©The Author(s) 2026.
World J Hepatol. Feb 27, 2026; 18(2): 111099
Published online Feb 27, 2026. doi: 10.4254/wjh.v18.i2.111099
Published online Feb 27, 2026. doi: 10.4254/wjh.v18.i2.111099
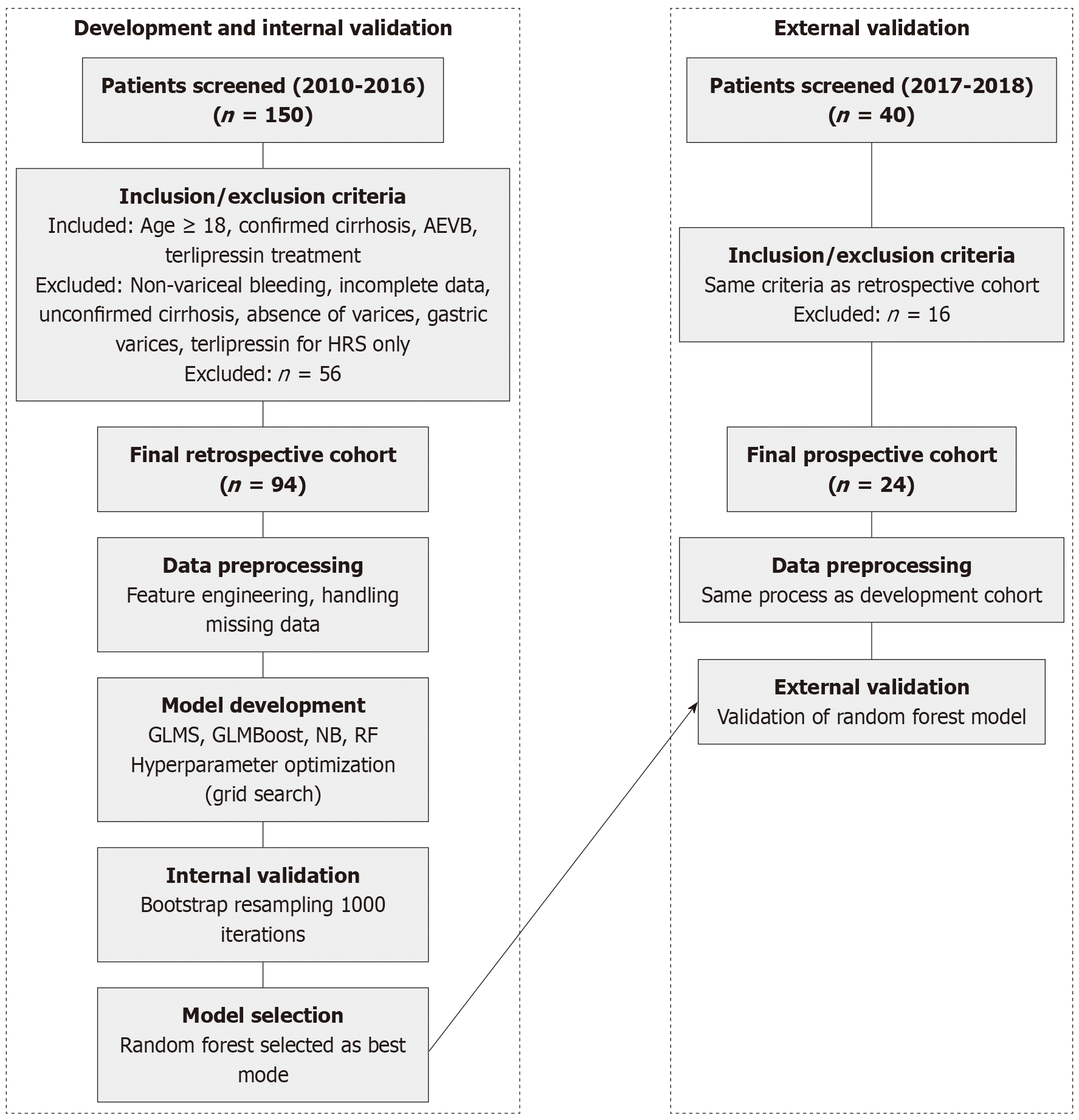
Figure 1 Study flowchart illustrating patient selection and analytical methodology.
