MODELS OF TUMOR CELL HETEROGENEITY
The concept of tumor heterogeneity has evolved in the last few decades into a complex picture of phenotypic, functional, and genetic heterogeneity not only within tumors but also between primary cancers and metastases. Adding to this multifaceted heterogeneity are the continuously changing extracellular influences that can modulate tumor development and progression including, but not limited to, inflammatory stimuli, microenvironmental signals, and immune cell interactions.
Recent technological advances have permitted in depth and rapid analysis of individual cancer genomes at the single-nucleotide level. These advances have shed light on intratumoral heterogeneity both within tumor biopsies and between spatially separated biopsies of the same tumor[1,2]. Sequential tumor sampling and analysis has also revealed that intratumoral heterogeneity temporally evolves during the course of disease[3]. This observed heterogeneity may present major challenges in the development of biomarkers, therapeutics, and personalized-medicine approaches.
CLONAL EVOLUTION, CANCER STEM CELLS, AND MOVEMENT TO A UNIFIED MODEL
The origins of intratumoral heterogeneity have been highly debated and different cellular mechanisms have been hypothesized to account for the diversity within a tumor. The clonal evolution theory, first introduced by Peter Nowell in a landmark article published almost four decades ago, proposed cancer to be an evolutionary process where most neoplasms arise from a single cell of origin, and tumor progression results from a stepwise acquisition of mutations within the original clone allowing sequential selection of more aggressive subclones. He hypothesized that cells in the dominant subclone populations would possess similar tumorigenic potential[4]. The second theoretically opposing hypothesis is the cancer stem cell (CSC) paradigm. Unlike clonal evolution, where subclones possess tumorigenic potential, the CSC hypothesis postulates that only small subpopulations of the tumor, the CSCs, are capable of self-renewal and the have potential to give rise to a tumor-the rest of the tumor consists of phenotypically diverse cells with limited proliferation and tumorigenic potential. The benchmark work that helped to establish the CSC theory in solid tumors also showed that no clear morphological distinction was apparent between tumorigenic and nontumorigenic breast cancer cells, with the two populations displaying equal cell kinetics-yet the tumors appeared to be hierarchically organized when tested functionally. The group identified the CD44+/CD24- cells as putative CSCs. When these isolated cells were injected into immunodeficient mice, tumors arose in 89% of cases-and only as few as 100 CD44+/CD24- were needed to recapitulate the tumor[5]. Further studies by other groups consistently verified that the frequencies of tumorigenic cells were low, variable, and were able to recapitulate some of the heterogeneity of the original tumors[6].
Proving that cancers strictly follow either the clonal evolution or CSC model is limited by experimental methodologies. The elucidation of a CSC model has been dependent on xenograft limiting dilution assays and tumor-specific CSC markers to isolate the tumor initiating cells. This assumes that the resulting increased tumorigenicity lies with an intrinsic trait of CSCs but does not rule out the that these cells are just somehow better suited to growth within an immunocompromised mouse that bears little resemblance to the normal human environment. It also relies heavily on the use of surface markers that may not adequately delineate the CSCs from non-CSCs. However, elegant lineage tracing experiments have clearly demonstrated that CSCs are a tumorigenic reservoir, capable of surviving chemotherapy, and drive relapse in mouse models of cancer[7-9]. Even with these experiments, direct evidence that unmanipulated human solid tumors harbor cells with self-renewing properties that fuel sustained tumor expansion is lacking. Further, the CSC model alone cannot account for functional heterogeneity in all tumors. Despite these complexities, recent studies illustrate that a stem cell-like gene expression signature is related to relapse in glioblastoma (GBM) and is predictive of patient outcome in human leukemia, breast cancer, GBM, ovarian cancer-lending support to the clinical relevance of cancer stem cells[10-13]. Furthermore, ample evidence exists that the prevalence of cells with a CSC phenotype can predict response and that these cells persist even after chemotherapy or radiotherapy. Despite experimental, analytical, and theoretical caveats for both the clonal evolution and CSC models, clinical studies in leukemia have shown that in a cancer distinctly driven by stem cells, such as in chronic myelogenous leukemia, clonal evolution is observed when tyrosine-kinase inhibitors are administered[14,15]. This initially successful therapy can result in the emergence of subclones that harbor mutations in the BCR-ABL fusion gene, the target of imatinib, giving rise to a tumor-resistant phenotype[15,16]. Moreover, it has also been demonstrated that CSCs harbor the BCR-ABL fusion gene but remain insensitive to imatinib. Instead, these CSCs revert to a normal dependence on cytokines for survival and proliferation[17]. These cells could therefore be the ones that survive the initial therapy and sustain further mutations giving rise to a fitter clone.
