©The Author(s) 2025.
World J Hepatol. May 27, 2025; 17(5): 104519
Published online May 27, 2025. doi: 10.4254/wjh.v17.i5.104519
Published online May 27, 2025. doi: 10.4254/wjh.v17.i5.104519
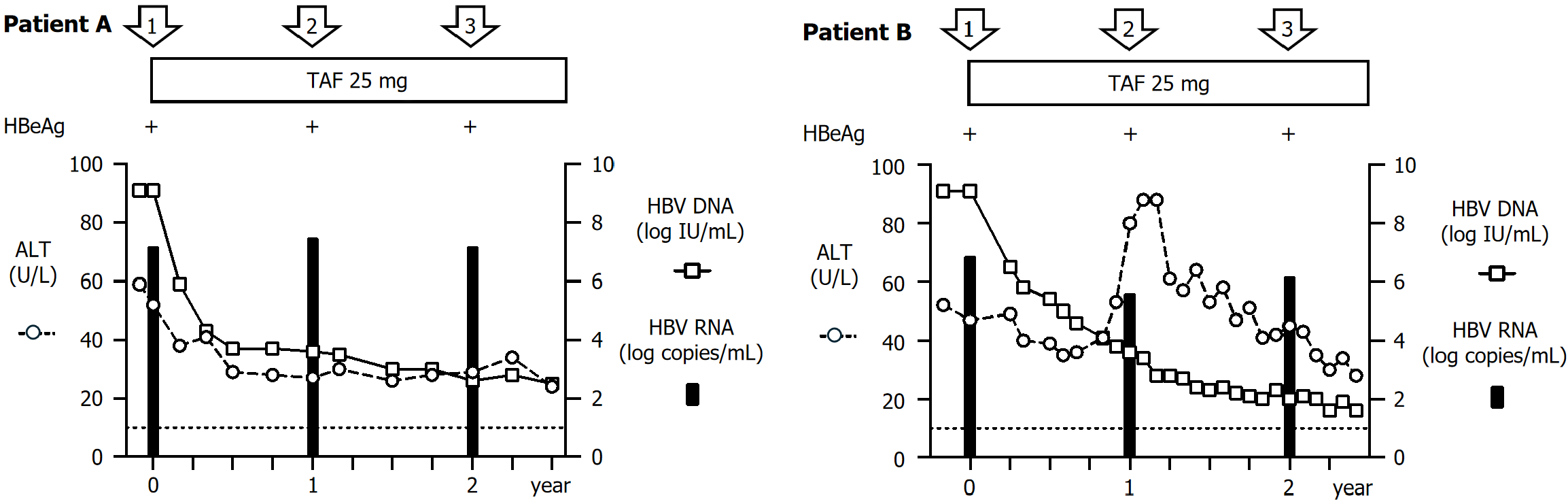
Figure 1 Clinical courses of two tenofovir alafenamide fumarate-incomplete responders.
Serum alanine aminotransferase levels (open circles), hepatitis B virus (HBV) DNA titers (open squares), HBV RNA titers (closed bars), and status of HBeAg are indicated. The lower limit of detection of HBV DNA is indicated by the dotted line. The serum samples were collected at the time points indicated by open arrows with numbers. ALT: Alanine aminotransferase; HBV: Hepatitis B virus; TAF: Tenofovir alafenamide fumarate; HBeAg: Hepatitis B e antigen.
