Published online Oct 26, 2013. doi: 10.4252/wjsc.v5.i4.172
Revised: July 23, 2013
Accepted: August 16, 2013
Published online: October 26, 2013
Processing time: 125 Days and 2.9 Hours
Mounting evidence in stem cell biology has shown that microRNAs (miRNAs) play a crucial role in cell fate specification, including stem cell self-renewal, lineage-specific differentiation, and somatic cell reprogramming. These functions are tightly regulated by specific gene expression patterns that involve miRNAs and transcription factors. To maintain stem cell pluripotency, specific miRNAs suppress transcription factors that promote differentiation, whereas to initiate differentiation, lineage-specific miRNAs are upregulated via the inhibition of transcription factors that promote self-renewal. Small molecules can be used in a similar manner as natural miRNAs, and a number of natural and synthetic small molecules have been isolated and developed to regulate stem cell fate. Using miRNAs as novel regulators of stem cell fate will provide insight into stem cell biology and aid in understanding the molecular mechanisms and crosstalk between miRNAs and stem cells. Ultimately, advances in the regulation of stem cell fate will contribute to the development of effective medical therapies for tissue repair and regeneration. This review summarizes the current insights into stem cell fate determination by miRNAs with a focus on stem cell self-renewal, differentiation, and reprogramming. Small molecules that control stem cell fate are also highlighted.
Core tip: Stem cells are important in regenerative medicine applications due to their capacity to self-renew and differentiate into specific cell types. MicroRNAs (miRNAs) are short non-coding RNAs that negatively regulate gene expression at the post-transcriptional level. Recent studies suggest that miRNAs are key molecules in the regulation of stem cell fate decisions; this regulation is manifested as the fine tuning of cell- and tissue-specific gene expression. This review summarizes the current insights into stem cell fate determination by miRNAs and focuses on stem cell self-renewal, differentiation, and reprogramming. Small molecules that control stem cell fate are also highlighted.
- Citation: Choi E, Choi E, Hwang KC. MicroRNAs as novel regulators of stem cell fate. World J Stem Cells 2013; 5(4): 172-187
- URL: https://www.wjgnet.com/1948-0210/full/v5/i4/172.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v5.i4.172
Stem cells are a potential source for regenerative medicine and tissue engineering applications. These cells have the dual capacity to self-renew and differentiate into multiple distinct cell lineages[1,2]. These cells are classified as embryonic stem cells (ESCs), non-embryonic adult stem cells, and induced pluripotent stem cells (iPSCs). ESCs are pluripotent cells produced within the inner cell mass of a blastocyst stage embryo 4-5 d post-fertilization and can differentiate into all three germ layers: ectoderm, endoderm, and mesoderm[3]. In contrast, adult stem cells are found in various tissues and organs, including the brain, bone marrow, peripheral blood, blood vessels, skeletal muscle, and skin[4]. Some adult stem cells are multipotent; they can produce a limited number of differentiated cell types from their specific tissue of origin. iPSCs are reprogrammed to be embryonic-like stem cells from adult somatic cells[5,6].
Stem cell fate is controlled by transcription factors, epigenetic regulation, and non-coding RNAs[7,8]. Transcription factors are well-known for regulating gene expression, by either directly or indirectly binding DNA elements, and for their role in epigenetic regulation, such as DNA methylation and histone modification. The control of gene expression also occurs during the post-transcription process. Recent findings have shown that small non-coding RNAs are involved in cell fate decisions, including the maintenance and differentiation of stem cells[7,9].
MicroRNAs (miRNAs) are single-stranded, small non-coding RNA molecules. miRNAs modulate gene expression by either inhibiting mRNA translation or inducing mRNA degradation, which results from the complete or incomplete binding to the 3’ untranslated region (3’-UTR) of specific mRNAs[10,11]. More than 1000 different mature miRNAs have been discovered in humans, and they regulate one third of all protein-coding genes[12,13]. Computational predictions of miRNA targets, functions, and expression, are accessible on multiple online prediction databases, such as TargetScan (http://targetscan.org), microRNA.org (http://www.microrna.org), miRBase (http://www.mirbase.org), PicTar (http://www.pictar.org), and miRWalk (http://mirwalk.uni-hd.de)[14,15]. One miRNA can target a large number of mRNAs, and/or many miRNAs can bind to one specific mRNA. This versatility may result in miRNAs mediating the effects of biological processes such as stem cell fate switches, proliferation, maintenance, and apoptosis. Interestingly, the first two miRNAs discovered, lin-4 and let-7, were characterized during the developmental stage transition in C. elegans[16,17]. By deleting enzymes involved in miRNA processing and maturation, namely, Dicer or Dgcr8, studies have shown that miRNAs are important in maintaining ESC pluripotency and differentiation capacity[18-20]. miRNAs also play a role in the differentiation and self-renewal of mesenchymal stem cells (MSCs)[21]. Many observations suggest that miRNAs critically regulate stem cell fate decisions, including self-renewal, differentiation into specific lineages, and reprogramming. Thus, this review focuses on miRNAs that are powerful regulators of stem cell fate. Furthermore, we discuss the potential of small molecules in regulating stem cell fate.
Self-renewal and differentiation potential are hallmarks of stem cells. Self-renewal is a process of symmetric division into two daughter cells. To self-renew, stem cells must proliferate without differentiating or becoming apoptotic to maintain their undifferentiated state[22,23].
Cell division during self-renewal is achieved through regulated cell cycle events, such as the alternating activities of various D-type cyclins, cyclin-dependent kinases (CDKs), and E2F transcription factors. These cell cycle modulators and miRNA molecules are regulated during post-transcriptional modification[10,24]. The transcription factors Oct4, Sox2, and Nanog are also important for the self-renewal of pluripotent cells[7,25,26]. Oct4 and Nanog were the first transcription factors to be identified as necessary for the development and maintenance of ESC pluripotency. The expression of these factors is limited to pluripotent cell lines[26-28]. Additionally, Oct4, Sox2, and Nanog have an autoregulatory feedback loop, which is an important feature of human ESCs[29], and Sox2 implicitly interacts with Oct4[30].
Oct4, Sox2, and Nanog may be upstream regulators of the miR-302-367 cluster of miRNA, which have been identified and differentially expressed in human ESCs[31-33]. Conversely, miR-302-367 is required for Oct4, Sox2, and Nanog expression. Thus, miR-302-367 and the transcription factors (Oct4, Sox2, and Nanog) are tightly linked through an autoregulatory positive loop in pluripotent cells[34,35]. Additionally, miR-302a promotes the G1/S transition by repressing the translation of cyclin D1 in human ESCs[36]. The inhibition of miR-302a causes an accumulation of pluripotent human ESCs in the G1 phase[36]. ESCs usually have a rapid G1/S transition, which results in an extremely rapid proliferation rate (-10 h) compared to that of differentiated cells (more than 18 h)[24]. The G1/S transition is regulated by the cyclin D-Cdk4, 6 and cyclin E-Cdk2 complexes. The cyclin D-Cdk4, 6 complex is not present in mouse ESCs; however, the cyclin E-Cdk2 complex that induces S phase and DNA replication is present and active[20,37]. In vivo experiments performed in a developing lung demonstrated that miR-302-367 decreased the expression of inhibitors of cdkn1a (p21) and Rbl2, inhibitors of the cyclin E-Cdk2 complex, which resulted in the formation of an undifferentiated multi-layered lung endoderm[38]. Furthermore, in Dicer- and Dgcr8-knockout mice, ESCs exhibited reduced cell proliferation and an extended G1 phase[18,19].
Similar to miR-302-367, the miR-290-295 cluster is highly expressed in mouse ESCs, is regulated by Oct4, and binds Oct4, Sox2, Nanog, and Tcf3 to its promoters[33,39]. The increased expression of the miR-290 family promotes the G1/S transition, which enables rapid ESC proliferation and mediates the suppression of cdkn1a, Rbl2, and Lats2[37]. Indeed, the miR-290 family functionally antagonizes differentiation-related miRNAs, such as the let-7 family. The miR-290-295 cluster is rapidly downregulated during differentiation, which occurs with the restoration of let-7 maturation. Increased let-7 expression promotes differentiation by directly targeting pluripotency factors and ESC-enriched genes[40].
Another important gene in stem cell maintenance is c-Myc, which is inhibited by let-7[41]. In addition, c-Myc binds to the promoters of miR-141, miR-200, and miR-429. These miRNAs inhibit differentiation in mouse ESCs[42]. Furthermore, c-Myc stimulates the expression of the miR-17-92 cluster in tumor cells[43]. These miRNAs reduce the expression of the cell cycle control gene Rb2, which plays an important role in stem cell self-renewal[44]. Moreover, miR-92b promotes the G1/S transition through the repression of cdkn1c (p57, Kip2) in human ESCs[45]. In fact, the miR-302-367, miR-290-295, and miR-17-92 clusters have been designated as ESC-specific cell cycle-regulating miRNAs (ESCC miRNAs) because they promote the G1/S transition and cellular proliferation in ESCs[37].
Compared to their role in ESCs, there is less evidence for the involvement of miRNAs in the self-renewal of somatic stem cells. The overexpression of miR-205 enhanced proliferation and expanded the population of progenitor cells by modulating PTEN, a tumor-suppressor gene[46].