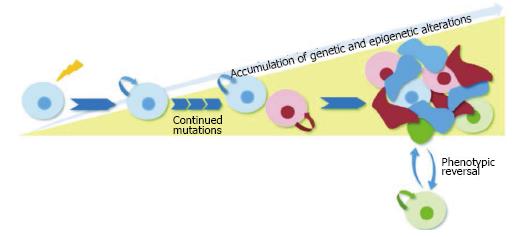
These observations highlight fact that the progression of tumor heterogeneity is a wildly complex process and support the possibility that clonal evolution and the CSC model are not mutually exclusive. A single tumor may comprise several cancer stem cell clones that have a common ancestor (the cell that sustained the first oncogenic mutation) yet are genetically distinct. These cancer stem subclones with self-renewal capabilities would persist over time and accumulate epigenetic and genetic changes required for cancer initiation and progression. Each different CSC subclone would give rise to intermediate progenitors as well as more differentiated, non tumorigenic cancer cells. The intermediate transit-amplifying cells would lack self-renewal capabilities, but could continue to accumulate genetic changes and possibly a mutation conferring self-renewal capabilities to the cell. Alternatively, these cells could follow a model of tumor cell plasticity in which microenvironmental stimuli could co-opt self-renewal mechanisms and acquire CSC characteristics-a process that is inherently transitory and would also allow the conversion from cancer stem cell to non-self-renewing progenitor (Figure 1).
Figure 1 Schematic of unified model of clonal evolution and cancer stem cells.
The proposed unified model depends on dynamic hierarchical organization and clonal mutations for tumor heterogeneity. In this depiction, the originating CSC that sustained the first oncogenic mutation gives rise to subclones with self-renewal capabilities that accumulate epigenetic and genetic changes over time. Each different CSC subclone gives rise to intermediate transit-amplifying progenitors that lack self-renewal capabilities. A subset of these progenitors (shown in green) follows a model of tumor cell plasticity and bidirectional conversion between non-CSC to CSC states. This phenotypic change is modulated by microenvironmental stimuli which confer CSC self-renewal capacities to the differentiated cell. CSC: Cancer stem cell.
The concept of cancer stem cell plasticity and bidirectional conversion between stem and non-stem cells has added additional complexity to the CSC and clonal evolution models and may help explain the tumor heterogeneity observed in solid tumors. Recent studies provide evidence that a select group of cancer cells can readily switch between non-tumorigenic and tumorigenic cell states in response to appropriate stimuli and that this conversion may be modulated by endogenous transcription factors[18-20]. This suggests that both CSCs and non-CSCs are highly adaptable populations capable of transient evolution and plasticity. This review will focus on the evidence for this transient state and discuss how these observations impact our understanding of the evolution of cancer and its treatment.