Left panel shows the development and internal validation process (2010-2016), including patient screening, selection criteria, data preprocessing, model development, and validation steps. Right panel demonstrates the external validation process (2017-2018) using an independent prospective cohort. AEVB: Acute esophageal variceal bleeding; GLMs: Generalized linear models; GLMBoost: Generalized linear models with boosting; HRS: Hepatorenal syndrome; NB: Naive Bayes; RF: Random forest.
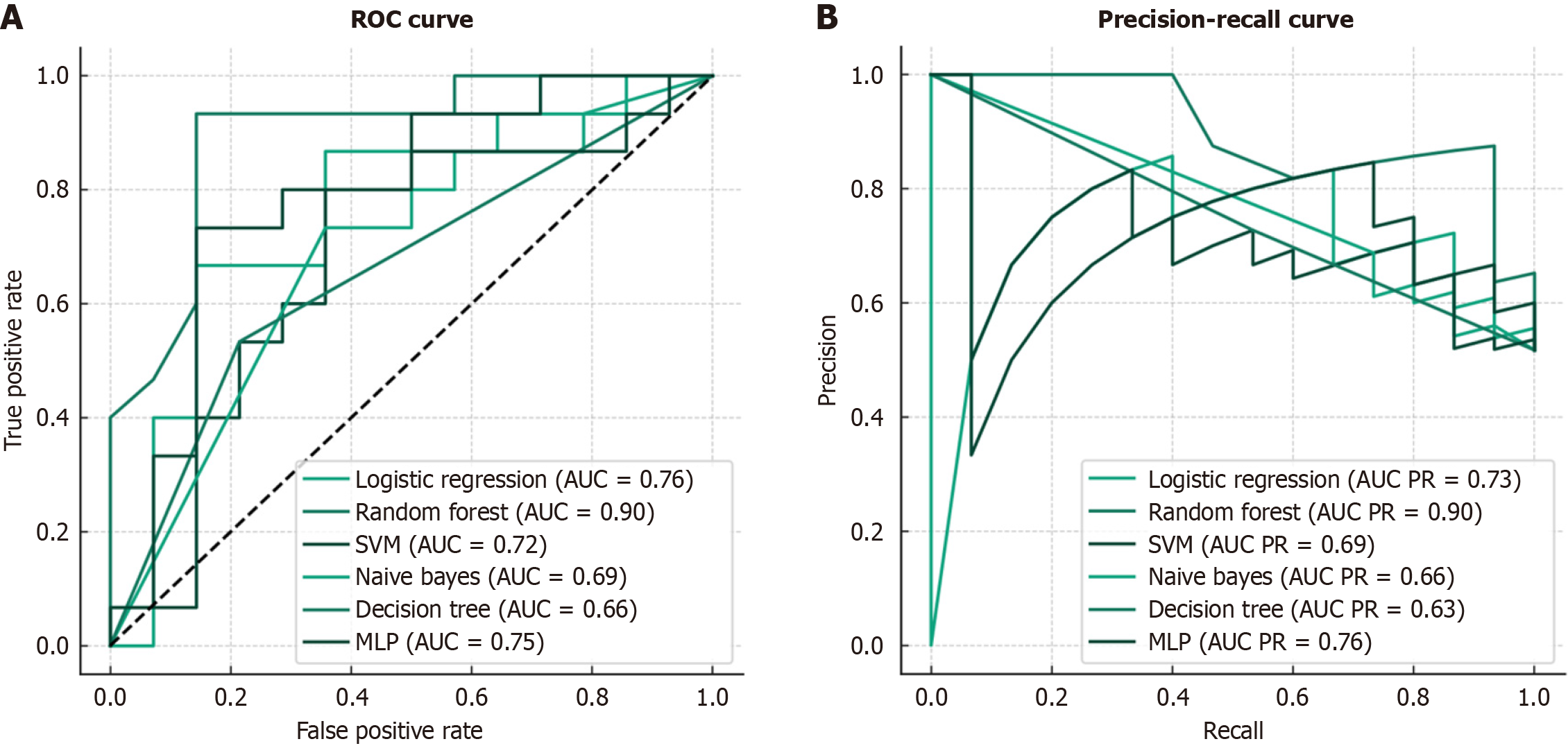
Figure 2 Performance comparison of machine learning models for predicting one-year mortality in patients with cirrhosis with acute esophageal variceal bleeding.