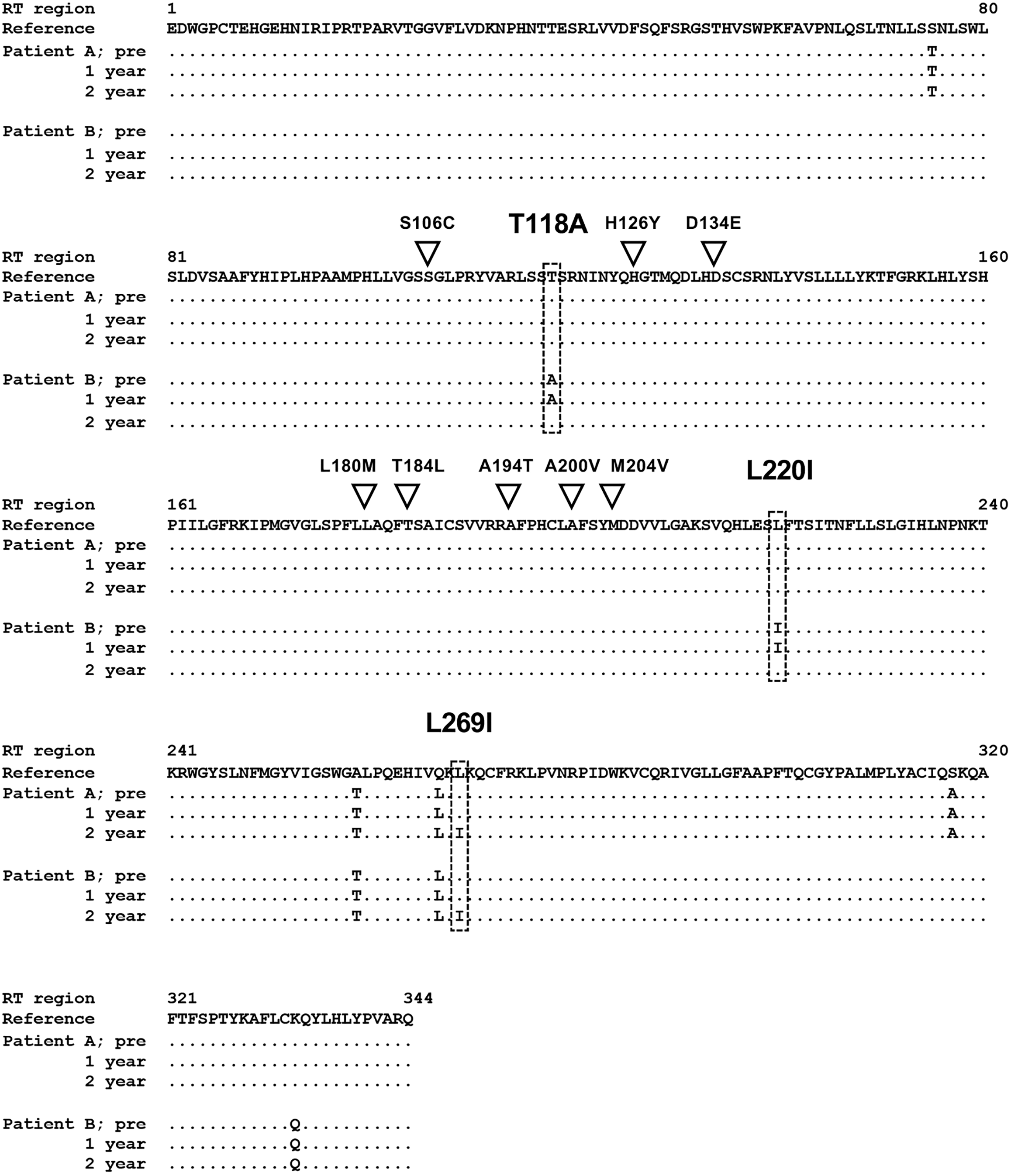
Figure 2 Alignment of reverse transcriptase regions of hepatitis B virus strains detected in tenofovir alafenamide fumarate-incomplete responders.
The reverse transcriptase regions of the hepatitis B virus strains detected in tenofovir alafenamide fumarate (TAF)-incomplete responders before treatment (pre), 1 year after TAF treatment (1 year), and 2 years after TAF treatment (2 year) were directly sequenced, and deduced amino acids were aligned with the reference sequence of genotype C (accession number: GQ872210). The inverted triangle indicates the positions of the reported resistance-associated mutations to tenofovir disoproxil fumarate and TAF. A box with a dotted line indicates the positions of the mutations detected in this study. RT: Reverse transcriptase.