EPITHELIAL-TO-MESENCHYMAL TRANSITION AND CANCER STEM CELLS
Epithelial-to-mesenchymal transition (EMT) is a process integral to early embryogenesis and development where epithelial cells transdifferentiate into motile mesenchymal cells[21]. During the process of conversion into mesenchymal cells, epithelial cells lose their cellular junctions and apico-basal polarity, reorganize their cytoskeleton, and reprogram their signaling patterns and gene expression to gain the ability to migrate, increase motility, and invade adjacent tissue[22,23]. During embryogenesis, EMT allows epithelial cells to travel through the embryo and participate in the formation of internal organs. The processes underlying EMT can also be reactivated for wound healing and cancer progression[24,25]. The mechanism of EMT is transient in nature and allows transformed mesenchymal cells the capability to reacquire their epithelial phenotype upon arriving at their organ or tissue of destination where they proliferate and differentiate into organs, a process termed mesenchymal to epithelial transition (MET)[22,23]. The ability of epithelial cells to transform into mesenchymal cells and back into epithelial cells illustrates the inherent plasticity of the epithelial phenotype. These EMT and MET programs are coordinated by pleiotropic EMT transcription factors (TFs) and a multitude of extracellular signals[26,27]. Three families of transcriptional regulators that are essential during EMT events have been identified as core EMT regulators: The zinc finger E-box binding homeobox members ZEB1 and ZEB2[28], the SNAIL zinc finger family[29], and the TWIST family of basic helix-loop-helix transcription factors[30].
Recent studies have elucidated molecular links between EMT/MET-TFs and self-renewal, one of the defining traits of cancer stem cells, suggesting that EMT processes play a role in the development of the CSC state. Several groups have demonstrated that EMT activation can induce CSC generation[18,19]. Using a mammary tumor progression model, Morel and others showed that EMT induction can drive mammary epithelial cells to acquire stem and tumorigenic characteristics of CSCs following the activation of the Ras-mitogen-activated protein kinase pathway[18]. In a correlative study, Mani et al[19] illustrated that cells that have undergone an EMT behave similarly to stem cells isolated from normal or neoplastic cell populations. The group induced EMT in nontumorigenic, immortalized human mammary epithelial cells (HMLEs) by ectopic expression of either the TWIST or SNAIL transcription factors (capable of inducing MET in epithelial cells). They found that most of the HMLEs that underwent MET acquired a CD44high/CD24low antigenic phenotype representative of mammary CSCs and that their mammosphere-forming ability was increased by 30-fold. Additionally, they demonstrated that CD44high stem-like cells are more mesenchymal than their CD44low counterparts[19].
More recently, a study using a model of basal breast carcinoma elucidated a new role of EMT transcription factors in regulating CSC plasticity during tumor progression. Chaffer and group showed that neoplastic non stem cells (CD44low) can readily convert to a stem-like state (CD44high) and that this transition is mediated by ZEB1, a well-characterized EMT-TF. Further, they demonstrated that TGFβ, a recognized EMT-inducing stimulus can efficiently promote non-CSC-to-CSC conversion in their basal cell model[20]. Taken together, these studies highlight the role of EMT regulators TWIST1, SNAIL2, and ZEB1 in regulating CSCs in some tumor models. Importantly, complementary studies by these groups indicate that EMT transcription factors and extracellular signals cannot universally induce a CSC phenotype in all models, highlighting the heterogeneity of tumor progression and its microenvironment.
A DYNAMIC CSC STATE
Cancer stem cells are characterized as having an intrinsically determined state of unrestricted proliferative potential that permit self-renewal and give rise to progenitors with limited proliferative potential[31]. Recent studies have proposed the concept of cancer stem cell plasticity in which these two states may not be definitive but instead have a transitionary capability of shifting from a non-CSC state to a CSC state and vice versa[20,32-36]. In experimental models of melanoma, tumorigenic cells display considerable plasticity transiently acquiring stemness properties depending on the tumor context. An early study revealed that both CD133+ and CD133- melanoma cells have the ability to form tumors, suggesting that CD133 is reversibly expressed by tumorigenic melanoma cells rather than identifying cells at a static level in a hierarchy[37]. This group further characterized 22 markers and found that none of them robustly distinguished tumorigenic from non tumorigenic cells; instead, they observed phenotypically distinct melanoma cells having the capacity to form tumors that recapitulated the parent tumor[33]. A complementary study further corroborates the plasticity of melanoma cells. The group showed that JARID1B histone demethylase is a regulator of tumorigenicity in melanoma cell lines and that JARID1B negative cells can become positive and acquire self-renewal potential. They proposed the hypothesis that the tumorigenicity of a single tumor-initiating cell is reversible and that some stem-like cells from solid tumors may not actually be static, but rather have a transient nature and acquire stem-cell-like properties depending on the tumor context[34]. These results are compatible with the concept that tumorigenic potential might reflect a reversible state in cancer.