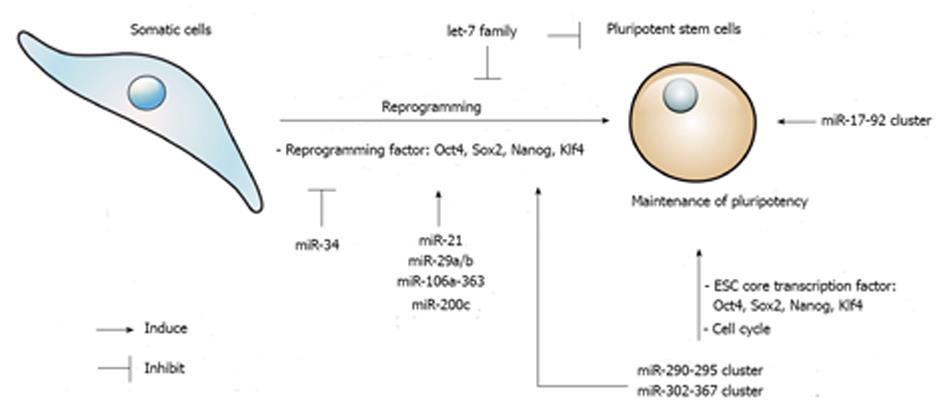
Therefore, stem cell self-renewal is tightly regulated through a complex network of core transcription factors, miRNAs, and the repression and/or promotion of differentiation mechanisms and pluripotent pathways, respectively (Figure 1).
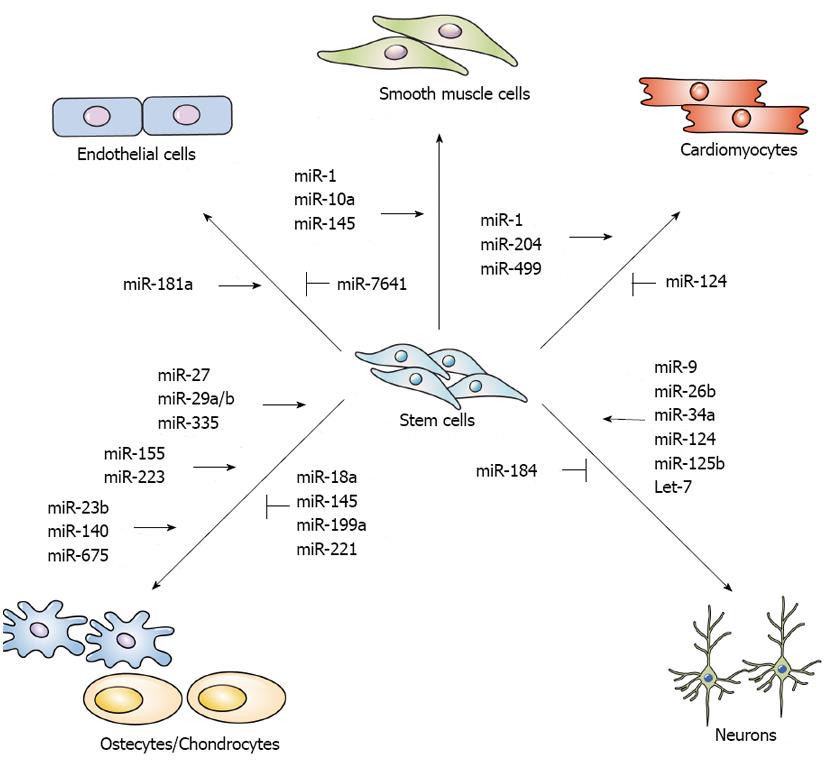
Some studies indicate that miRNAs affect the vascular development or differentiation of stem cells, and others provide detailed reviews of the effect of miRNAs on endothelial cells (ECs), vascular smooth muscle cells (VSMCs), and cardiomyocytes[47-49] (Figure 2).
The first evidence for the regulation of endothelial cell functions by miRNAs came from observations that dicer knockout mice displayed defects in embryos and yolk sacs during vasculogenesis and early angiogenesis[50]. Dicer, accompanied by the altered expression of vascular endothelial growth factor (VEGF), fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor; FLT1), kinase insert domain receptor (a type III receptor tyrosine kinase; KDR), and tyrosine kinase with immunoglobulin-like and EGF-like domains 1 (Tie-1), plays and essential role in endothelial development.
Increased expression of miR-126 was first identified in Flk-1+ mesoderm populations derived from mouse ESCs[51]. Two additional studies, performed with zebrafish and mice, demonstrated that miR-126 is essential for vessel integrity and endothelial function regulation but that it is not required to control the differentiation of ESCs to ECs[52,53].
The expression of miRNAs associated with angiogenesis (let-7b, let-7f, miR-126, miR-130a, miR-133a, miR-133b, miR-210, and miR-296) was enhanced in day 10 differentiated cells compared to pluripotent human ESCs[54]. Increased expression of the let-7 family during differentiation occurred by directly targeting pluripotency factors and ESC-enriched genes[40]. Specifically, let-7f contributed to the angiogenic sprouting of ECs in vitro[55]. The other upregulated miRNAs, miR-130a, enhanced angiogenesis by modulating GAX (growth arrest-specific homeobox) and HOXA5 (homeobox protein Hox-A5), which are anti-angiogenic homeobox transcription factors[56]. Additionally, miR-210 was shown to be required for angiogenesis by targeting EphinA3[57], and miR-146b, miR-197, and miR-625 expression was enriched in CD31+ endothelial populations derived from mouse ESCs[52]. Although the function of these miRNAs has been studied in cancer cells[58-60], their role in the differentiation and functionality of ECs remains unknown.
The miRNA miR-181a promotes the reprogramming of lymphatic ECs toward a blood vascular phenotype[61]. The binding of miR-181a, to the 3’UTR of Prox1 (prospero homeobox 1, a key gene involved in lymphatic EC identity) results in inhibited expression. In human ESCs, miR-99b, miR-181a and miR-181b regulated the mRNA and protein expression of EC-specific markers, increased nitric oxide production, and improved therapeutic neovascularization in vivo[62]. In addition, the expression of miR-7641 was downregulated during the endothelial differentiation of human ESCs. The overexpression of this miRNA significantly suppressed the expression of CXCL1 (a member of the CXC chemokine family)[63]. CXCL1, which is involved in EC biogenesis and angiogenesis, is known to promote neovascularization by binding G-protein-coupled receptors[64,65].
The miRNAs miR-143 and miR-145 are abundantly expressed in smooth muscle tissue. These miRNAs promote smooth muscle cell (SMC) differentiation from neural crest stem cells and are upregulated during differentiation, which is consistent with early expression patterns in the aorta of developing mouse embryos[66-68]. Recently, it was discovered that miR-145 also promotes SMC differentiation from human ESCs[69]. The expression of miR-143 and miR-145 is controlled by serum response factor (SRF), myocardin (MYOCD), and the following miRNA target transcription factors: KLF4, ELK1, and angiotensin-converting enzyme (ACE)[66-68]. Other targets of miR-145 are Oct4, Sox2, and Klf4, which are transcription factors for the self-renewal of pluripotent cells. These miRNAs are involved in regulating cell fate decisions across different lineages[70]. A loss of miR-145 induced a different SMC phenotype, which was similar to the proliferating SMCs found in vascular lesions, but did not affect SMC differentiation[66,67]. A reduction in neointima formation after vessel injury was observed in miR-145-/-mice and, to a lesser extent, in miR-143-/-mice[68]. However, the overexpression of miR-143 and miR-145 also decreased neointima formation in a rat model of acute vascular injury[71]. These data suggest that miR-143 and miR-145 are vital to SMC differentiation in vitro, but are not essential for SMC differentiation during embryonic development in vivo.
Another study showed that the increased expression of miR-10a during the in vitro differentiation of mouse ESCs to SMCs occurred via the post-transcriptional inhibition of histone deacetylase 4 (HDAC4)[72]. The inhibition of miR-10a impairs SMC differentiation.
The miRNA miR-1 is involved in cardiomyocyte differentiation, cardiac hypertrophy, and apoptosis; however, recent studies suggest that it also plays a role in SMC differentiation[73]. During the differentiation of mouse ESCs to SMCs, the expression of miR-1 steadily increased. Loss-of-function approaches using inhibitors against miR-1 resulted in the downregulation of SMC-specific markers and a decrease in the population of derived SMCs, indicating that miR-1 is required for the SMC lineage differentiation of ESC cultures. Previously identified as a miR-145 target, KLF is a target for miR-1.
The miRNAs miR-1 and miR-133 were first described as critical regulators for muscle proliferation and skeletal muscle[74] and cardiac muscle[51] differentiation. Both miR-1 and miR-133 promote mesoderm formation from ESCs; however, these miRNAs have opposing functions during differentiation to cardiac muscle progenitors[51,74-76].
