Published online Jan 26, 2015. doi: 10.4252/wjsc.v7.i1.1
Peer-review started: July 24, 2014
First decision: August 28, 2014
Revised: September 3, 2014
Accepted: September 17, 2014
Article in press: December 16, 2014
Published online: January 26, 2015
Processing time: 172 Days and 0.3 Hours
Alternative splicing (AS) is an essential mechanism in post-transcriptional regulation and leads to protein diversity. It has been shown that AS is prevalent in metazoan genomes, and the splicing pattern is dynamically regulated in different tissues and cell types, including embryonic stem cells. These observations suggest that AS may play critical roles in stem cell biology. Since embryonic stem cells and induced pluripotent stem cells have the ability to give rise to all types of cells and tissues, they hold the promise of future cell-based therapy. Many efforts have been devoted to understanding the mechanisms underlying stem cell self-renewal and differentiation. However, most of the studies focused on the expression of a core set of transcription factors and regulatory RNAs. The role of AS in stem cell differentiation was not clear. Recent advances in high-throughput technologies have allowed the profiling of dynamic splicing patterns and cis-motifs that are responsible for AS at a genome-wide scale, and provided novel insights in a number of studies. In this review, we discuss some recent findings involving AS and stem cells. An emerging picture from these findings is that AS is integrated in the transcriptional and post-transcriptional networks and together they control pluripotency maintenance and differentiation of stem cells.
Core tip: Alternative splicing (AS) produces multiple transcript isoforms from a single gene, and the regulation of cell-type-specific splicing pattern is crucial for the properties and functions of cells, including pluripotent stem cells. A better understanding of the role of AS in stem cell pluripotency maintenance and differentiation will offer potential new approaches for enhancing the production of induced pluripotent stem cells and/or better control of cell differentiation for research or therapeutic purposes. In this brief review, we provide a timely update of recent studies related to stem cell regulation and splicing in a genome-wide scale.
- Citation: Chen K, Dai X, Wu J. Alternative splicing: An important mechanism in stem cell biology. World J Stem Cells 2015; 7(1): 1-10
- URL: https://www.wjgnet.com/1948-0210/full/v7/i1/1.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v7.i1.1
The splicing of messenger RNA precursors, namely the precise removal of introns and the joining of exons, is a crucial yet highly dynamic and flexible process in the synthesis of mature eukaryotic mRNAs. Alternative splicing (AS), the inclusion of different exons in mature mRNA by selecting different splice sites in pre-mRNA, can result in different transcript isoforms from a single gene, and give rise to a much larger number of proteins compared to the number of genes encoded in metazoan genomes[1-3]. AS regulation plays an important role in almost every aspect of eukaryotic biological processes, including cell growth, death, pluripotency maintenance, differentiation, development, circadian rhythms, response to external changes, and disease[4,5]. Recent advances in high-throughput RNA sequencing technology revealed that a greater number of multi-exon genes can produce alternatively spliced transcripts than previously thought[6,7]. In humans, more than 90% of genes were estimated to undergo AS in different tissues and/or cell types. Compared with other RNA processing mechanisms such as alternative transcription initiation, RNA editing and alternative poly(A) site selection, AS is the most prominent in generating mRNA complexity. In addition, AS events can introduce premature termination codons in mature mRNAs, triggering mRNA degradation by the process of nonsense-mediated mRNA decay (NMD)[8,9]. AS events can also cause mRNA un-translated region (UTR) variation, which affects mRNA translation efficiency, stability and localization[10-12].
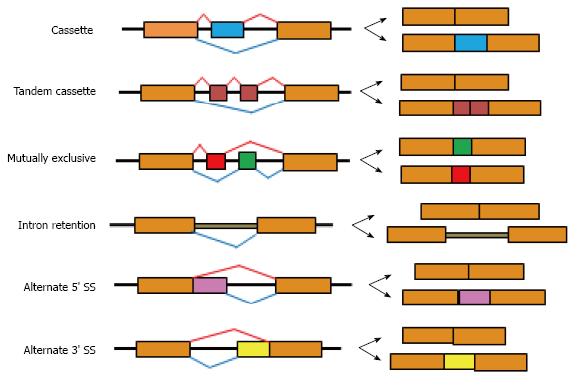
Splicing of pre-mRNA involves the formation of active splicing complexes on pre-mRNAs via a stepwise assembly process. The basal splicing machinery (spliceosome) is comprised of five small nuclear ribonucleoprotein particles (snRNP), such as U1, U2, U4/U6 and U5 in the case of the major spliceosome, and U11, U12, U4atac/U6atac and U5 in the case of the minor spliceosome. AS is primarily regulated by approximately 200 RNA-binding proteins (splicing factors) together with a basal spliceosome through direct recognition of short sequence motifs near exon/intron boundaries[13]. Depending on the pattern of exon inclusion/skipping, AS events can be categorized into at least six major types, including cassette exon skipping, mutually exclusive exons, alternative 5’ splice site selection, alternative 3’ splice site selection, alternative retained intron, and tandem cassette (Figure 1). There are more complex patterns but they are much fewer in number than these major types, therefore most analyses of AS events focus on these six types, particularly cassette exon skipping which represents the majority of AS events.
The knowledge of the crosstalk between splicing and other layers of gene regulatory network is fundamentally important for understanding biological processes such as cell differentiation, development, and pluripotency maintenance. In this review, we will highlight recent progress related to these themes, with an emphasis on studies involving both AS and stem cell research, to provide timely insight into AS regulation and its important roles in the determination of cell fate. The general principles of splicing regulation have been covered in detail in a number of excellent reviews, and readers who are interested in the mechanisms of splicing regulation can refer to these[9-11,14-21].