There is additional evidence supporting the model of dynamic stemness in breast cancer studies[20,32,35]. Gupta and group developed and validated a theoretical quantitative Markov model of phenotypic transitions that predicts evolution toward equilibrium in cancer cell populations. Their model predicts that cancer cells could interconvert between different states in a manner that maintains equilibrium in the proportions of cellular states; more specifically, it predicts that cancer stem-like cells can arise from non-stem-like cells. To test this prediction, they evaluated the ability of stem-like, basal, and luminal cells of seeding tumors. They demonstrated that the luminal and basal fractions could indeed generate functional stem-like cells in vivo supporting their hypothesis that convergence toward equilibrium cell-state proportions could be occurring due to cell-state interconversion within tumors[35]. Additional work by the Weinberg group proposes that not all cancers adhere to a unidirectional model of cancer stem cell hierarchy. They identified subpopulations of non-cancer stem cells that could readily switch from a non-CSC to a CSC state. They found that the non-CSC population could give rise to aggressive CSCs[32]. Further, they found that this plasticity is enabled by the transcription factor ZEB1 (a well-characterized EMT transcription factor) and that microenvironmental stimuli could enhance the rate of non-CSC to CSC conversion. Importantly, they note that this plasticity and stimulus-mediated conversion is readily observed in basal-type cells, but not in luminal-type cells[20]. Together, these studies suggest that certain cancer types may be following a bi-directional CSC model; however, they are not without limitations. The experiments described above rely on cell sorting to isolate and characterize cells, yet definitive surface markers are lacking for CSCs and their progenitors in undisturbed human tumors. Additionally, the results are obtained using artificial experimental systems, however delineation of functional plasticity will require minimally manipulated human cells or clinically relevant models of human disease. Further, if CSCs are present in a dynamic equilibrium, their respective isolation will be confounded by this fluctuating phenotype.
Two recent investigations in glioblastoma and colon cancer further validate the model of dynamic stemness and elucidate potential reprogramming factors responsible for the plastic phenotype[36,38]. Suva and colleagues identified a set of core neurodevelopmental transcription factors (POU3F2, SOX2, SALL2, and OLIG2) that are essential for GBM propagation. They introduced each transcription factor individually into differentiated glioblastoma cells (DGCs) and monitored enhanced sphere formation ability. Next, they coinfected DGCs with different combinations of TFs in a stepwise fashion and observed that the combined induction of POU3F2 + SOX2 + SALL2 + OLIG2 yielded cells capable of tumor initiation in 100% of animals. They showed that this set of TFs coordinately bind and activate CSC-specific regulatory elements and are sufficient to fully reprogram differentiated GBM cells to ‘‘induced’’ stem-like cells, recapitulating the epigenetic landscape and phenotype of native CSCs[39]. Similarly, a study in colorectal cancer introduced a set of defined factors (OCT3/4, SOX2 and KLF4) into human colon cancer cells and observed an enhancement in CSC properties such as sphere formation capability, marker gene expression, chemoresistance, and tumorigenicity. The tumors derived from these induced CSCs had immunohistological similarities to human colon cancer tissue and their phenotypes were reproducible in serial transplantation experiments[38].
Tumor suppressors have also been shown to play a role in CSC maintenance and plasticity. A study using an in vivo model of breast cancer showed knockdown of the PTEN tumor suppressor in normal mammary epithelial cells enriched for the stem/progenitor compartment leading to the generation of premalignant lesions[40]. In cell lines, overexpression of MiR-7, a micro-RNA with tumor suppressor-like characteristics, decreased the population of breast cancer stem cells and partially reversed EMT by directly targeting the oncogene SETDB1[41]. In prostate, in vitro analyses showed that PTEN and TP53 play a role in regulating self-renewal and differentiation of prostate stem/progenitor cells[42]. Further, using clonal epithelial cell lines the researchers showed that prostate epithelial PTEN/TP53 loss led to transformation of multipotential progenitors, EMT, and increased plasticity of tumor cells[43].