A: Receiver operating characteristic (ROC) curves showing the trade-off between true-positive rate and false-positive rate for each model; B: Precision-recall (PR) curves demonstrating the relationship between precision and recall across different classification thresholds. AUC: Area under the curve; MLP: Multilayer perceptron; SVM: Support vector machine.
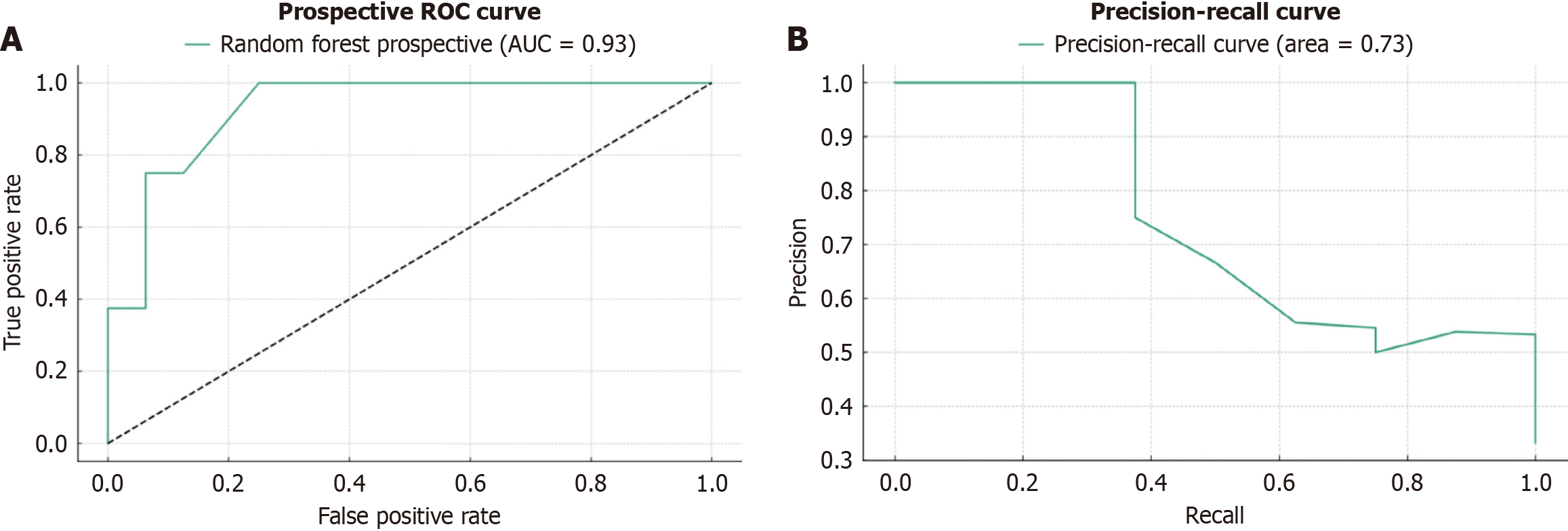
Figure 3 Performance evaluation of the random forest model in the prospective validation cohort.
A: Receiver operating characteristic (ROC) curve showing the relationship between true-positive rate and false-positive rate. The dashed line represents random prediction; B: Precision-recall curve demonstrating the model’s predictive performance across different classification thresholds. AUC: Area under the curve.