Our understanding and knowledge about AS has increased rapidly during last decade, thanks to the advancement of several high-throughput technologies. To better understand AS regulation, it is necessary to be familiar with the basic principles of these technologies. Here, we summarized some of the technologies that were applied to study AS in a genome-wide scale.
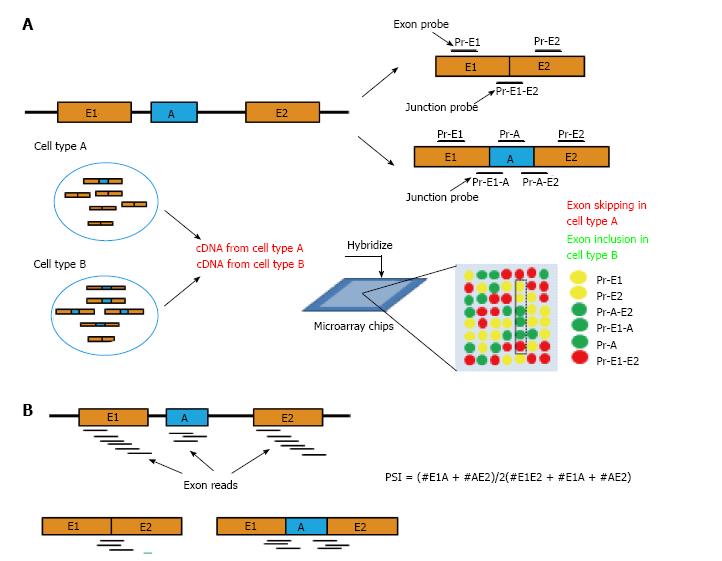
The first genome-scale AS study was carried out using microarray platform. Traditional microarrays have been designed to measure the total level of expression of a gene, without discrimination of its different isoforms[22-24]. To probe AS events, several splicing-sensitive microarray platforms have been developed[3,25,26]. Although there are variations between them, these splicing-sensitive microarrays all utilize short oligonucleotide probes designed to cross exon-exon junctions. cDNA samples were derived from mRNA and hybridized to the probes (Figure 2A). The signal intensity of these junction probes can then be used to infer exon inclusion ratios by sophisticated algorithms[27-37]. These microarrays have been applied in a number of studies to generate genome-scale profiling of AS, and provided quantitative measurements of AS at different time points of development, across tissues, and upon perturbation of interesting splicing factors[28,31,32,34,35,37]. From these pioneering studies, genome-level regulatory mechanisms of AS have been better understood, and have largely transformed our view about AS in every aspect including their evolution, dynamic regulation, and their organization in global transcription networks[2,19].
Recently, RNA sequencing technology has been evolving rapidly, and has become the method of choice for genome-wide AS analysis. In RNA sequencing methodology, cDNA fragments derived from poly(A) selected RNA population are sequenced from the ends and generate a large number of short sequence tags (reads). These reads can then be mapped (aligned) back to the reference transcriptome and splice-mapped reads can reveal the exon-exon junctions (Figure 2B)[38,39].
Compared with microarrays, RNA sequencing (RNA-Seq) does not rely on probes pre-designed across exon-exon junctions based on prior knowledge about the transcriptome under study, thus novel exons and splice junctions can be detected in an unbiased manner. RNA sequencing also has other advantages such as no cross-hybridization issues, higher sensitivity and broader dynamic range[40-46]. As the technology keeps improving and costs continue to decline, longer read length and more extensive sequencing coverage can lead to more accurate AS detection at a reasonable price.
High-throughput reverse transcription-polymerase chain reaction (RT-PCR) has also been developed and used for monitoring AS changes[47-49]. Although the number of AS events monitored is limited by prior knowledge from the reference AS database, in theory, it has the advantage of avoiding bias towards the highly expressed genes, and can quantify AS of medium- and extremely low-expressed genes[50]. There is also a very good correlation between percent spliced-in (PSI, the percent of transcripts that include a specific AS exon; Figure 2) values obtained with RNA-Seq data and the PCR-based method for events in which RNA-Seq data had enough coverage to produce confident PSI estimates, suggesting that the PCR-based method is consistent with RNA-Seq and these two methods can complement each other[48,51].
Methods for directly mapping RNA-binding protein (RBP) and the mRNA interaction transcriptome-wide in vivo have also been developed, complementing AS event profiling to decipher the regulatory network of splicing by RBP. To identify binding targets, a specific RBP together with its associated RBP complex is immunoprecipitated from cell lysate, and bound RNA transcripts are then purified and subjected to high-throughput sequencing[52-54]. After mapping the reads back to the reference genome sequence, potential binding locations of RBPs can then be inferred by computer algorithms. RBP complexes can be immunoprecipitated under native condition; however, this can increase the risk of losing low-affinity yet specific in vivo binding or of obtaining artificial binding following cell extraction[55]. A cross-linking step is usually performed to circumvent these problems. Several methods have been developed in this area. The CLIP-Seq (cross-linking immunoprecipitation and high-throughput sequencing, or HITS-CLIP) method uses UV light to crosslink proteins with RNAs[56]. In PAR-CLIP (photoactivatable-ribonucleoside-enhanced crosslinking immunoprecipitation), photoreactive ribonucleotide analogs are used to treat cells and are incorporated into RNAs before UV treatment[57]. And individual-nucleotide resolution cross-linking and immunoprecipitation (iCLIP) employs a self-circularization strategy to achieve individual-nucleotide resolution[58]. RBP mapping combined with AS profiling can be used for constructing “RNA maps” which correlate binding site positions with splicing regulatory differences upon perturbation of specific splicing factors.
Advanced technologies have recently been adopted to profile AS in stem cells. Extensive AS patterns were observed in stem cells and their contribution to pluripotency maintenance and differentiation has been noted.