TUMOR MICROENVIRONMENT AND CSC PLASTICITY
The tumor niche or microenvironment is a complex network of cells, signaling molecules, soluble factors, and the extracellular matrix that plays a crucial role in tumor development, metastasis, and response to therapy[44]. During embryonic development, niche factors affect stem cell gene expression and regulate cellular differentiation for fetal development. After birth, stem cells continue to be highly regulated for tissue homeostasis and during response to injury by the surrounding microenvironment[45]. A tumor is comprised of neoplastic cancer cells together with microenvironmental factors that include hematopoietic cells, stroma, inflammatory cells, vasculature, and the extracellular matrix (a network of polysaccharides and proteins secreted by cells that serves as a structural tissue element and influences development and physiology)[46,47]. Each of these cellular and non-cellular factors contributes to the transformation of tumor cells and may also affect response to chemotherapeutic and radiation treatments by providing protection from these agents[46]. Similarly, cancer stem cell function and plasticity may be induced by specific microenvironmental signals and cellular interactions arising in the tumor niche[48].
As discussed in the EMT section above, stromal cells secrete signaling factors that are received by epithelial cells and generate a signaling cascade that can orchestrate an epithelial to mesenchymal transition. A correlative interplay is observed between neoplastic cancer cells, CSCs, and the stroma in pancreas, breast, and colon cancer models. In the pancreas, stellate cells (myofibroblast-like cells found in the stromal compartment) remain activated after chemotherapy and radiation and impart a protective effect on cancer cells[49-51]. Stellate cells have been shown to enhance spheroid-forming ability of CSCs through induced expression of CSC-related genes and to promote the cancer stem cell phenotype through paracrine Nodal/Activin signaling at the tumor-stromal interface[52,53]. Additionally, pancreatic stellate cells isolated from patients and co-cultured with pancreatic cancer cells enhanced cancer cell migration, and increased mesenchymal gene expression suggestive of an EMT phenotype[54]. Parallel studies in breast and colon cancer revealed that CSC plasticity can be regulated by cytokine networks and growth factors[55,56]. Vermeulen and group showed that hepatocyte growth factor can activate β-catenin-dependent transcription, CSC clonogenicity, and restore the CSC phenotype in more differentiated tumor cells both in vitro and in vivo[56].
The interaction between immune cells and cancer cells has been shown to promote tumor development and progression and results in tumor immune evasion[57]. In colorectal cancer, analysis of the type, density, and location of tumor-infiltrating immune cells within patient tumor samples revealed that this immunological data was a better predictor of patient survival than current histopathology methodologies used for staging that disease type[58]. Some tumor cells escape immune system detection by decreasing the expression of specific antigen-presenting proteins on their cell surface, allowing them to evade cytotoxic T lymphocytes. Tumor cells can also secrete factors that inhibit effector T cell activity and promote the production of regulatory T cells that suppress immune responses[57,59]. Similarly, stem-like cells in melanoma have been shown to preferentially inhibit T-cell activation and to support induction of regulatory T cells in order to evade immune system recognition[60]. In GBM, CSCs suppress T cell response by producing immunosuppressive cytokines- they inhibit T cells through the STAT3 pathway, and induce T cell apoptosis mediated by inhibitory molecules[61,62]. A recent study has elucidated a novel role for how the immune system affects CSCs through paracrine signaling in colorectal cancer. This study revealed that CD4+ T cells secreted interleukin (IL)-22 which acted on cancer cells through STAT3 and induced the core stem cell genes NANOG, SOX2, and POU5F1, resulting in increased cancer stemness and tumorigenic potential[63].