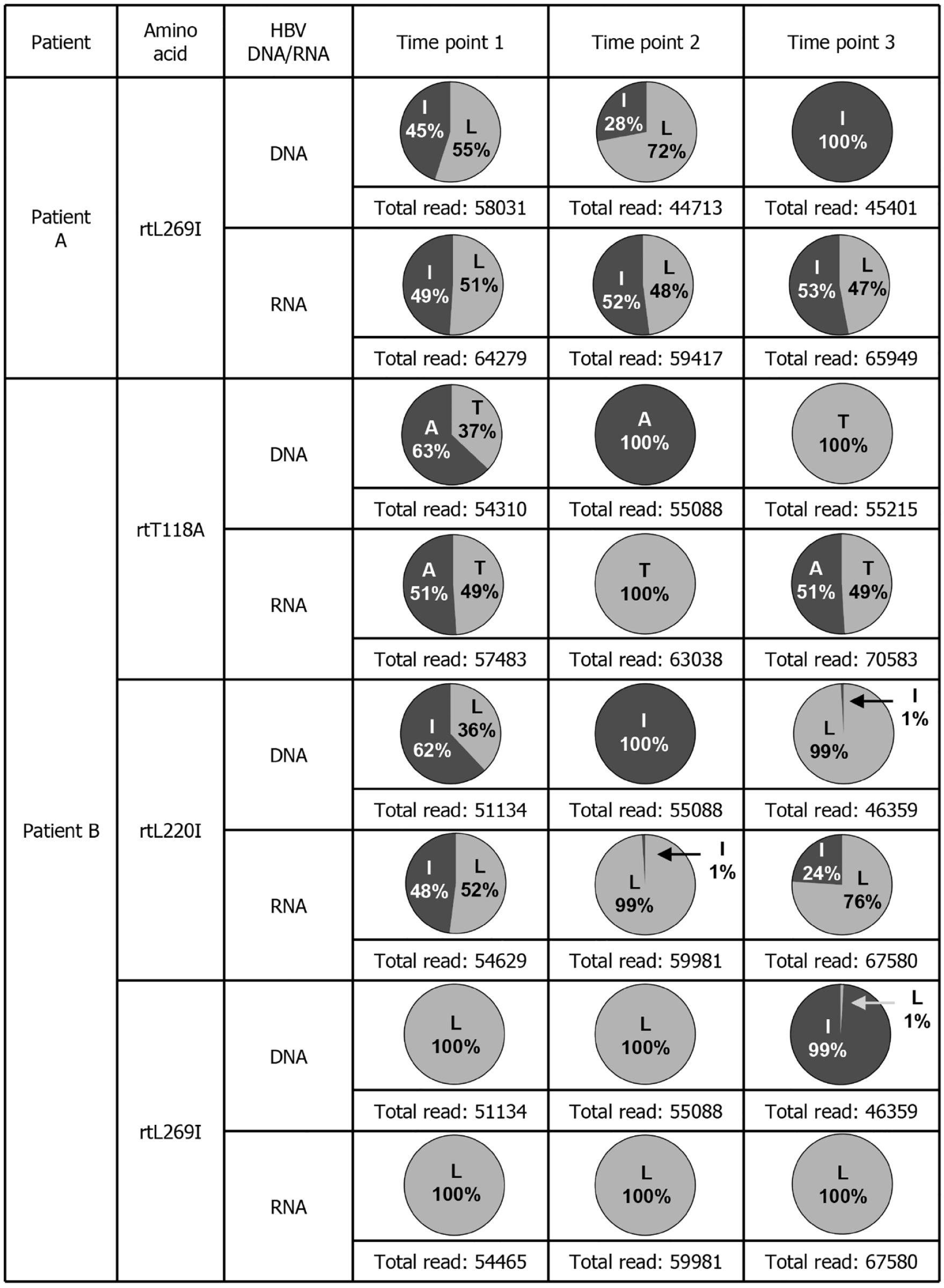
Figure 3 Deep sequencing data of serum hepatitis B virus DNA and RNA from tenofovir alafenamide fumarate-incomplete responders.
The amino acid mutations that changed in the clinical course of each patient are shown. The time points at which the serum samples were collected were the same as those indicated in Figure 1. The detection rates at which mutations were detected in hepatitis B virus DNA and RNA and the number of total reads are indicated. HBV: Hepatitis B virus.
- Citation: Yamada N, Igarashi H, Murayama A, Suzuki M, Yasuda K, Saito M, Isogawa M, Kato T. Deep sequencing analysis of hepatitis B virus in patients with incomplete response to tenofovir alafenamide fumarate treatment. World J Hepatol 2025; 17(5): 104519
- URL: https://www.wjgnet.com/1948-5182/full/v17/i5/104519.htm
- DOI: https://dx.doi.org/10.4254/wjh.v17.i5.104519