Embryonic stem cells (ESCs) are pluripotent cells which can self-renew and has the ability to differentiate into all three germ layers[59,60]. As ESCs can generate most if not all of the cell types of a human body, they serve as an excellent model for studying early embryonic development. ESC is also a valuable source for producing differentiated cells for potential cell therapeutic purposes[61,62]. Thus, intensive efforts have been devoted to stem cell gene expression profiling, and genes associated with pluripotency were discovered[63,64]. However, only recent advances in next generation sequencing technology made it possible to profile the AS pattern of a given cell/tissue in a global scale. A number of genome-wide studies showed specific transcriptome changes during the differentiation of ESCs into different lineages[65-70].
In 2005, Pritsker et al[65] started using expressed sequence tag collections derived from stem cells to identify splice variants in ESCs and hematopoietic stem cells, and this was one of the first AS analyses in stem cells on a genome-wide scale. AS was detected in > 1000 genes. Although the technology is outdated nowadays, it showed that AS generates a large diversity in the stem cell molecular repertoire.
Further studies using advanced technologies confirmed the pervasive AS in ESC. A study by Wu et al[66] adopted three types of RNA sequencing technologies and profiled the transcriptome changes during the differentiation of human ESCs (hESCs) into the neural lineage. The authors combined Illumina single and paired-end reads (sequence reads from both ends of cDNA fragments; 35 bp reads) and longer Roche 454 FLX and titanium sequencing reads (250-450 bp reads) to discern transcript structure and analyze transcriptome complexity. Transcriptome profiles of cells in the ESC stage, N1 (early neural initiation) stage, N2 (neural progenitor) stage, and N3 (early glial-like) stage were reconstructed from mapped sequencing reads. Utilizing the unique spliced junction reads detected from each gene across all four stages, the authors then calculated a “junction complexity index” and found that splicing isoform diversity is highest in undifferentiated hESCs and decreases upon differentiation, a phenomenon they named “isoform specialization”. Observations like this can only be achieved with a genome-scale study, demonstrating the power and potential of RNA sequencing in AS research. In 2010, Revil et al[71] applied splicing-sensitive exon microarray technology to profile alternative isoform expression in embryonic day 8.5, 9.5 and 11.5 embryos and placenta. Although the profiling was not performed using pure ESCs, their results revealed frequent AS during embryonic developmental stages. Intriguingly, a number of RBPs, including putative splicing factors, are differentially expressed and spliced across developmental stages, suggesting these RBPs may be involved in regulating tissue and temporal variations in isoform expression.
It is well known that when somatic cells are reprogrammed to pluripotent stem cells (PSCs), the transcription of most genes reverted to an ESC-like state. An interesting question is whether this is also true for AS?
Several recent studies answered this question by profiling both induced PSC (iPSC) and ESC AS patterns in a genome-wide scale. Ohta et al[72] combined RNA-seq and high-throughput absolute qRT-PCR to analyze splicing pattern changes during the reprogramming process. The somatic cell splicing profiles reverted to a pluripotent-like state during reprogramming. In addition, to determine whether alterations in splicing patterns are specific for PSCs, the authors identified 27 genes which undergo alterations during the reprogramming process, and profiled the splicing pattern of these genes across multiple tissues by qRT-PCR. Interestingly, the splicing patterns in iPSCs were most similar in the testes compared with other tissues, suggesting an intriguing hypothesis that PSCs regulate AS using the same mechanism as the testes does. Other work also showed that the splicing pattern is similar in iPSCs and ESCs[48,51]. These observations raised the possibility that manipulating specific splicing regulators can potentially fine tune the reprogramming process.
In addition to investigating AS patterns during ESC differentiation, efforts have also been made to determine the functional impact of AS in ESCs[65]. In the study of Pritsker et al[65], splicing complexity in ESCs was observed, and they also found that AS can modify multiple components of signaling pathways which are important for stem cell function. The distribution of splice variants across different classes of genes indicated that tissue-specific genes have a higher tendency to undergo AS than ubiquitously expressed genes. Comparisons between all orthologous genes which undergo AS in human and mouse transcriptomes showed that the patterns of AS are only weakly conserved, supporting that AS patterns evolve fast[73,74]. Because multiple genes in stem cells undergo AS and these genes are enriched in regulatory proteins, stem cell molecular networks are highly dependent on AS.