These miRNAs, miR-1-1 and miR-1-2, are specifically expressed in cardiac and skeletal muscle precursor cells and direct transcriptional targets, such as SRF, myogenic differentiation 1 (MyoD), and myocyte enhancement factor 2 (Mef2)[77]. An increased expression of miR-1 in mice led to embryonic developmental arrest at day 13.5, which resulted in a decreased population of proliferating ventricular cardiomyocytes[77]. Hand2, a transcription factor that regulates ventricular cardiomyocyte expansion, is a direct target of miR-1[77]. However, the targeted deletion of one of the two miR-1 genes (miR-1-2) located in muscle-specific miRNAs revealed numerous dysfunctions in the heart, including defective morphogenesis, electrical conduction, and unregulated cell-cycle control[76]. Additionally, Drosophila melanogaster miR-1 modulates cardiogenesis and muscle-gene expression[75]. Ivey et al[51] described that miR-1 acts as a repressor of non-muscle genes and that the overexpression of miR-1 upregulates Nkx2.5, an early cardiac marker, to promote cardiac differentiation. Notch ligand Delta-like 1 (Dll-1) is a target of miR-1[51]. In human ESC-derived embryoid bodies, miR-1 also increased the expression of myosin heavy chain (MHC) genes[78]. Additionally, miR-1 increased the expression of cardiomyocyte-specific genes and enhanced cardiomyocyte differentiation from human-derived cardiomyocyte progenitor cells by targeting HDAC4[79]. Interestingly, the transplantation of murine ESCs overexpressing miR-1 into the border zone of infarcted mouse hearts prevented ischemia-induced apoptosis[80]. In addition, miR-1 facilitates the electrophysiological maturation of ESCs[81]. Furthermore, when miR-1 was transfected into fibroblast cells, gene expression profiles shifted toward that of muscle-like cells[82]. Recently, miR-1 induced the expression of several cardiomyocyte markers, including Nkx2.5, GATA-4, cTnT, and CX43, via the downregulation of Hes-1, the downstream target molecule of the Notch pathway in MSCs[83].
Although miR-1 and miR-133 are bicistronic[76,84], they have opposing actions. The deletion of miR-133a genes causes lethal ventricular-septal defects, and results in the ectopic expression of smooth muscle genes. Therefore, miR-133a regulates the proliferation of cardiomyocytes by SRF and cyclin D2 activity[84]. Specific cardiac markers were downregulated in miR-133-overexpressed mouse and human ESCs[51,85], and miR-133 induced the proliferation of myoblasts by repressing SRF[74]. A recent study revealed that miR-133 inhibited the proliferation of the prostate cancer cell lines PC3 and DU145 by targeting the epidermal growth factor receptor (EGFR)[86]. Concurrently, our group also discovered that miR-133a expression increased during differentiation and that the overexpression of miR-133a promoted cardiac differentiation in human MSCs by targeting EGFR[87].
Increased miR-499 expression was discovered in adult cardiac progenitor cells and human ESCs[78,79]. This miRNA is encoded by an intron of MHC[88] and shares many predicted targets with miR-208, which plays a crucial role in the stress-adaptation of the adult heart. The overexpression of miR-499 reduced the proliferation and enhanced the differentiation of human cardiomyocyte progenitor cells and ESCs through targeting Sox6, which is expressed in heart and skeletal muscle[79]. The miRNA miR-499 has also been shown to play a role in myocyte lineage differentiation and the generation of mature working cardiomyocytes in vitro and after infarction in vivo[89]. Both Sox6 and regulator of differentiation 1 (Rod1) are targets of miR-499. In addition to ESCs, cardiac stem cells, and cardiomyocyte progenitor cells, a recent study showed that the overexpression of miR-499 in rat MSCs induced cardiac differentiation through the Wnt/β-catenin signaling pathway[90].
Additionally, miR-204 is required for human cardiomyocyte progenitor cell differentiation, which occurs through targeting ATF-2[91], whereas miR-124 inhibits the cardiomyocyte differentiation of MSCs by targeting STAT3[92]. Finally, the deletion of the miR-17-92 cluster led to very specific defects in the development of the heart[93]; however, the function of the miR-17-92 cluster in cardiac differentiation and development remains unclear.
Neural stem cells (NSCs) give rise to neurons, astrocytes, and oligodendrocytes and play an important role in embryonic development and the maintenance of the adult central nervous system (CNS)[94]. The differentiation of NSCs is tightly associated with multiple signaling pathways: the Wnt signaling pathway regulates NSC proliferation and differentiation[95], the transcription factors Neurog2 and Tbr2 are linked to NSC differentiation[96]; the orphan nuclear receptor TLX is necessary for adult NSC proliferation[97]; and the methyl CpG binding protein 2 (MeCP2), methyl-CpG binding protein 1 (MBD1), and histone-lysine N-methyltransferase Ezh2 are related to adult neurogenesis[98,99]. In the mammalian brain, some miRNAs expression is tissue-specific, such as the let-7 family, miR-124, and miR-9, which regulate neurogenesis[100,101]. Brain-specific miR-124 is upregulated during CNS development and the neuronal differentiation of the adult subventricular zone (SVZ)[102,103]. During neurogenesis, the suppression of RE-1-silencing transcription repressor (REST) induces the expression of miR-124, which represses JAG1, Dlx2, and Sox9. In addition, laminin γ1 and integrin β1, which are expressed in neural progenitors but inhibit neuronal differentiation, are also targeted by miR-124 and lead to neurogenesis[104]. The miRNA miR-9 is also highly expressed in the brain and is involved in modulating the balance between NSC self-renewal and differentiation via negative TLX expression[105]. The overexpression of miR-9 promotes neural differentiation but downregulates TLX. Let-7d, a member of the let-7 family, also targets TLX, promotes neurogenesis, and reduces NSC proliferation[106]. Let-7a is a downstream molecule of tripartite motif-containing protein 32 (TRIM32); therefore, let-7a is also required to induce NSC differentiation[107]. The overexpression of TRIM32 induces neuronal differentiation, whereas the inhibition of TRIM32 preserves the self-renewal capability of neural progenitor cells. The miRNA miR-137 is essential for embryonic NSC fate decisions; the overexpression of miR-137 inhibits NSC proliferation and induces accelerated differentiation by suppressing histone lysine-specific demethylase 1 (LSD1), a co-transcription factor of TLX[108]. Additionally, miR-137, which mediates epigenetic proteins such as MeCP2 (a DNA methyl-CpG-binding protein), Ezh2, and Polycomb group (PcG) protein, regulates the balance of NSC proliferation and differentiation in adult neurogenesis. A reduction of miR-137 expression promotes differentiation, whereas the overexpression of miR-137 increases adult NSCs proliferation[98]. Similar to miR-137, miR-184 is associated with controlling the balance between the proliferation and differentiation of adult NSCs. Upregulated miR-184 targets methyl-CpG binding protein 1 (MBD1) and Numblike (Numbl), which are related to NSC differentiation in the adult brain, to induce cell proliferation and reduce the differentiation of adult NSCs[99]. In neural stem/progenitor cells (NSPCs) isolated from adult mice, the miR-106b-25 cluster (miR-106b, miR-93, and miR-25) regulates NSPC proliferation and differentiation. The miRNA miR-25 targets insulin/insulin-like growth factor-1 (IGF) signaling pathways. The expression of miR-106b-25 is mediated by FoxO3, a member of the FoxO family of transcription factors that is important for the maintenance and differentiation of NSCs[109]. Recently, it was determined that miR-34a is involved in NSC differentiation; miR-34a promotes Notch signaling by repressing Numbl, a negative regulator of Notch signaling that inhibits neuronal differentiation[110]. Additionally, miR-26b activates neurogenesis by suppressing Ctdsp2 protein expression[111,112]. By targeting Nestin, miR-125b promotes NSPC differentiation and migration while inhibiting NSPC proliferation[113] (Figure 2).
The skeleton consists of osteoblasts and osteoclasts in bone tissue and chondrocytes in cartilage tissue[114]. Increasing evidences suggests that miRNAs are an integral part of regulating bone and cartilage formation, metabolism, homeostasis, osteogenesis, and chondrogensis[115,116].
Osteoblast differentiation from bone marrow stromal cells undergoes three stages: pre-osteoblast (proliferation), osteoblast/pre-osteocyte (matrix maturation), and osteocyte (mineralization)[117]. Each cell type expresses different genes and factors; therefore, miRNAs may be selectively expressed in particular stages during osteogenesis. At different stages of osteoblast differentiation, miR-29 has multiple distinct functions. For example, miR-29b initiates the osteogenic pathway by repressing anti-osteogenic factors, such as HDAC4, TGF-β3, activin A receptor type IIA (ACVR2A), beta-catenin-interacting protein 1 (CTNNBIP1), and dual-specific phosphatase (DUSP2). Collagen type I (COL1A1) directly targets miR-29b. During mineralization, when collagen accumulation is at a steady state, high endogenous levels of miR-29b downregulation the mRNA expression of COL1A1[118]. In addition, miR-29 suppresses osteonectin (secreted protein acidic and rich in cysteine, SPARC) during matrix maturation and the mineralization phase during late differentiation[119]. Although collagens and osteonectin play an important role in bone mass and osteogenesis, the inhibition of these proteins by miR-29b prevents sclerotic bone formation and increases bone structure stability[117]. Moreover, canonical Wnt signaling is involved in osteoblast differentiation; a high level of β-catenin is required for osteogenesis. Therefore, targeting the Wnt pathway by miRNAs has been shown to contribute to osteogenesis[120]. The miR-29 family also targets Wnt signaling-mediated proteins; the expression of miR-29 is increased by Wnt activation during osteoblast differentiation. Additionally, miR-29a negatively regulates the Wnt receptor complex Dickkopf-related protein 1 (Dkk1), Kremen2, and secreted frizzled related protein 2 (sFRP2)[121], whereas miR-29b downregulates the β-catenin inhibitor CTNNBIP1[118]. Both miR-27 and miR-335 are upregulated during osteogenesis and target the APC gene and Dkk1, a negative regulator of Wnt signaling, respectively, which leads to osteoblast differentiation[122,123].