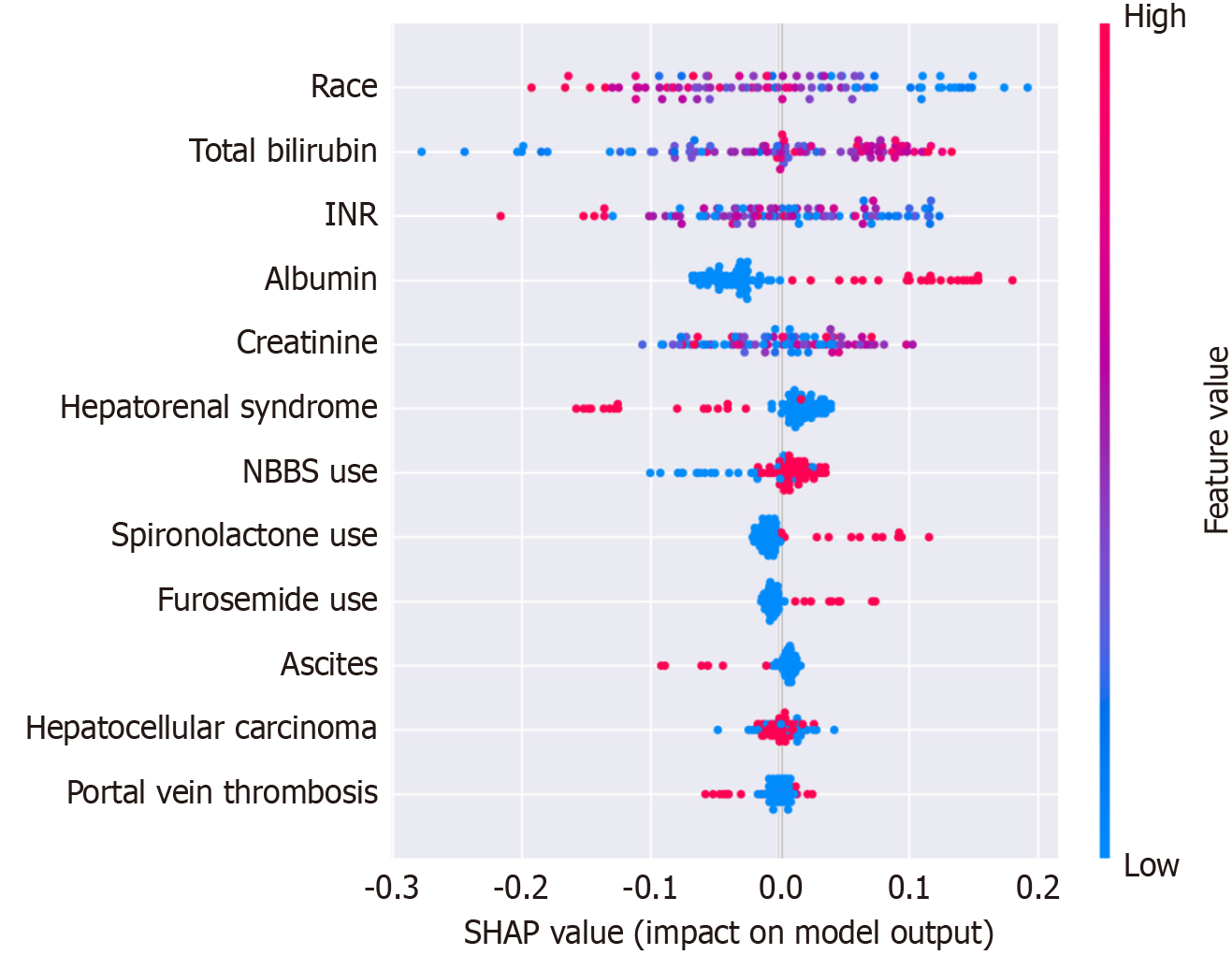
Figure 4 SHapley Additive exPlanations value distribution for predictor variables in the random forest model.
The X-axis represents SHapley Additive exPlanations values, indicating the magnitude and direction of each feature's impact on model output. Each point represents an individual observation, with color intensity indicating the feature value (blue: Low, red: High). Features are ordered by mean absolute SHapley Additive exPlanations value, with race showing the highest overall impact on model predictions. The vertical line at X = 0 represents no effect on model output. INR: International normalized ratio; NBBS: Non-selective beta-blockers; SHAP: SHapley Additive exPlanations.