Salomonis et al[67] investigated the roles of AS and alternative promoter selection in differentiating mouse ESCs using the Affymetrix exon-exon junction microarray. Among approximately 7500 genes and 40000 putative exon-exon junctions represented in the microarray, the authors identified 170 unique alternative exons (AEs) corresponding to 144 genes. It was predicted that 67% of these AEs altered the protein sequence and domain composition. Pathway analysis of these genes showed enrichment in genes associated with Wnt and transforming growth factor-beta receptor signaling pathways, the actin cytoskeleton, lipid transport, muscle contraction, mRNA metabolism, and embryonic development. Most of these 170 AEs were conserved between mouse and human, suggesting their functional importance. In order to examine the functional impact of AS, two genes, SERCA2 and Tcf3 that showed large differences in the expression level of alternative isoforms, were selected for examination. SERCA2 is a Ca2+ pump that hydrolyzes ATP during the translocation of calcium from the cytosol to sarco/endoplasmic reticulum[75]. PCR confirmed that one of its isoform (SERCA2b), with an additional 44 amino acids and a longer 3’UTR region, was expressed in both ESCs and embryoid bodies (EBs), whereas another isoform with a shorter alternative 3’UTR (SERCA2a) was mainly expressed in EBs. Interestingly, the 3’UTR of SERCA2b was predicted to be targeted by many microRNAs (miR-200b, miR-214, etc.) but not SERCA2a in both mice and humans. This is consistent with the observation that SERCA2b mRNA is more degraded than SERCA2a in an experiment in vitro[76], indicating 3’UTR of SERCA2b can inhibit protein expression. Using a library of miRNA mimics, the authors further confirmed that miR-200b and miR-214 among other miRNAs were indeed targeting SERCA2b. Bioinformatics analysis showed that miR-214 and miR-200b binding sites were enriched for inhibitor genes of cardiac differentiation, indicating they play functional roles in cardiac development by repressing cardiac inhibitor gene expression. In addition, previous studies showed that miR-200b and SERCA2a are both highly induced upon cardiac differentiation[75,77,78]. Taken together, this study suggests that SERCA2 can avoid direct repression by miRNAs through selectively expressing one of its isoforms (SERCA2a) which has no miRNA target sites in differentiated ESCs. This observation implies the ability of AS to regulate protein expression, without affecting gene or miRNA transcription. The other gene studied is Tcf3 (TCF7L1 in humans), a Wnt signaling transcription factor and a repressor of ESC self-renewal. Tcf3 inhibits ESC self-renewal through repression of Nanog and Oct4 transcription[79,80]. The authors identified a longer isoform of Tcf3 [Tcf3(l)] which is enriched in ESCs but downregulated upon differentiation. Tcf3(l) includes a 42-bp cassette exon which encodes an additional 14 amino acids overlapping with the Groucho binding domain[81,82]. This domain is necessary for Tcf3 to repress Nanog expression[79]. Tcf3(l) is upregulated in ESCs compared with EBs, while a shorter isoform of Tcf3, Tcf3(s), is expressed at a constant level. Selective knockdown of Tcf3(l) in ESCs revealed several distinct targets of transcriptional repression compared with Tcf3(s). For example, knocking down any Tcf3 isoform increased Nanog expression whereas Oct4 was upregulated only during knockdown of Tcf3(l). Knockdown of one or both Tcf3 isoforms can lead to delayed differentiation in ESCs. Interestingly, Tcf3(l) knockdown and Tcf3(s) knockdown inhibit distinct differentiation pathways, raising the intriguing hypothesis that isoform-specific regulation of Tcf3 targets affects distinct lineage commitment decisions. In short, these examples demonstrate that specific AS events can modulate transcriptional networks involved in pluripotency maintenance vs differentiation.
During the last decade, a core set of transcription factors (TFs), including OCT4 (POU5F1), NANOG and SOX2 among others which control the pluripotency of ESCs, has been uncovered[83]. Together with specific microRNAs and long non-coding RNAs, these TFs control the expression of gene cohorts required for establishment and maintenance of ESC pluripotency[84-86].
The AS of the core TFs can directly influence pluripotency control. One classic example is the pluripotency gene OCT4. The OCT4 gene was identified to encode three isoforms which were named OCT4A, OCT4B and OCT4B1[87]. Two isoforms, OCT4A and OCT4B, were shown to encode different binding domains which resulted in different target genes. While OCT4A can regulate genes that are responsible for stemness[83,88], OCT4B does not have the ability to maintain ESC self-renewal and it regulates genes that are responsive to cell stress.
Not only can the AS of core TFs affect pluripotency control, but the AS of several other genes is also linked to stem cell self-renewal and lineage specification[89-93]. Genes that have ESC-specific isoforms are particularly intriguing. A study conducted by Gabut et al[94] used microarray profiling to compare patterns of AS in undifferentiated and differentiated hESCs, and identified an evolutionarily conserved ESC-specific AS event of the gene FOXP1 (Forkhead box transcription factor 1). Experimental validation showed that inclusion of FOXP1 exon 18b is specific to self-renewing, pluripotent hESCs, thus this transcript isoform was named “FOXP1-ES”. The inclusion of exon 18b within FOXP1-ES changes the DNA-binding specificity of FOXP1, and allows FOXP1-ES to regulate distinct programs of gene expression in hESCs. Knockdown of FOXP1-ES results in a significant decrease in the expression of the pluripotency genes OCT4, NANOG, NR5A2, GDF3, and TDGF1, and an increase in the expression of differentiation-associated genes, including GAS1, HESX1, SFRP4. Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-Seq) was performed to identify genes that are directly regulated by FOXP1-ES and FOXP1 in hESCs. The FOXP1-ES binding target genes significantly overlap with the set of genes that are dependent on FOXP1-ES expression in hESCs and a set of genes that are regulated by OCT4 in hESCs. Overexpression of FOXP1-ES in mESC also promotes mESC self-renewal and pluripotency. Collectively, this study provided evidence that an AS switch regulating the FOXP1-ES isoform is integrated into the core circuit of the transcriptional regulatory network required for ESC pluripotency and iPSC reprogramming.
Compared with transcription factors, little is known about splicing factors that may contribute to stem-cell self-renew and lineage specification. Technology advancement allows the identification of functional RNA cis-elements related to AS and splicing factors in stem cells recently.
In 2007, Yeo et al[68] studied the AS events in hESCs and neural progenitors using exon array analysis combined with sophisticated algorithms to identify exons undergoing AS. The analysis showed that RBFOX binding motif GCAUG was enriched proximal to a set of exons that are alternatively spliced in hESCs, suggesting that RBFOX splicing factors may play a critical role in hESCs. Following this study, the same group constructed an RNA map for RBP RBFOX2 to identify functional RNA elements in the human genome in hESCs[91]. RBFOX2 is expressed abundantly in hESC cell lines, whereas RBFOX1 is not. Using CLIP-seq technology, thousands of RBFOX2 RNA targets were uncovered, representing approximately 7% of human genes in hESCs. Many RBFOX2 targets are themselves splicing factors, suggesting that RBFOX2 might act as an upstream regulator of many splicing factors. Interestingly, RBFOX2 pre-mRNA is also the target of itself, supporting the autoregulation of RBFOX2. It is possible that AS of RBFOX2 pre-mRNA may result in distinct proteins that can target different pre-mRNAs. RBFOX2 depletion in hESCs led to rapid cell death, indicating that RBFOX2 is important in maintaining hESC viability. However, RBFOX2 depletion in neural progenitor cells or primary human fetal neural stem cells did not cause cell death, suggesting that RBFOX2 has a different set of targets in different cell types.