Only a few miRNAs contribute to osteoclast differentiation. In particular, miR-223 is regulated by transcription factor PU.1. An increased expression of miR-223 and receptor activator of nuclear factor-κB (RANK) is observed in bone marrow derived osteoclast precursors after induction by M-CSF[124]. miR-223 regulates NFIA, a suppressor of osteoclastogenesis, which leads to the upregulation of the M-CSF receptor[125]. A key regulator in the maturation of hematopoietic cells to macrophages, miR-155 has been studied as another osteoclastogenic miRNA[126]. miR-155 represses MITF, a necessary transcription factor for osteoclast differentiation, to inhibit osteoclastogenesis[127].
Cartilages tissue forms bone via the endochondral process of ossification. The loss of miRNAs in cartilage accelerates the differentiation of mature hypertrophic chondrocytes and abnormal bone growth[128]. Cartilage-specific miR-140[129] is related to palatogenesis, which mediates platelet-derived growth factor D (PDGFD) signaling in zebrafish[130], craniofacial development and endochondral bone formation via targeting HDAC4[131] and inhibits BMP signaling in mouse models[132]. HDAC4 and BMP signaling pathways contribute to chondrocyte hypertrophy and osteoblast differentiation and can be negative effector of osteogenesis. The miRNA miR-675 can promote chondrogenic differentiation by inducing the expression of cartilage-specific collagen type IIa through the positive regulation of cartilage-specific Sox9[133]. The chondrogenic differentiation of MSCs is induced by miR-23b, which negatively inhibits of protein kinase A signaling[134]. In addition, miR-18a, miR-199a, miR-145, and miR-221 have been identified as negative regulators of chondrogenesis. To repress chondrogenesis, miR-18a directly targets the CCN family protein 2/connective tissue growth factor (CCN2/CTGF)[135]. Similarly, miR-199a, a bone morphogenic protein 2-responsive miRNA, significantly inhibits early chondrogenesis by targeting Smad1[136]. In addition, miR-145 targets Sox9, a key transcription factor for chondrogenic differentiation[137,138], and miR-221 negatively regulates Mdm2 and therefore prevents the degradation of Slug protein, which is involved in chondrogenesis inhibition[139] (Figure 2).
Despite the multi-lineage differentiation potential of stem cells, little is known about the differentiation of stem cells to other cell types than those described above. For example, the hepatic differentiation of human umbilical cord lining-derived MSCs (hUC-MSCs) and liver-derived progenitor cells (LDPCs) is regulated by miR-542-5p and miR-146a[140]. The miRNA miR-182 is involved in the differentiation of inner ear stem/progenitor cells into hair-like cells via the repression of Tbx1[141]. Pancreatic transcription factor Ptf1a is specifically expressed at different stages during pancreatic development; low levels of Ptf1a enhance the differentiation of pancreatic progenitor cells to endocrine cells, whereas high levels of Ptf1a are involved in exocrine cell differentiation. The endogenous expression of Ptf1a is regulated by miR-18a[142]. During the adipogenic differentiation of mouse ESCs, the expression of miR-10b, miR-15, miR-26a, miR-30a-5p, miR-30c, miR-98, miR-99a, miR-103, miR-143, miR-148a, miR-152, miR-224, miR-422b, and miR-let-7b increased, whereas the expression of the miR-17-92 cluster was downregulated[143]. Myeloid differentiation is promoted by PU.1 transcription factor, and the overexpression of the miR-23a cluster in hematopoietic progenitor cells suppresses B-cell development[144]. Furthermore, miRNAs are involved in the differentiation of diploid spermatogonia to haploid spermatozoa. The miRNA miR-34c is highly expressed in the late stages of spermatogenesis, which induces the upregulation of germ cell-specific genes[145].
In 2006, the astonishing research of Yamanaka demonstrated that somatic cells such as mouse fibroblasts, can be reprogrammed to a pluripotent state using only four transcription factors: Oct4, Sox2, Klf4, and c-Myc[5]. These reprogrammed fibroblasts are referred to as iPSCs, and they are functionally and molecularly similar to ESCs. After one year, the same group induced human iPSCs in a similar manner as the mouse iPSCs[146]. These initial studies introduced the somatic cell reprogramming strategy. Although the method of transcription factor-mediated reprogramming is simple, problems such as time, low efficiency, and the possibility of tumorigenesis remain unsolved[147]. To improve the quality of generated iPSCs, researchers have focused on using miRNAs, which are associated with regulating the epigenome. Because the ectopic expression of transcription factors during reprogramming is related to epigenetic changes, miRNAs are considered an attractive alternative for somatic cell reprogramming[35] (Figure 1).
To improve the efficiency of iPSC generation, reprogramming barriers must be overcome. The reprogramming process undergoes two phases: the early phase (initiation phase) and the late phase[8]. The early phase is a pre-pluripotent state involving increased cell proliferation and a change into an epithelial-like cellular state called the mesenchymal-epithelial transition (MET)[148]. This phase is regulated by p53-induced cell-cycle repression and the TGF-β-accelerated epithelial-mesenchymal transition (EMT). The late phase is the transition of pre-iPSCs by inducing pluripotency-related genes, such as Nanog, Sox2, and Lin28, and establishing the pluripotency network[8]. Thus, reducing these barriers by utilizing miRNA-mediated epigenetic and transcriptional regulation enhances reprogramming efficiency and generates functional cells that resemble ESCs[8,148,149].
The first attempt to reprogram focused on miRNAs that were highly expressed in ESCs and governed pluripotency but were absent in fibroblasts. Among members of the miR-290-295 family, miR-291-3p, miR-294, and miR-295, in combination with Oct4, Sox2, and Klf4, increased the reprogramming efficiency of mouse fibroblasts[150]. In human somatic cells, miR-302a-367 and/or miR-371-373 (mouse homolog miR-290-295), in combination with Oct4, Sox2, Klf4, and c-Myc, enhanced the efficiency of reprogramming by inhibiting TGF-β-induced EMT[151]. During the early reprogramming stage, miR-17-92, miR-106b-25, and miR-106a-363 clusters, which share the seed sequences of the miR-302 cluster, were shown to be highly induced[152]. The overexpression of the miR-106a-363 and miR-302-367 clusters promoted a distinct increase in iPSCs generated from mouse fibroblasts. This increase was achieved by targeting TGF-β type II receptor with Sox2, Klf4, and Oct4, which accelerated MET[153]. In addition, the activation of BMP signaling induced the expression of the miR-205 and miR-200 family and enhanced the MET[154]. Therefore, the TGF-β and BMP signaling pathways are critical mechanisms that induce MET and promote reprogramming. Further investigation of somatic reprogramming is possible using only miRNAs to directly promote reprogramming events. Recently, Anokye-Danso and coworkers reported that the transfection of miR-302 and miR-367 clusters successfully reprogramed mouse and human somatic cells to iPSCs without the use of exogenous transcription factors[155]. Interestingly, the direct transfection of mature biomimetic miRNAs, such as miR-200c and the miR-302-369 family, promoted the reprogramming of mouse and human somatic cells. This method does not require lentiviral vectors for gene transfer[156].
Contrary to the aforementioned examples, some miRNAs must be suppressed to enhance reprogramming. For example, let-7 miRNAs are negative regulators of the potent reprogramming factor Lin28. The inhibition of let-7 miRNAs leads to the dedifferentiation of somatic cells to iPSCs, which induces cell proliferation and pluripotency genes[40]. Another important miRNA barrier for reprogramming is the p53-mediated pathway. The p53-mediated pathway induces the expression of the miR-34 family and the suppression of the pluripotency factors Nanog and Sox2[157]. The genetic deletion of miR-34a increased the efficiency and kinetics of reprogramming and established pluripotency at a late stage. Additionally, the suppression of p53, by overexpressing miR-138[158] or repressing miR-21 and miR-29a, enhanced reprogramming[159]. The expression of endogenous miRNAs is regulated by transcription factors[160]. The expression of miR-29b is directly regulated by Sox2 during iPSC generation and miR-29b is an essential facilitator for Oct4, Klf4, Sox2, and c-Myc (or Oct4, Klf4, and Sox2) mediated reprogramming[161].