The reciprocal communication and interplay between cancer stem cells and the immune niche can also induce CSC plasticity. This bidirectional phenotypic change may be driven by pro-inflammatory mediators such as tumor necrosis factor (TNF) and IL-6 that are secreted by various immune cells in the tumor microenvironment. Examples of cytokine-driven tumor cell plasticity have been demonstrated in melanoma, breast, and lung cancer where TNF and IL-6 can affect the differentiation state of tumor cells by the upregulation of mesenchymal genes-resembling an EMT-type switch. Similar changes have been observed in a study using an experimental model of colon cancer where elevated inflammatory nuclear factor kappa B signaling enhanced Wnt activation and induced dedifferentiation of non-stem cells that acquire tumor-initiating capacity. In an important complementary study, a group at the Mayo Clinic observed that CD8+ T cells promoted EMT in an intact in vivo model of breast cancer. These transformed cells had characteristics of cancer stem cells that included potent tumorigenicity, the ability to reestablish an epithelial tumor, and enhanced resistance to drugs and radiation[64]. In support of the hypothesis that cancer stem cell plasticity is driven by pro-inflammatory mediators induced by therapy regimens, gene expression signatures and histological studies in patients revealed an increase in mesenchymal markers in chemotherapy-resistant tumors[65-67]. Taken together, these results suggest that immune cells and their related cytokines and growth factors can directly modulate and enhance the CSC phenotype. However, the niche effect likely will differ for different tumor subtypes with CSCs displaying diverse levels of dependency on these extrinsic microenvironmental interactions.
These experimental observations underscore the role of the tumor microenvironment, including immune cells, the stromal compartment, growth factors, and cytokines in phenotypic plasticity of tumor cells and CSCs and highlight potential novel targets to exploit therapeutic synergies to target multiple factors in the tumor niche.
CSC TARGETED THERAPIES
The data reviewed herein describe the hierarchical, molecular, extracellular, and plastic complexities of cancer and highlight the need for more comprehensive therapeutic strategies that address and target the multiple stages and branches of a single tumor. The renewed interest and deeper understanding of the CSC field have led to new ideas about therapeutic strategies including CSC signaling pathway inhibitors, immune mediated therapies targeting CSCs, and combination treatments targeting the microenvironment. However, CSC plasticity induced by microenvironmental and immune-related signals may limit the effect of CSC treatments. Therapies targeting CSCs would only be temporarily effective in eliminating this population since new CSCs may arise from non-CSCs left untargeted. Therefore, targeting each subpopulation or subclone within the tumor will be the most effective way of achieving a successful clinical response.
Data from high-throughput screening studies revealed that EMT programs rely on classic embryonal signaling pathways such as Notch, Wnt, Shh, and TGFbeta[68]. As reviewed, EMT processes play a role in the development of the CSC state and may be an attractive therapeutic target for inhibiting CSC plasticity. Because aberrant signaling in these pathways has been recognized to cause tumorigenesis in a broad variety of tissues, significant investment has been made by the industry to develop inhibitors towards them[48]. It is possible that these agents already in development could be used to induce differentiation of CSCs in combination with drugs that would target the newly-differentiated cells. A number of experimental agents are being evaluated in the clinic for their ability to inhibit Wnt signaling that could be studied for this purpose[69-72]. Novartis is currently testing a Wnt-specific acetyltransferase inhibitor, LGK974, in a phase I study on malignancies dependent on Wnt ligands[70]. Interestingly, their pre-clinical studies showed that cell lines with loss-of-function mutations in the Notch signaling pathway responded well to this inhibitor, suggesting an underlying mechanism of action for increased sensitivity to LGK974 in the Notch1 mutant carcinomas and highlighting the potential to target a subset of patients[73]. One caveat of these therapies is the fact that the pathways they target are not exclusive to CSCs but are shared with normal stem cells. Identifying a threshold of killing activity of normal cells will be necessary to prevent deleterious effects. Additionally, it is unlikely that a single pathway will be operative in all of the CSCs in a given tumor; therefore, concurrent use of drugs that can affect multiple pathways that are essential to CSCs and EMT-mediated plasticity are likely to be obligatory to achieve effective therapies.