Several recent studies have also used CLIP-seq to map binding sites of specific splicing factors. Combined with AS profiling, these studies revealed several splicing factors that are potentially associated with pluripotency maintenance. The work of Han et al[51] demonstrated a systematic strategy to study the function of specific splicing factors. They combined RNA-seq, CLIP-seq datasets, and “splicing code” analysis (a computational method which predicts cis-elements that promote or repress specific splicing events) to identify splicing regulators that are differentially expressed between stem cells and differentiated cells and control cell-specific AS. In particular, MBNL1 and MBNL2 were found to have the lowest relative expression levels in stem cells (ESCs and iPSCs) compared with differentiated cells in both humans and mice, suggesting that these proteins performed their function by repressing ESC-specific AS events. The authors tested the hypothesis using the FOXP1 transcription factor. As mentioned previously, isoform FOXP1-ES contains an ESC-specific exon which allows FOXP1 to bind and activate genes (OCT4 and NANOG, etc.) required for pluripotency. Supporting this hypothesis, the experiments showed that the FOXP1-ES-specific exon was retained in differentiated cells in which MBNL1 and MBNL2 were knocked down, whereas, over-expression of MBNL1 and MBNL2 in ESCs promoted differentiated cell-like splicing patterns. Furthermore, MBNL knockdown enhanced the efficiency of reprogramming from fibroblasts into iPSCs by about 2-fold. Taken together, the study revealed that MBNL protein expression plays a functional role in differentiation by promoting FOXP1-ES specific exon skipping, and their knockdown can facilitate reprogramming of somatic cell into iPSCs.
Another study by Venables et al[48] found that splicing factors MBNL1 and RBFOX2 cooperate to control pluripotency in stem cells. The authors adopted high-throughput RT-PCR technology to monitor splicing changes of more than 3000 AS events annotated in the RefSeq database during the induction of fibroblasts into iPSCs, and their subsequent re-differentiation. Comparing the AS profiles in fibroblasts and iPSCs, and fibroblasts re-differentiated from iPSCs, the authors observed that AS changes are reversible during these processes. Their finding uncovered a program of concerted AS changes involved in late mesoderm differentiation. The authors selected 47 AS regions (in different genes) whose splicing profiles showed an equivalent near-perfect anti-correlation in reversible stem cell induction and re-differentiation, and used these AS regions to investigate splicing mechanisms involved in stemness and maintenance of pluripotency. To identify the splicing factors involved in pluripotency and reprogramming, the authors knocked down 81 potential splicing factors in various cell lines and monitored these 47 AS regions using RT-PCR. The differences in PSI values (between iPSCs and original fibroblasts used for inducing pluripotent cells) of these 47 AS events were then compared with the differences in PSI values before and after 81 splicing factors were knocked down individually during reprogramming of fibroblasts into iPSCs. They found that MBNL1 knockdown correlated most strongly with the splicing profile change in the induction of pluripotency. Splicing factor RBFOX2 knockdown showed the second highest correlation with the induction of pluripotency. By knocking down both MBNL1 and RBFOX2 in fibroblasts, a significant correlation between splicing changes and the induction of pluripotency was observed, and the correlation was even higher than knocking down MBNL1 and RBFOX2 individually, suggesting that MBNL1 and RBFOX2 cooperate to establish the splicing program involved in stem cell differentiation.
Ohta et al[72] used siRNA screening in PSCs to identify RBPs involved in the reprogramming process by enhancing stem cell specific AS. After a screen of 92 RBPs, 9 RBP-coding genes that had a major effect on the splicing patterns were examined to assess the impact on somatic cell reprogramming using shRNA knockdown. The downregulation of U2af1 and Srsf3 was found to suppress both the efficiency of alkaline phosphatase-positive colony formation and ESC marker gene expression.
AS can also affect RNA stability through NMD. Most recently, Jangi et al[95] performed a genome-wide analysis of RBFOX2 activity in mESCs by mapping RBFOX2 binding sites to transcriptome changes after loss of RBFOX2. Using iCLIP and RNA-seq technologies, the authors identified more than 200 AS-NMD (AS-coupled nonsense mediated decay) splicing events that were mediated by RBFOX2 in mESCs. These events showed minimal splicing changes but appreciable changes in gene expression upon RBFOX2 knockdown due to the degradation of the NMD-inducing isoform. About 70 of these AS-NMD events were within genes encoding RBPs. Many of these RBPs were also autoregulated. A large fraction of bound but apparently unregulated events likely generated the NMD isoforms. This led to the hypothesis that RBFOX2 can control the gene expression level by regulating AS-NMD. The authors further demonstrated that RBFOX2 determines a threshold for the ratio of NMD to non-NMD isoforms for several of these RBPs. These findings uncovered an unexpectedly broad multilayered regulatory network controlled by RBFOX2, and established a model for how autoregulatory splicing networks are tuned.