Reported reprogramming factors Oct4, Klf4, Sox2, and c-Myc have demonstrated that miRNAs play a crucial role in regulating stem cell fate events, such as reprogramming, differentiation, and self-renewal. However, some questions pertaining to the mechanisms of reprogramming remain unresolved. Addressing these questions will provide further understanding of reprogramming and will promote the development of iPSC generation technologies and stem cell therapies.
Stem cell fate is regulated by both intrinsic/extrinsic regulators and the extracellular niche. Because these regulators have limitations, such as efficiency and selectivity for controlling stem cell fate, a new strategy is to use of small molecules[162] (Table 1). Compared to genetic manipulations, small-molecule approaches have a number of advantages: 1) the biological effects of small molecules are rapid, reversible, and dose-dependent; 2) small molecules have specific targets in signaling pathways or epigenetic mechanisms; and 3) a variety of chemical libraries provide data for the functional optimization of small molecules[163]. Recently, many small molecules have been identified and characterized that can manipulate stem cell fate, including self-renewal, lineage-specific differentiation, and somatic cell reprogramming[35,164].
| Chemical | Effect (Target) | Result | Ref. |
| PD0325901 | MEK inhibitor | Promotes mouse ESC self-renewal | [161] |
| CHIR9902 | GSK-3 inhibitor | ||
| Y27632 | ROCK inhibitor | Enhances human ESC survival | [161-164] |
| Thiazovivin | |||
| SB431542 | TGF-β receptor inhibitor (SMAD signaling inhibitor) | Induces human ESC differentiation into endothelial cells and neural tissues | [167,168] |
| VPA | HDAC inhibitor | Somatic cell reprogramming | [170-175] |
| BIX-01294 | HMT inhibitor | ||
| RSC133 | DNMT inhibitor | ||
| 5-Aza | |||
| SB431542 | TGF-β receptor inhibitor | ||
| PD0325901 | MEK inhibitor | ||
| TSA | HDAC inhibitor | Promote HSC self- renewal | [177-179] |
| Trapoxin | |||
| Chlamydocin | |||
| SR1 | AHR antagonist | ||
| PGE2 | PG pathway | ||
| Pyrvinium | Wnt inhibitor | Promote MSC self- renewal | [191,192] |
| SKL2001 | |||
| H-89 | PKC inhibitor | Induces human MSC differentiation into chondrocytes | [130,180] |
| Katogenin | Filamin A | ||
| Purmorphamine | RUNX2 activator | Induces human MSC differentiation into osteoblasts | [181,182] |
| CW008 | cAMP/PKA/CREP pathway agonist | ||
| SJA710-6 | Induces rat MSC differentiation into hepatocytes | [183] | |
| PMA | PKC activator | Induces rat MSC differentiation into cardiomyocytes | [184,185] |
| LY294002 | PI3K/AKTinhibitor | Inhibits mouse MSC differentiation into adipocytes | [186,187] |
| CHIR9902 | GSK-3 inhibitor | ||
| Troglitazone | PPARγ agonist | Induces human MSC differentiation into adipocytes | |
| SB431542 | SMAD inhibitor | Induces human MSC differentiation into neural-like cells | [188-190] |
| LY94002 | PI3K/AKT inhibitor |
The self-renewal capacity of mouse ESCs is maintained by PD0325901 (MEK inhibitor) and CHIR99021 (GSK3 inhibitor) without feeder cells or exogenous cytokines[165]. The molecules Y-27632 and thiazovivin (ROCK inhibitor) enhance the survival of human ESCs[166-168], whereas a combination of PD0325901, CHIR99021, and Y-27632 supplemented with bFGF supports the maintenance of human ESCs[169]. Because the lineage-specific commitment of stem cells provides possible therapeutic applications, studies that control stem cell differentiation have been consistently reported. Wnt signaling modulators promote cardiomyocyte generation in zebrafish embryos and murine ESCs[170], and the inhibition of TGF-β receptor by SB431542 induces the endothelial cell differentiation of human ESCs[171]. The inhibition of SMAD signaling by noggin and SB431542 directs the differentiation of human ESCs to neural tissues[172].
ESCs have the ability to propagate indefinitely and to differentiate into any cell type; however, ethical issues regarding the use of ESCs still remain. Therefore, tissue-specific adult stem cells and the ability to reprogram somatic cells have fascinated researchers[164,173]. Ever since Yamanaka demonstrated that Oct4, Sox2, Klf4, and c-Myc can convert mouse fibroblasts into induced pluripotent stem cells (iPSCs)[5], the study of reprogramming has accelerated with the use of epigenetic process modulators, which target histone deacetylase (HDAC)[174,175], histone acetyltransferase (HMT)[176,177], and DNA methyltransferase (DNMT)[176,178]. Recently, a chemical cocktail including HDAC inhibitors and other kinase inhibitors enhanced the reprogramming efficiency of human fibroblasts[175,179].
Hematopoietic stem cells (HSCs) are related to the hematopoietic lineage, and cell phenotypes include macrophages, erythrocytes, dendritic cells, T-cells, B-cells, and NK-cells[180]. The fate of HSCs is regulated by small molecules that promote self-renewal[181-183]. Using small molecules, multipotent MSCs can differentiate into various non-hematopoietic cells, such as chondrocytes[134,184], osteoblasts[185,186], hepatocytes[187], cardiomyocytes[188,189], adipocytes[190,191] and neuronal-like cells[192-194]. Additionally, the maintenance of MSCs is associated with the Wnt signaling pathway[195,196].
Although chemical approaches are a very young field in stem cell research, these small molecules exhibit a similar biological outcome to that achieved with the use of miRNAs in stem cell fate regulation[35]. Recently, small molecules have been correlated with endogenous miRNA expression and function[197-202]. Therefore, identifying the relationship between miRNAs and small molecules could provide new insights for drug development for regenerative medicine and elucidate detailed mechanisms of miRNA expression and function in the control of stem cell fate.
Increasing evidence has demonstrated that miRNAs are promising regulators of stem cell fate. The current strategy in stem cell biology can elucidate the links between miRNAs and stem cell fate determination. Although miRNAs strictly regulate the multiple molecular signaling pathways and transcription factors that control stem cell fate, some significant issues have not received adequate attention. Current challenges focus on verifying the downstream targets of miRNA; however, the study of miRNA upstream targets is virtually nonexistent. In addition, the correlation between miRNAs is not well understood. Small molecules not only modulate stem cell fate but also regulate miRNA synthesis and the function of transcription factors and miRNAs. The challenge of identifying the relationship between miRNAs and small molecules is still at an initial stage. Complementary to conventional and interdisciplinary strategies, including miRNAs and/or chemical manipulation techniques in the regulation of stem cell self-renewal, tissue- or organ-specific differentiation, and iPSC generation provides a powerful tool to identify the underlying cellular mechanisms of stem cell biology and isolate the therapeutic agents required for clinical applications such as cell therapy and regenerative medicine.
The authors thank Mr. Dong-Su Jang, Research Assistant, Department of Anatomy, Yonsei University College of Medicine, Seoul South Korea, for his help with the figures.