It is well-accepted that tumors deregulate certain immune-checkpoint pathways as a major mechanism of evasion and immune-mediated therapies (IMT) have been developed that utilize the immune system to target cancer cells. Cancer cells that undergo EMT develop a mesenchymal phenotype yet retain some epithelial characteristics[64]. A recent study demonstrated that mesenchymal cells overexpress genes for immune inhibitory molecules including programmed cell death protein 1 (PD1)/PD-L1 and cytotoxic T lymphocyte-associated antigen-4 (CTLA-4)[74]. It is also possible that mesenchymal cells resulting from EMT during bidirectional CSC conversion also express these molecules and that IMTs may have the potential to target this subgroup of dynamic cells. The two immune-checkpoint receptors that have been most actively studied in the context of clinical cancer immunotherapy are CTLA-4 and PD1. Both are inhibitory receptors and regulate immune responses at different levels and through different mechanisms[75]. In 2010, a crucial Phase III trial using an anti-CTLA4 antibody, ipilimumab, demonstrated statistical survival benefit in patients with melanoma[76]. This was the first immune-mediated therapy to demonstrate survival benefit in patients with advanced melanoma in a randomized trial. Other drug companies have accelerated preclinical studies to advance their immune-mediated therapies to the clinic[77-79]. The observations that mesenchymal cells in non-small-cell lung cancer are associated with distinct immune phenotypes with increased expression of inhibitory molecules provide a potential mechanism for EMT-associated immunosuppression utilizing the therapies that are currently being tested in the clinic. Therefore, the IMTs described above may have the potential to target newly transformed CSCs as well as cancer cells that have not undergone a transformation thereby simultaneously eliminating both subpopulations leading to a more complete therapeutic response.
Cancer stem cells are characterized by immune suppressive activity and a low immunogenicity that enhance their ability to survive. CSCs need to evade immune reactions mounted by the host and adapt to the dynamic microenvironment altered by radiation and chemotherapy[80]. Phenotypic analysis in GBM, colorectal cancer, and melanoma revealed a potential impairment of antigen presenting function by CSCs helping to shield them from T-cells[81]. Additionally, CSCs enhance immunosuppression by down-regulation of major histocompatibility complex MHC-I and -II (a milieu of proteins that regulate cell-mediated adaptive response) and through the release of immunosuppressive cytokines such as IL-10 and TGFβ[60,61,81,82]. These cytokines can also affect the differentiation state of tumor cells by upregulating EMT genes. Therefore, Identifying targets that will inhibit and reverse the CSC escape from immunosurveillance may also elucidate targets for CSC plasticity and could prove necessary to successfully eliminate tumors.
PERSPECTIVE AND FUTURE DIRECTIONS
Additional understanding of the role of tumor cell dynamics will continue to inform our therapeutic strategies. Answers to important questions remain to be determined in the characterization of patient tumors and advances in technologies like single cell analysis are likely needed to identify a resolution. Perhaps the most important questions that remain with respect to therapeutic targeting are (1) whether plasticity causing non-CSCs to develop the tumorigenic properties ascribed to a CSC truly exists in human patient tumors and, even more importantly; (2) whether this interconversion results in a CSC clone that is different from the original clone and requires a different therapeutic approach. Additionally, (3) whether the extent of heterogeneity found within CSCs results in the need for independent therapies targeting of multiple CSC clones. Despite these questions, an overwhelming body of literature points to a need to develop therapeutics specifically targeted to CSCs as they are clearly implicated in disease relapse.
Continued research is needed to identify CSC-specific targets and to understand the effects of the stem cell niche on plasticity, survival, and tumorigenicity in order to develop novel therapeutic regimens. Identification of additional immuno-modulating therapies that can inhibit or reverse the ability of CSCs and their dynamic subclones to escape from immunosurveillance will contribute to the comprehensive cancer therapy approach that is likely necessary to achieve successful eradication of a tumor. Further, in order to achieve more complete therapeutic responses it will be necessary to pursue treatments against CSCs in each of their dynamic states or to first halt the interconversion between non-CSC to CSC and then eliminate each subpopulation therapeutically, and ultimately to combine these specialized CSC therapies with strategies that target the bulk population.