Induced PSCs hold the promise of future cell-based therapy. A thorough understanding of the mechanisms underlying stem cell pluripotency and differentiation is critical for harnessing the cell reprogramming process. In this review, we have summarized recent progress in the field of AS and its role in stem cell pluripotency maintenance and differentiation. It was found that AS is pervasive in stem cells, and reprogramming reverts the splicing pattern to an ESC-like state. AS has a fundamental impact on stem cell differentiation by regulating different isoforms of the core pluripotency transcription factors. AS of genes other than the core factors is also linked to stem cell self-renewal and lineage specification. Additionally, splicing factors can regulate pluripotency by affecting stem cell-specific AS. In summary, these findings present a picture in which AS is integrated in the transcriptional and post-transcriptional networks, and the crosstalk between AS and other layers of the gene regulatory network have a fundamental effect on stem cell pluripotency maintenance and differentiation. These findings can lead to novel approaches for improving iPSC development and a better control of cell differentiation.
P- Reviewer: Eyras E S- Editor: Song XX L- Editor: A E- Editor: Lu YJ
| 1. | Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 909] [Cited by in RCA: 977] [Article Influence: 46.5] [Reference Citation Analysis (0)] |
| 2. | Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 791] [Cited by in RCA: 849] [Article Influence: 42.5] [Reference Citation Analysis (0)] |
| 3. | Ben-Dov C, Hartmann B, Lundgren J, Valcárcel J. Genome-wide analysis of alternative pre-mRNA splicing. J Biol Chem. 2008;283:1229-1233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 111] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 4. | Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587-1593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 728] [Cited by in RCA: 785] [Article Influence: 60.4] [Reference Citation Analysis (0)] |
| 5. | Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12:715-729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 488] [Cited by in RCA: 541] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 6. | Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413-1415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2521] [Cited by in RCA: 2898] [Article Influence: 161.0] [Reference Citation Analysis (0)] |
| 7. | Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470-476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4232] [Cited by in RCA: 3960] [Article Influence: 220.0] [Reference Citation Analysis (0)] |
| 8. | Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA. 2003;100:189-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 791] [Cited by in RCA: 813] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 9. | Wu J. Characterize mammalian transcriptome complexity: from genome-wide rt-pcr cdna cloning, to novel mammalian gene discovery, and transcriptome complexity. Germany: LAP LAMBERT Academic Publishing 2011; . |
| 10. | Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 508] [Cited by in RCA: 567] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 11. | Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1406] [Cited by in RCA: 1624] [Article Influence: 101.5] [Reference Citation Analysis (0)] |
| 12. | Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet. 2008;9:699-712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 250] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 13. | Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701-718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1837] [Cited by in RCA: 2039] [Article Influence: 119.9] [Reference Citation Analysis (0)] |
| 14. | Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16-26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 658] [Cited by in RCA: 619] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 15. | Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499-506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 862] [Cited by in RCA: 863] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 16. | Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741-754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 850] [Cited by in RCA: 985] [Article Influence: 57.9] [Reference Citation Analysis (0)] |
| 17. | Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1877] [Cited by in RCA: 1988] [Article Influence: 86.4] [Reference Citation Analysis (0)] |
| 18. | Luco RF, Misteli T. More than a splicing code: integrating the role of RNA, chromatin and non-coding RNA in alternative splicing regulation. Curr Opin Genet Dev. 2011;21:366-372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 19. | Calarco J, Zhen M, Blencowe B. Networking in a global world: establishing functional connections between neural splicing regulators and their target transcripts. RNA. 2011;17:775-791. [RCA] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 59] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 20. | König J, Zarnack K, Luscombe N, Ule J. Protein-RNA interactions: new genomic technologies and perspectives. Nat Rev Genet. 2012;13:77-83. |
| 21. | Tenney AE, Wu JQ, Langton L, Klueh P, Quatrano R, Brent MR. A tale of two templates: automatically resolving double traces has many applications, including efficient PCR-based elucidation of alternative splices. Genome Res. 2007;17:212-218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 22. | Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6477] [Cited by in RCA: 5129] [Article Influence: 165.5] [Reference Citation Analysis (0)] |
| 23. | DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3178] [Cited by in RCA: 2818] [Article Influence: 97.2] [Reference Citation Analysis (3)] |
| 24. | Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863-14868. [PubMed] |
| 25. | Calarco JA, Saltzman AL, Ip JY, Blencowe BJ. Technologies for the global discovery and analysis of alternative splicing. Adv Exp Med Biol. 2007;623:64-84. [PubMed] |
| 26. | Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688-700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 661] [Cited by in RCA: 666] [Article Influence: 39.2] [Reference Citation Analysis (0)] |
| 27. | Yeakley JM, Fan JB, Doucet D, Luo L, Wickham E, Ye Z, Chee MS, Fu XD. Profiling alternative splicing on fiber-optic arrays. Nat Biotechnol. 2002;20:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 147] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 28. | Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141-2144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1028] [Cited by in RCA: 1064] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 29. | Wang H, Hubbell E, Hu JS, Mei G, Cline M, Lu G, Clark T, Siani-Rose MA, Ares M, Kulp DC. Gene structure-based splice variant deconvolution using a microarray platform. Bioinformatics. 2003;19 Suppl 1:i315-i322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 70] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 30. | Le K, Mitsouras K, Roy M, Wang Q, Xu Q, Nelson SF, Lee C. Detecting tissue-specific regulation of alternative splicing as a qualitative change in microarray data. Nucleic Acids Res. 2004;32:e180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 71] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 31. | Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, Babak T, Siu H, Hughes TR, Morris QD. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol Cell. 2004;16:929-941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 236] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 32. | Stolc V, Gauhar Z, Mason C, Halasz G, van Batenburg MF, Rifkin SA, Hua S, Herreman T, Tongprasit W, Barbano PE. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 238] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 33. | Watahiki A, Waki K, Hayatsu N, Shiraki T, Kondo S, Nakamura M, Sasaki D, Arakawa T, Kawai J, Harbers M. Libraries enriched for alternatively spliced exons reveal splicing patterns in melanocytes and melanomas. Nat Methods. 