| 1. | Bajada S, Mazakova I, Richardson JB, Ashammakhi N. Updates on stem cells and their applications in regenerative medicine. J Tissue Eng Regen Med. 2008;2:169-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 216] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 2. | Nava MM, Raimondi MT, Pietrabissa R. Controlling self-renewal and differentiation of stem cells via mechanical cues. J Biomed Biotechnol. 2012;2012:797410. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 140] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 3. | Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145-1147. [PubMed] |
| 4. | Li D, Zhou J, Chowdhury F, Cheng J, Wang N, Wang F. Role of mechanical factors in fate decisions of stem cells. Regen Med. 2011;6:229-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 141] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 5. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17989] [Cited by in RCA: 18590] [Article Influence: 929.5] [Reference Citation Analysis (1)] |
| 6. | Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917-1920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7589] [Cited by in RCA: 7335] [Article Influence: 386.1] [Reference Citation Analysis (0)] |
| 7. | Yu Z, Li Y, Fan H, Liu Z, Pestell RG. miRNAs regulate stem cell self-renewal and differentiation. Front Genet. 2012;3:191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 8. | Li MA, He L. microRNAs as novel regulators of stem cell pluripotency and somatic cell reprogramming. Bioessays. 2012;34:670-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 9. | Guan D, Zhang W, Zhang W, Liu GH, Belmonte JC. Switching cell fate, ncRNAs coming to play. Cell Death Dis. 2013;4:e464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 10. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [PubMed] |
| 11. | Ambros V. The functions of animal microRNAs. Nature. 2004;431:350-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7919] [Cited by in RCA: 8668] [Article Influence: 394.0] [Reference Citation Analysis (3)] |
| 12. | Porrello ER. microRNAs in cardiac development and regeneration. Clin Sci (Lond). 2013;125:151-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 13. | Merlet E, Atassi F, Motiani RK, Mougenot N, Jacquet A, Nadaud S, Capiod T, Trebak M, Lompré AM, Marchand A. miR-424/322 regulates vascular smooth muscle cell phenotype and neointimal formation in the rat. Cardiovasc Res. 2013;98:458-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 14. | Dweep H, Sticht C, Pandey P, Gretz N. miRWalk--database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44:839-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1257] [Cited by in RCA: 1389] [Article Influence: 92.6] [Reference Citation Analysis (0)] |
| 15. | Shirdel EA, Xie W, Mak TW, Jurisica I. NAViGaTing the micronome--using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS One. 2011;6:e17429. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 178] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 16. | Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8672] [Cited by in RCA: 9045] [Article Influence: 274.1] [Reference Citation Analysis (0)] |
| 17. | Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901-906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3462] [Cited by in RCA: 3438] [Article Influence: 132.2] [Reference Citation Analysis (10)] |
| 18. | Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:12135-12140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 609] [Cited by in RCA: 613] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 19. | Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380-385. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 836] [Cited by in RCA: 775] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 20. | Berardi E, Pues M, Thorrez L, Sampaolesi M. miRNAs in ESC differentiation. Am J Physiol Heart Circ Physiol. 2012;303:H931-H939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 21. | Guo L, Zhao RC, Wu Y. The role of microRNAs in self-renewal and differentiation of mesenchymal stem cells. Exp Hematol. 2011;39:608-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 106] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 22. | Satija NK, Gurudutta GU, Sharma S, Afrin F, Gupta P, Verma YK, Singh VK, Tripathi RP. Mesenchymal stem cells: molecular targets for tissue engineering. Stem Cells Dev. 2007;16:7-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 124] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 23. | Molofsky AV, Pardal R, Morrison SJ. Diverse mechanisms regulate stem cell self-renewal. Curr Opin Cell Biol. 2004;16:700-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 216] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 24. | Wang Y, Blelloch R. Cell cycle regulation by microRNAs in stem cells. Results Probl Cell Differ. 2011;53:459-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 25. | Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 856] [Cited by in RCA: 881] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 26. | Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2459] [Cited by in RCA: 2406] [Article Influence: 104.6] [Reference Citation Analysis (0)] |
| 27. | Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Schöler H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2554] [Cited by in RCA: 2512] [Article Influence: 89.7] [Reference Citation Analysis (0)] |
| 28. | Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2410] [Cited by in RCA: 2344] [Article Influence: 101.9] [Reference Citation Analysis (0)] |
| 29. | Catena R, Tiveron C, Ronchi A, Porta S, Ferri A, Tatangelo L, Cavallaro M, Favaro R, Ottolenghi S, Reinbold R. Conserved POU binding DNA sites in the Sox2 upstream enhancer regulate gene expression in embryonic and neural stem cells. J Biol Chem. 2004;279:41846-41857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 121] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 30. | Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947-956. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3597] [Cited by in RCA: 3416] [Article Influence: 162.7] [Reference Citation Analysis (15)] |
| 31. | Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 740] [Cited by in RCA: 728] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 32. | Barroso-delJesus A, Romero-López C, Lucena-Aguilar G, Melen GJ, Sanchez L, Ligero G, Berzal-Herranz A, Menendez P. Embryonic stem cell-specific miR302-367 cluster: human gene structure and functional characterization of its core promoter. Mol Cell Biol. 2008;28:6609-6619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 167] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 33. | Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521-533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1207] [Cited by in RCA: 1144] [Article Influence: 63.6] [Reference Citation Analysis (0)] |
| 34. | Lin SL, Chang DC, Lin CH, Ying SY, Leu D, Wu DT. Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res. 2011;39:1054-1065. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 255] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 35. | Anokye-Danso F, Snitow M, Morrisey EE. How microRNAs facilitate reprogramming to pluripotency. J Cell Sci. 2012;125:4179-4187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 36. | Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426-6438. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 407] [Cited by in RCA: 402] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 37. | Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478-1483. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 549] [Cited by in RCA: 520] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 38. | Tian Y, Zhang Y, Hurd L, Hannenhalli S, Liu F, Lu MM, Morrisey EE. Regulation of lung endoderm progenitor cell behavior by miR302/367. Development. 2011;138:1235-1245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 75] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 39. | Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 835] [Cited by in RCA: 826] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 40. | Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621-626. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 523] [Cited by in RCA: 542] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 41. | Melton C, Blelloch R. MicroRNA Regulation of Embryonic Stem Cell Self-Renewal and Differentiation. Adv Exp Med Biol. 2010;695:105-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 42. | Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 2009;28:3157-3170. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 145] [Cited by in RCA: 151] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 43. | O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839-843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2140] [Cited by in RCA: 2176] [Article Influence: 103.6] [Reference Citation Analysis (1)] |
| 44. | Smith KN, Singh AM, Dalton S. Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell. 2010;7:343-354. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 148] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 45. | Sengupta S, Nie J, Wagner RJ, Yang C, Stewart R, Thomson JA. MicroRNA 92b controls the G1/S checkpoint gene p57 in human embryonic stem cells. Stem Cells. 2009;27:1524-1528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 93] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 46. | Greene SB, Gunaratne PH, Hammond SM, Rosen JM. A putative role for microRNA-205 in mammary epithelial cell progenitors. J Cell Sci. 2010;123:606-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 117] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 47. | Howard L, Kane NM, Milligan G, Baker AH. MicroRNAs regulating cell pluripotency and vascular differentiation. Vascul Pharmacol. 2011;55:69-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 48. | Heinrich EM, Dimmeler S. MicroRNAs and stem cells: control of pluripotency, reprogramming, and lineage commitment. Circ Res. 2012;110:1014-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 112] [Article Influence: 8.0] [Reference Citation Analysis (1)] |
| 49. | Ohtani K, Dimmeler S. Control of cardiovascular differentiation by microRNAs. Basic Res Cardiol. 2011;106:5-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 88] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 50. | Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280:9330-9335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 388] [Cited by in RCA: 397] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 51. | Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, Hsiao EC, Schwartz RJ, Conklin BR, Bernstein HS. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219-229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 495] [Cited by in RCA: 466] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 52. | Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272-284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1380] [Cited by in RCA: 1368] [Article Influence: 76.0] [Reference Citation Analysis (0)] |
| 53. | Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261-271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1493] [Cited by in RCA: 1494] [Article Influence: 83.0] [Reference Citation Analysis (0)] |