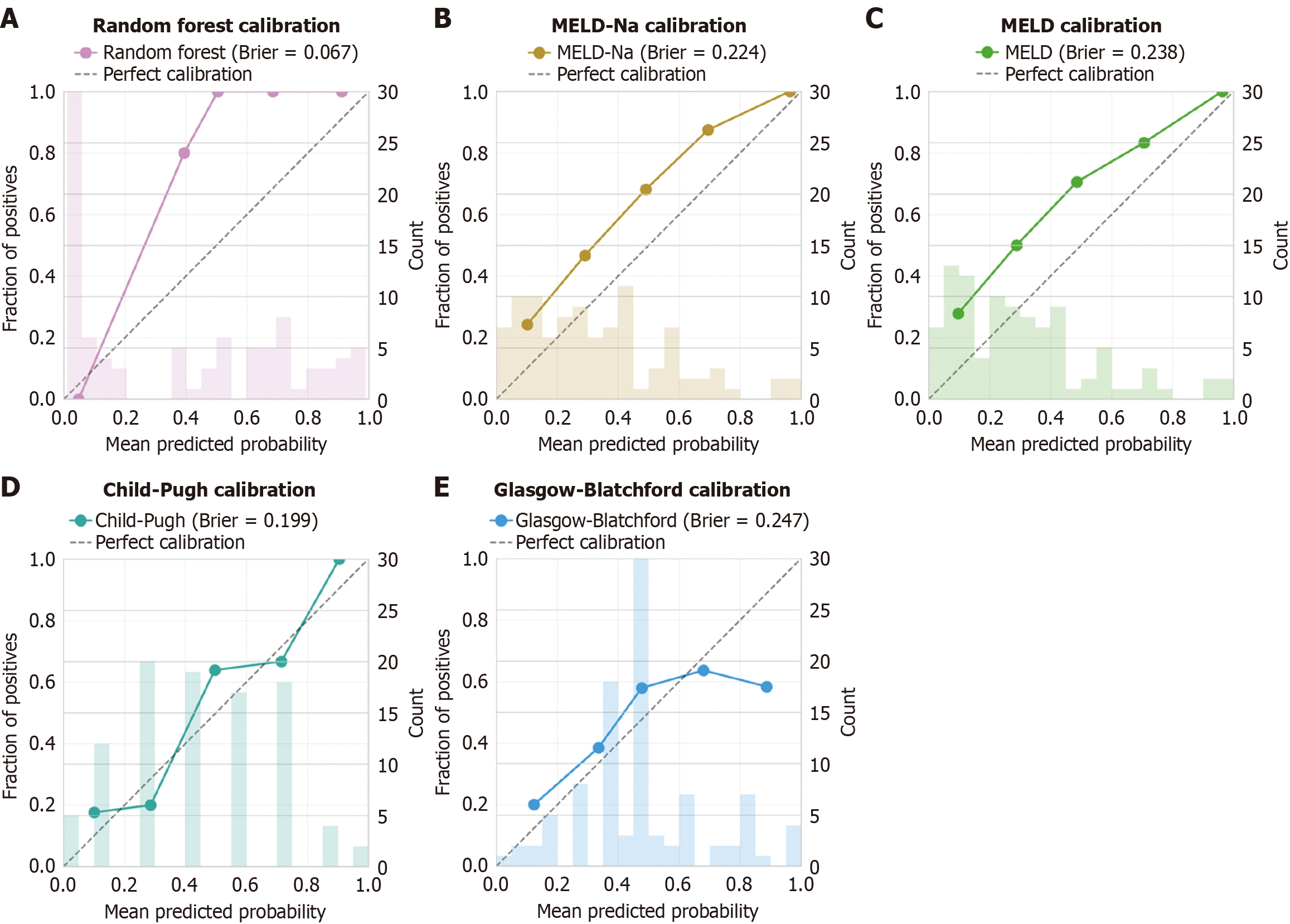
Figure 5 Calibration plots for mortality prediction models.
A: Random Forest; B: Model for end-stage liver disease-serum sodium; C: Model for end-stage liver disease; D: Child-Pugh; E: Glasgow-Blatchford scores. Calibration curves (solid lines) compare predicted probabilities (X-axis) with observed mortality rates (Y-axis). Perfect calibration is shown by the diagonal dashed line. Histograms display the distribution of predictions. The random forest model shows excellent calibration (Brier score = 0.067), while traditional scores show systematically poorer calibration (Brier scores 0.199-0.247). n = 97 patients from the retrospective cohort. MELD: Model for end-stage liver disease; MELD-Na: Model for end-stage liver disease-serum sodium.
- Citation: Rech MM, Corso LL, Dal Bó EF, Ferraza AD, Tomé F, Terres AZ, Balbinot RS, Balbinot RA, Balbinot SS, Soldera J. Development and prospective validation of a machine learning model to predict mortality in cirrhosis with esophageal variceal bleeding. World J Hepatol 2026; 18(2): 111099
- URL: https://www.wjgnet.com/1948-5182/full/v18/i2/111099.htm
- DOI: https://dx.doi.org/10.4254/wjh.v18.i2.111099