2004;1:233-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 34. | Blanchette M, Green RE, Brenner SE, Rio DC. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes Dev. 2005;19:1306-1314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 100] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 35. | Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, Wang H, Clark T, Fraser C, Ruggiu M. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37:844-852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 376] [Cited by in RCA: 408] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 36. | Shai O, Morris QD, Blencowe BJ, Frey BJ. Inferring global levels of alternative splicing isoforms using a generative model of microarray data. Bioinformatics. 2006;22:606-613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 44] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 37. | Sugnet CW, Srinivasan K, Clark TA, O’Brien G, Cline MS, Wang H, Williams A, Kulp D, Blume JE, Haussler D. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput Biol. 2006;2:e4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 152] [Cited by in RCA: 165] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 38. | Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat Methods. 2009;6:S22-S32. [RCA] [DOI] [Full Text] [Cited by in Crossref: 416] [Cited by in RCA: 408] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 39. | Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. 2011;12:671-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 860] [Cited by in RCA: 821] [Article Influence: 54.7] [Reference Citation Analysis (0)] |
| 40. | Chen K, Deng S, Lu H, Zheng Y, Yang G, Kim D, Cao Q, Wu JQ. RNA-seq characterization of spinal cord injury transcriptome in acute/subacute phases: a resource for understanding the pathology at the systems level. PLoS One. 2013;8:e72567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 66] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 41. | Kahvejian A, Quackenbush J, Thompson JF. What would you do if you could sequence everything? Nat Biotechnol. 2008;26:1125-1133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 130] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 42. | Malone JH, Oliver B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol. 2011;9:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 398] [Cited by in RCA: 348] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 43. | Fu X, Fu N, Guo S, Yan Z, Xu Y, Hu H, Menzel C, Chen W, Li Y, Zeng R. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics. 2009;10:161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 205] [Cited by in RCA: 212] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 44. | Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509-1517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2021] [Cited by in RCA: 2012] [Article Influence: 111.8] [Reference Citation Analysis (0)] |
| 45. | Soneson C, Delorenzi M. A comparison of methods for differential expression analysis of RNA-seq data. BMC Bioinformatics. 2013;14:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 589] [Cited by in RCA: 566] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 46. | Nookaew I, Papini M, Pornputtapong N, Scalcinati G, Fagerberg L, Uhlén M, Nielsen J. A comprehensive comparison of RNA-Seq-based transcriptome analysis from reads to differential gene expression and cross-comparison with microarrays: a case study in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40:10084-10097. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 223] [Cited by in RCA: 218] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 47. | Brosseau JP, Lucier JF, Lapointe E, Durand M, Gendron D, Gervais-Bird J, Tremblay K, Perreault JP, Elela SA. High-throughput quantification of splicing isoforms. RNA. 2010;16:442-449. [DOI] [Full Text] |
| 48. | Venables JP, Lapasset L, Gadea G, Fort P, Klinck R, Irimia M, Vignal E, Thibault P, Prinos P, Chabot B. MBNL1 and RBFOX2 cooperate to establish a splicing programme involved in pluripotent stem cell differentiation. Nat Commun. 2013;4:2480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 110] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 49. | Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, Durand M, Couture S, Froehlich U, Lapointe E. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol. 2009;16:670-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 281] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 50. | Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5228] [Cited by in RCA: 4973] [Article Influence: 331.5] [Reference Citation Analysis (0)] |
| 51. | Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B, David L, Golipour A, Gabut M, Michael IP, Nachman EN. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498:241-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 261] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 52. | Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci USA. 2000;97:14085-14090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 321] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 53. | Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212-1215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 833] [Cited by in RCA: 876] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 54. | Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 457] [Cited by in RCA: 468] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 55. | Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: Implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692-1694. [DOI] [Full Text] |
| 56. | Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA. 2010;1:266-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 314] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 57. | Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129-141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2523] [Cited by in RCA: 2291] [Article Influence: 143.2] [Reference Citation Analysis (0)] |
| 58. | König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909-915. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1037] [Cited by in RCA: 914] [Article Influence: 57.1] [Reference Citation Analysis (0)] |
| 59. | Ramirez JM, Gerbal-Chaloin S, Milhavet O, Qiang B, Becker F, Assou S, Lemaître JM, Hamamah S, De Vos J. Brief report: benchmarking human pluripotent stem cell markers during differentiation into the three germ layers unveils a striking heterogeneity: all markers are not equal. Stem Cells. 2011;29:1469-1474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 60. | Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, Eliazer S, Young H, Richardson M, Smart NG, Cunningham J. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26:443-452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1331] [Cited by in RCA: 1315] [Article Influence: 73.1] [Reference Citation Analysis (0)] |
| 61. | Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 825] [Cited by in RCA: 802] [Article Influence: 57.3] [Reference Citation Analysis (0)] |
| 62. | Wu SM, Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat Cell Biol. 2011;13:497-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 379] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 63. | Raya A, Rodríguez-Pizà I, Guenechea G, Vassena R, Navarro S, Barrero MJ, Consiglio A, Castellà M, Río P, Sleep E. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53-59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 566] [Cited by in RCA: 513] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 64. | Evseenko D, Zhu Y, Schenke-Layland K, Kuo J, Latour B, Ge S, Scholes J, Dravid G, Li X, MacLellan WR. Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proc Natl Acad Sci USA. 2010;107:13742-13747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 203] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 65. | Pritsker M, Doniger T. Diversification of stem cell molecular repertoire by alternative splicing. Proc Natl Acad Sci USA. 2005;102:14290-14295. [PubMed] |