| 54. | Kane NM, Meloni M, Spencer HL, Craig MA, Strehl R, Milligan G, Houslay MD, Mountford JC, Emanueli C, Baker AH. Derivation of endothelial cells from human embryonic stem cells by directed differentiation: analysis of microRNA and angiogenesis in vitro and in vivo. Arterioscler Thromb Vasc Biol. 2010;30:1389-1397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 122] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 55. | Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 597] [Cited by in RCA: 631] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 56. | Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217-1226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 369] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 57. | Fasanaro P, Greco S, Lorenzi M, Pescatori M, Brioschi M, Kulshreshtha R, Banfi C, Stubbs A, Calin GA, Ivan M. An integrated approach for experimental target identification of hypoxia-induced miR-210. J Biol Chem. 2009;284:35134-35143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 239] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 58. | Wang M, Li C, Nie H, Lv X, Qu Y, Yu B, Su L, Li J, Chen X, Ju J. Down-regulated miR-625 suppresses invasion and metastasis of gastric cancer by targeting ILK. FEBS Lett. 2012;586:2382-2388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 59. | Geraldo MV, Yamashita AS, Kimura ET. MicroRNA miR-146b-5p regulates signal transduction of TGF-β by repressing SMAD4 in thyroid cancer. Oncogene. 2012;31:1910-1922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 171] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 60. | Du L, Schageman JJ, Subauste MC, Saber B, Hammond SM, Prudkin L, Wistuba II, Ji L, Roth JA, Minna JD. miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol Cancer Res. 2009;7:1234-1243. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 193] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 61. | Kazenwadel J, Michael MZ, Harvey NL. Prox1 expression is negatively regulated by miR-181 in endothelial cells. Blood. 2010;116:2395-2401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 134] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 62. | Kane NM, Howard L, Descamps B, Meloni M, McClure J, Lu R, McCahill A, Breen C, Mackenzie RM, Delles C. Role of microRNAs 99b, 181a, and 181b in the differentiation of human embryonic stem cells to vascular endothelial cells. Stem Cells. 2012;30:643-654. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 81] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 63. | Yoo JK, Jung HY, Kim CH, Son WS, Kim JK. miR-7641 modulates the expression of CXCL1 during endothelial differentiation derived from human embryonic stem cells. Arch Pharm Res. 2013;36:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 64. | Mohsenin A, Burdick MD, Molina JG, Keane MP, Blackburn MR. Enhanced CXCL1 production and angiogenesis in adenosine-mediated lung disease. FASEB J. 2007;21:1026-1036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 65. | Raghuwanshi SK, Su Y, Singh V, Haynes K, Richmond A, Richardson RM. The chemokine receptors CXCR1 and CXCR2 couple to distinct G protein-coupled receptor kinases to mediate and regulate leukocyte functions. J Immunol. 2012;189:2824-2832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 131] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 66. | Boettger T, Beetz N, Kostin S, Schneider J, Krüger M, Hein L, Braun T. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119:2634-2647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 555] [Cited by in RCA: 560] [Article Influence: 32.9] [Reference Citation Analysis (8)] |
| 67. | Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705-710. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1310] [Cited by in RCA: 1328] [Article Influence: 78.1] [Reference Citation Analysis (0)] |
| 68. | Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, Richardson JA, Bassel-Duby R, Olson EN. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166-2178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 530] [Cited by in RCA: 547] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 69. | Yamaguchi S, Yamahara K, Homma K, Suzuki S, Fujii S, Morizane R, Monkawa T, Matsuzaki Y, Kangawa K, Itoh H. The role of microRNA-145 in human embryonic stem cell differentiation into vascular cells. Atherosclerosis. 2011;219:468-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 52] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 70. | Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647-658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 837] [Cited by in RCA: 885] [Article Influence: 52.1] [Reference Citation Analysis (0)] |
| 71. | Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MV, Peterson KL, Indolfi C, Catalucci D, Chen J. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590-1598. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 439] [Cited by in RCA: 467] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 72. | Huang H, Xie C, Sun X, Ritchie RP, Zhang J, Chen YE. miR-10a contributes to retinoid acid-induced smooth muscle cell differentiation. J Biol Chem. 2010;285:9383-9389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 106] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 73. | Xie C, Huang H, Sun X, Guo Y, Hamblin M, Ritchie RP, Garcia-Barrio MT, Zhang J, Chen YE. MicroRNA-1 regulates smooth muscle cell differentiation by repressing Kruppel-like factor 4. Stem Cells Dev. 2011;20:205-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 129] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 74. | Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2026] [Cited by in RCA: 2197] [Article Influence: 104.6] [Reference Citation Analysis (0)] |
| 75. | Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci USA. 2005;102:18986-18991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 321] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 76. | Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1078] [Cited by in RCA: 1084] [Article Influence: 57.1] [Reference Citation Analysis (0)] |
| 77. | Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1242] [Cited by in RCA: 1251] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 78. | Wilson KD, Hu S, Venkatasubrahmanyam S, Fu JD, Sun N, Abilez OJ, Baugh JJ, Jia F, Ghosh Z, Li RA. Dynamic microRNA expression programs during cardiac differentiation of human embryonic stem cells: role for miR-499. Circ Cardiovasc Genet. 2010;3:426-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 150] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 79. | Sluijter JP, van Mil A, van Vliet P, Metz CH, Liu J, Doevendans PA, Goumans MJ. MicroRNA-1 and -499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler Thromb Vasc Biol. 2010;30:859-868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 262] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 80. | Glass C, Singla DK. MicroRNA-1 transfected embryonic stem cells enhance cardiac myocyte differentiation and inhibit apoptosis by modulating the PTEN/Akt pathway in the infarcted heart. Am J Physiol Heart Circ Physiol. 2011;301:H2038-H2049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 94] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 81. | Fu JD, Rushing SN, Lieu DK, Chan CW, Kong CW, Geng L, Wilson KD, Chiamvimonvat N, Boheler KR, Wu JC. Distinct roles of microRNA-1 and -499 in ventricular specification and functional maturation of human embryonic stem cell-derived cardiomyocytes. PLoS One. 2011;6:e27417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 145] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 82. | Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769-773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3585] [Cited by in RCA: 3759] [Article Influence: 179.0] [Reference Citation Analysis (3)] |
| 83. | Huang F, Tang L, Fang ZF, Hu XQ, Pan JY, Zhou SH. miR-1-mediated induction of cardiogenesis in mesenchymal stem cells via downregulation of Hes-1. Biomed Res Int. 2013;2013:216286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 84. | Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242-3254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 617] [Cited by in RCA: 635] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 85. | Takaya T, Ono K, Kawamura T, Takanabe R, Kaichi S, Morimoto T, Wada H, Kita T, Shimatsu A, Hasegawa K. MicroRNA-1 and MicroRNA-133 in spontaneous myocardial differentiation of mouse embryonic stem cells. Circ J. 2009;73:1492-1497. [PubMed] |
| 86. | Tao J, Wu D, Xu B, Qian W, Li P, Lu Q, Yin C, Zhang W. microRNA-133 inhibits cell proliferation, migration and invasion in prostate cancer cells by targeting the epidermal growth factor receptor. Oncol Rep. 2012;27:1967-1975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 68] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 87. | Lee SY, Ham O, Cha MJ, Song BW, Choi E, Kim IK, Chang W, Lim S, Lee CY, Park JH. The promotion of cardiogenic differentiation of hMSCs by targeting epidermal growth factor receptor using microRNA-133a. Biomaterials. 2013;34:92-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 88. | van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1265] [Cited by in RCA: 1260] [Article Influence: 66.3] [Reference Citation Analysis (0)] |
| 89. | Hosoda T, Zheng H, Cabral-da-Silva M, Sanada F, Ide-Iwata N, Ogórek B, Ferreira-Martins J, Arranto C, D’Amario D, del Monte F. Human cardiac stem cell differentiation is regulated by a mircrine mechanism. Circulation. 2011;123:1287-1296. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 154] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 90. | Zhang LL, Liu JJ, Liu F, Liu WH, Wang YS, Zhu B, Yu B. MiR-499 induces cardiac differentiation of rat mesenchymal stem cells through wnt/β-catenin signaling pathway. Biochem Biophys Res Commun. 2012;420:875-881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 91. | Xiao J, Liang D, Zhang H, Liu Y, Zhang D, Liu Y, Pan L, Chen X, Doevendans PA, Sun Y. MicroRNA-204 is required for differentiation of human-derived cardiomyocyte progenitor cells. J Mol Cell Cardiol. 2012;53:751-759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 92. | Cai B, Li J, Wang J, Luo X, Ai J, Liu Y, Wang N, Liang H, Zhang M, Chen N. microRNA-124 regulates cardiomyocyte differentiation of bone marrow-derived mesenchymal stem cells via targeting STAT3 signaling. Stem Cells. 2012;30:1746-1755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 77] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 93. | Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875-886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1370] [Cited by in RCA: 1319] [Article Influence: 73.3] [Reference Citation Analysis (1)] |
| 94. | Ji F, Lv X, Jiao J. The role of microRNAs in neural stem cells and neurogenesis. J Genet Genomics. 2013;40:61-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 95. | Atlasi Y, Noori R, Gaspar C, Franken P, Sacchetti A, Rafati H, Mahmoudi T, Decraene C, Calin GA, Merrill BJ. Wnt signaling regulates the lineage differentiation potential of mouse embryonic stem cells through Tcf3 down-regulation. PLoS Genet. 2013;9:e1003424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 67] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 96. | Brill MS, Ninkovic J, Winpenny E, Hodge RD, Ozen I, Yang R, Lepier A, Gascón S, Erdelyi F, Szabo G. Adult generation of glutamatergic olfactory bulb interneurons. Nat Neurosci. 2009;12:1524-1533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 249] [Cited by in RCA: 274] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 97. | Zou Y, Niu W, Qin S, Downes M, Burns DK, Zhang CL. The nuclear receptor TLX is required for gliomagenesis within the adult neurogenic niche. Mol Cell Biol. 2012;32:4811-4820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 98. | Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, Santistevan NJ, Li W, Zhao X, Jin P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189:127-141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 409] [Cited by in RCA: 404] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 99. | Liu C, Teng ZQ, Santistevan NJ, Szulwach KE, Guo W, Jin P, Zhao X. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell. 2010;6:433-444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 265] [Cited by in RCA: 262] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 100. | Kawahara H, Imai T, Okano H. MicroRNAs in Neural Stem Cells and Neurogenesis. Front Neurosci. 2012;6:30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 87] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 101. | Lang MF, Shi Y. Dynamic Roles of microRNAs in Neurogenesis. Front Neurosci. 2012;6:71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 102. | Papagiannakopoulos T, Kosik KS. MicroRNA-124: micromanager of neurogenesis. Cell Stem Cell. 2009;4:375-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 62] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 103. | Åkerblom M, Sachdeva R, Barde I, Verp S, Gentner B, Trono D, Jakobsson J. MicroRNA-124 is a subventricular zone neuronal fate determinant. J Neurosci. 2012;32:8879-8889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 166] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 104. | Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 277] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 105. | Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16:365-371. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 487] [Cited by in RCA: 462] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 106. | Zhao C, Sun G, Ye P, Li S, Shi Y. MicroRNA let-7d regulates the TLX/microRNA-9 cascade to control neural cell fate and neurogenesis. Sci Rep. 2013;3:1329. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 89] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 107. | Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 2009;136:913-925. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 349] [Cited by in RCA: 335] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 108. | Sun G, Ye P, Murai K, Lang MF, Li S, Zhang H, Li W, Fu C, Yin J, Wang A. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat Commun. 2011;2:529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 252] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 109. | Brett JO, Renault VM, Rafalski VA, Webb AE, Brunet A. The microRNA cluster miR-106b~25 regulates adult neural stem/progenitor cell proliferation and neuronal differentiation. Aging (Albany NY). 2011;3:108-124. [PubMed] |
