IL-36R antibodies, as antagonists of IL-36, may block the inflammatory cascade initiated by IL-36 cytokines. In addition, the use of monoclonal antibodies targeting IL-36R may be an effective approach to modulate liver inflammation and fibrosis. However, it should be noted that systemic IL-36R blockade may impair mucosal immunity, altering the composition of the gut microbiota. Thus, future studies should assess whether localized liver-targeted delivery mitigates such risks.
The IL-36 subfamily could be crucial in the development of liver diseases, particularly in modulating immune responses and promoting inflammation. Although much remains to be elucidated regarding the precise mechanisms by which IL-36 cytokines contribute to liver damage, emerging evidence indicates that targeting IL-36 signaling may represent a novel therapeutic strategy for the management of chronic liver diseases, including inflammation, cirrhosis, and fibrosis. Further research, such as longitudinal patient sampling and interventional animal studies, is imperative to investigate the potential value of IL-36 as both a biomarker and a treatment target in clinical practice.
| 1. | Pan YZ, Chen WT, Jin HR, Liu Z, Gu YY, Wang XR, Wang J, Lin JJ, Zhou Y, Xu LM. Correlation between the interleukin-36 subfamily and gut microbiota in patients with liver cirrhosis: Implications for gut-liver axis imbalance. World J Hepatol. 2025;17:105660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 2. | Hu YC, Ding XC, Liu HJ, Ma WL, Feng XY, Ma LN. Effects of Lactobacillus paracasei N1115 on gut microbial imbalance and liver function in patients with hepatitis B-related cirrhosis. World J Gastroenterol. 2024;30:1556-1571. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (1)] |
| 3. | Dou JY, Zhou MJ, Xuan MY, Guo J, Liu SH, Lian LH, Cui ZY, Nan JX, Wu YL. Astilbin alleviates hepatic fibrosis through PXR-PINK1/Parkin pathway: A new strategy by regulating hepatic stellate cells-macrophage crosstalk. Phytomedicine. 2024;135:156144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 4. | Barbier L, Ferhat M, Salamé E, Robin A, Herbelin A, Gombert JM, Silvain C, Barbarin A. Interleukin-1 Family Cytokines: Keystones in Liver Inflammatory Diseases. Front Immunol. 2019;10:2014. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 123] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 5. | Tsutsui H, Cai X, Hayashi S. Interleukin-1 Family Cytokines in Liver Diseases. Mediators Inflamm. 2015;2015:630265. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 6. | Li R, Mukherjee MB, Jin Z, Liu H, Lin K, Liu Q, Dilger JP, Lin J. The Potential Effect of General Anesthetics in Cancer Surgery: Meta-Analysis of Postoperative Metastasis and Inflammatory Cytokines. Cancers (Basel). 2023;15:2759. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 38] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 7. | Jin Z, Zhang W, Liu H, Ding A, Lin Y, Wu SX, Lin J. Potential Therapeutic Application of Local Anesthetics in Cancer Treatment. Recent Pat Anticancer Drug Discov. 2022;17:326-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 8. | Bamias G, Cominelli F. Cytokines and intestinal inflammation. Curr Opin Gastroenterol. 2016;32:437-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 9. | Neurath MF. IL-36 in chronic inflammation and cancer. Cytokine Growth Factor Rev. 2020;55:70-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 10. | Teshima R, Saito-Sasaki N, Sawada Y. Generalized Pustular Psoriasis and Systemic Organ Dysfunctions. Int J Mol Sci. 2024;25:6270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 11. | Ge Y, Huang M, Yao YM. Autophagy and proinflammatory cytokines: Interactions and clinical implications. Cytokine Growth Factor Rev. 2018;43:38-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 149] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 12. | Cao J, Liu JH, Wise SG, Fan J, Bao S, Zheng GS. The role of IL-36 and 37 in hepatocellular carcinoma. Front Immunol. 2024;15:1281121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 13. | Liu H, Tang T. MAPK signaling pathway-based glioma subtypes, machine-learning risk model, and key hub proteins identification. Sci Rep. 2023;13:19055. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 62] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/