| 66. | Wu JQ, Habegger L, Noisa P, Szekely A, Qiu C, Hutchison S, Raha D, Egholm M, Lin H, Weissman S. Dynamic transcriptomes during neural differentiation of human embryonic stem cells revealed by short, long, and paired-end sequencing. Proc Natl Acad Sci USA. 2010;107:5254-5259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 155] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 67. | Salomonis N, Schlieve CR, Pereira L, Wahlquist C, Colas A, Zambon AC, Vranizan K, Spindler MJ, Pico AR, Cline MS. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci USA. 2010;107:10514-10519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 186] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 68. | Yeo GW, Xu X, Liang TY, Muotri AR, Carson CT, Coufal NG, Gage FH. Alternative splicing events identified in human embryonic stem cells and neural progenitors. PLoS Comput Biol. 2007;3:1951-1967. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 97] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 69. | Brandenberger R, Wei H, Zhang S, Lei S, Murage J, Fisk GJ, Li Y, Xu C, Fang R, Guegler K. Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation. Nat Biotechnol. 2004;22:707-716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 247] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 70. | Cloonan N, Forrest ARR, Kolle G, Gardiner BBA, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nature Methods. 2008;5:613-619. [RCA] [DOI] [Full Text] [Cited by in Crossref: 765] [Cited by in RCA: 752] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 71. | Revil T, Gaffney D, Dias C, Majewski J, Jerome-Majewska LA. Alternative splicing is frequent during early embryonic development in mouse. BMC Genomics. 2010;11:399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 65] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 72. | Ohta S, Nishida E, Yamanaka S, Yamamoto T. Global splicing pattern reversion during somatic cell reprogramming. Cell Rep. 2013;5:357-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 73. | Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 378] [Cited by in RCA: 439] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 74. | Modrek B, Lee CJ. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat Genet. 2003;34:177-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 379] [Cited by in RCA: 401] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 75. | Greene AL, Lalli MJ, Ji Y, Babu GJ, Grupp I, Sussman M, Periasamy M. Overexpression of SERCA2b in the heart leads to an increase in sarcoplasmic reticulum calcium transport function and increased cardiac contractility. J Biol Chem. 2000;275:24722-24727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 48] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 76. | Misquitta CM, Mwanjewe J, Nie L, Grover AK. Sarcoplasmic reticulum Ca(2+) pump mRNA stability in cardiac and smooth muscle: role of the 3’-untranslated region. Am J Physiol Cell Physiol. 2002;283:C560-C568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 77. | Domian IJ, Chiravuri M, van der Meer P, Feinberg AW, Shi X, Shao Y, Wu SM, Parker KK, Chien KR. Generation of functional ventricular heart muscle from mouse ventricular progenitor cells. Science. 2009;326:426-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 159] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 78. | Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, Hsiao EC, Schwartz RJ, Conklin BR, Bernstein HS. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219-229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 495] [Cited by in RCA: 466] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 79. | Pereira L, Yi F, Merrill BJ. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol Cell Biol. 2006;26:7479-7491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 238] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 80. | Tam WL, Lim CY, Han J, Zhang J, Ang YS, Ng HH, Yang H, Lim B. T-cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells. 2008;26:2019-2031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 141] [Article Influence: 7.8] [Reference Citation Analysis (1)] |
| 81. | Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 545] [Cited by in RCA: 542] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 82. | Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destrée O, Clevers H. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 522] [Cited by in RCA: 531] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 83. | Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947-956. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3597] [Cited by in RCA: 3411] [Article Influence: 162.4] [Reference Citation Analysis (13)] |
| 84. | Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106-1117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1957] [Cited by in RCA: 1970] [Article Influence: 109.4] [Reference Citation Analysis (0)] |
| 85. | Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049-1061. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1124] [Cited by in RCA: 1065] [Article Influence: 59.2] [Reference Citation Analysis (0)] |
| 86. | Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722-737. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 859] [Cited by in RCA: 791] [Article Influence: 46.5] [Reference Citation Analysis (0)] |
| 87. | Wang X, Dai J. Concise review: isoforms of OCT4 contribute to the confusing diversity in stem cell biology. Stem Cells. 2010;28:885-893. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 166] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 88. | Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Schöler H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2554] [Cited by in RCA: 2512] [Article Influence: 89.7] [Reference Citation Analysis (0)] |
| 89. | Mayshar Y, Rom E, Chumakov I, Kronman A, Yayon A, Benvenisty N. Fibroblast growth factor 4 and its novel splice isoform have opposing effects on the maintenance of human embryonic stem cell self-renewal. Stem Cells. 2008;26:767-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 50] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 90. | Lin H, Shabbir A, Molnar M, Yang J, Marion S, Canty JM, Lee T. Adenoviral expression of vascular endothelial growth factor splice variants differentially regulate bone marrow-derived mesenchymal stem cells. J Cell Physiol. 2008;216:458-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 49] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 91. | Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130-137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 509] [Cited by in RCA: 474] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 92. | Rao S, Zhen S, Roumiantsev S, McDonald LT, Yuan GC, Orkin SH. Differential roles of Sall4 isoforms in embryonic stem cell pluripotency. Mol Cell Biol. 2010;30:5364-5380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 139] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 93. | Rosa A, Brivanlou AH. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 2011;30:237-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 157] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 94. | Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O’Hanlon D, Sung HK, Alvarez M, Talukder S, Pan Q, Mazzoni EO. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147:132-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 303] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 95. | Jangi M, Boutz PL, Paul P, Sharp PA. Rbfox2 controls autoregulation in RNA-binding protein networks. Genes Dev. 2014;28:637-651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 110] [Article Influence: 9.2] [Reference Citation Analysis (0)] |