| 110. | Fineberg SK, Datta P, Stein CS, Davidson BL. MiR-34a represses Numbl in murine neural progenitor cells and antagonizes neuronal differentiation. PLoS One. 2012;7:e38562. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 111. | Dill H, Linder B, Fehr A, Fischer U. Intronic miR-26b controls neuronal differentiation by repressing its host transcript, ctdsp2. Genes Dev. 2012;26:25-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 113] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 112. | Han J, Denli AM, Gage FH. The enemy within: intronic miR-26b represses its host gene, ctdsp2, to regulate neurogenesis. Genes Dev. 2012;26:6-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 113. | Cui Y, Xiao Z, Han J, Sun J, Ding W, Zhao Y, Chen B, Li X, Dai J. MiR-125b orchestrates cell proliferation, differentiation and migration in neural stem/progenitor cells by targeting Nestin. BMC Neurosci. 2012;13:116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 114. | Dong S, Yang B, Guo H, Kang F. MicroRNAs regulate osteogenesis and chondrogenesis. Biochem Biophys Res Commun. 2012;418:587-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 102] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 115. | Hong E, Reddi AH. MicroRNAs in chondrogenesis, articular cartilage, and osteoarthritis: implications for tissue engineering. Tissue Eng Part B Rev. 2012;18:445-453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 116. | van Wijnen AJ, van de Peppel J, van Leeuwen JP, Lian JB, Stein GS, Westendorf JJ, Oursler MJ, Im HJ, Taipaleenmäki H, Hesse E. MicroRNA functions in osteogenesis and dysfunctions in osteoporosis. Curr Osteoporos Rep. 2013;11:72-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 181] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 117. | Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, Zhang Y. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol. 2012;8:212-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 467] [Cited by in RCA: 477] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 118. | Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284:15676-15684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 437] [Cited by in RCA: 475] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 119. | Kapinas K, Kessler CB, Delany AM. miR-29 suppression of osteonectin in osteoblasts: regulation during differentiation and by canonical Wnt signaling. J Cell Biochem. 2009;108:216-224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 206] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 120. | Zhou H, Mak W, Zheng Y, Dunstan CR, Seibel MJ. Osteoblasts directly control lineage commitment of mesenchymal progenitor cells through Wnt signaling. J Biol Chem. 2008;283:1936-1945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 109] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 121. | Kapinas K, Kessler C, Ricks T, Gronowicz G, Delany AM. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem. 2010;285:25221-25231. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 324] [Cited by in RCA: 326] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 122. | Wang T, Xu Z. miR-27 promotes osteoblast differentiation by modulating Wnt signaling. Biochem Biophys Res Commun. 2010;402:186-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 135] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 123. | Zhang J, Tu Q, Bonewald LF, He X, Stein G, Lian J, Chen J. Effects of miR-335-5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J Bone Miner Res. 2011;26:1953-1963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 239] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 124. | Sugatani T, Hruska KA. MicroRNA-223 is a key factor in osteoclast differentiation. J Cell Biochem. 2007;101:996-999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 155] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 125. | Sugatani T, Hruska KA. Impaired micro-RNA pathways diminish osteoclast differentiation and function. J Biol Chem. 2009;284:4667-4678. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 177] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 126. | O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604-1609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1365] [Cited by in RCA: 1507] [Article Influence: 79.3] [Reference Citation Analysis (0)] |
| 127. | Mann M, Barad O, Agami R, Geiger B, Hornstein E. miRNA-based mechanism for the commitment of multipotent progenitors to a single cellular fate. Proc Natl Acad Sci USA. 2010;107:15804-15809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 90] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 128. | Kobayashi T, Lu J, Cobb BS, Rodda SJ, McMahon AP, Schipani E, Merkenschlager M, Kronenberg HM. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci USA. 2008;105:1949-1954. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 289] [Cited by in RCA: 290] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 129. | Miyaki S, Sato T, Inoue A, Otsuki S, Ito Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, Yamashita S. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24:1173-1185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 492] [Cited by in RCA: 487] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 130. | Eberhart JK, He X, Swartz ME, Yan YL, Song H, Boling TC, Kunerth AK, Walker MB, Kimmel CB, Postlethwait JH. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet. 2008;40:290-298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 280] [Cited by in RCA: 275] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 131. | Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 589] [Cited by in RCA: 616] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 132. | Nakamura Y, Inloes JB, Katagiri T, Kobayashi T. Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling. Mol Cell Biol. 2011;31:3019-3028. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 145] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 133. | Dudek KA, Lafont JE, Martinez-Sanchez A, Murphy CL. Type II collagen expression is regulated by tissue-specific miR-675 in human articular chondrocytes. J Biol Chem. 2010;285:24381-24387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 120] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 134. | Ham O, Song BW, Lee SY, Choi E, Cha MJ, Lee CY, Park JH, Kim IK, Chang W, Lim S. The role of microRNA-23b in the differentiation of MSC into chondrocyte by targeting protein kinase A signaling. Biomaterials. 2012;33:4500-4507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 92] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 135. | Ohgawara T, Kubota S, Kawaki H, Kondo S, Eguchi T, Kurio N, Aoyama E, Sasaki A, Takigawa M. Regulation of chondrocytic phenotype by micro RNA 18a: involvement of Ccn2/Ctgf as a major target gene. FEBS Lett. 2009;583:1006-1010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 67] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 136. | Lin EA, Kong L, Bai XH, Luan Y, Liu CJ. miR-199a, a bone morphogenic protein 2-responsive MicroRNA, regulates chondrogenesis via direct targeting to Smad1. J Biol Chem. 2009;284:11326-11335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 202] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 137. | Yang B, Guo H, Zhang Y, Chen L, Ying D, Dong S. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS One. 2011;6:e21679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 190] [Cited by in RCA: 213] [Article Influence: 14.2] [Reference Citation Analysis (11)] |
| 138. | Martinez-Sanchez A, Dudek KA, Murphy CL. Regulation of human chondrocyte function through direct inhibition of cartilage master regulator SOX9 by microRNA-145 (miRNA-145). J Biol Chem. 2012;287:916-924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 156] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 139. | Kim D, Song J, Jin EJ. MicroRNA-221 regulates chondrogenic differentiation through promoting proteosomal degradation of slug by targeting Mdm2. J Biol Chem. 2010;285:26900-26907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 77] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 140. | Cui L, Zhou X, Li J, Wang L, Wang J, Li Q, Chu J, Zheng L, Wu Q, Han Z. Dynamic microRNA profiles of hepatic differentiated human umbilical cord lining-derived mesenchymal stem cells. PLoS One. 2012;7:e44737. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 141. | Wang XR, Zhang XM, Du J, Jiang H. MicroRNA-182 regulates otocyst-derived cell differentiation and targets T-box1 gene. Hear Res. 2012;286:55-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 142. | Yang Y, Ding L, An Y, Zhang ZW, Lang Y, Tai S, Guo F, Teng CB. MiR-18a regulates expression of the pancreatic transcription factor Ptf1a in pancreatic progenitor and acinar cells. FEBS Lett. 2012;586:422-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 143. | Knelangen JM, van der Hoek MB, Kong WC, Owens JA, Fischer B, Santos AN. MicroRNA expression profile during adipogenic differentiation in mouse embryonic stem cells. Physiol Genomics. 2011;43:611-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 144. | Kong KY, Owens KS, Rogers JH, Mullenix J, Velu CS, Grimes HL, Dahl R. MIR-23A microRNA cluster inhibits B-cell development. Exp Hematol. 2010;38:629-640.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 86] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 145. | Bouhallier F, Allioli N, Lavial F, Chalmel F, Perrard MH, Durand P, Samarut J, Pain B, Rouault JP. Role of miR-34c microRNA in the late steps of spermatogenesis. RNA. 2010;16:720-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 230] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 146. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14327] [Cited by in RCA: 14570] [Article Influence: 809.4] [Reference Citation Analysis (0)] |
| 147. | Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet. 2013;14:427-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 336] [Cited by in RCA: 359] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 148. | Bao X, Zhu X, Liao B, Benda C, Zhuang Q, Pei D, Qin B, Esteban MA. MicroRNAs in somatic cell reprogramming. Curr Opin Cell Biol. 2013;25:208-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 149. | Lamouille S, Subramanyam D, Blelloch R, Derynck R. Regulation of epithelial-mesenchymal and mesenchymal-epithelial transitions by microRNAs. Curr Opin Cell Biol. 2013;25:200-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 226] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 150. | Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459-461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 593] [Cited by in RCA: 536] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 151. | Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29:443-448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 505] [Cited by in RCA: 473] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 152. | Li Z, Yang CS, Nakashima K, Rana TM. Small RNA-mediated regulation of iPS cell generation. EMBO J. 2011;30:823-834. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 252] [Cited by in RCA: 244] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 153. | Liao B, Bao X, Liu L, Feng S, Zovoilis A, Liu W, Xue Y, Cai J, Guo X, Qin B. MicroRNA cluster 302-367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J Biol Chem. 2011;286:17359-17364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 211] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 154. | Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 792] [Cited by in RCA: 808] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 155. | Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376-388. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 978] [Cited by in RCA: 903] [Article Influence: 60.2] [Reference Citation Analysis (0)] |
| 156. | Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 565] [Cited by in RCA: 557] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 157. | Choi YJ, Lin CP, Ho JJ, He X, Okada N, Bu P, Zhong Y, Kim SY, Bennett MJ, Chen C. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol. 2011;13:1353-1360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 299] [Cited by in RCA: 329] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 158. | Ye D, Wang G, Liu Y, Huang W, Wu M, Zhu S, Jia W, Deng AM, Liu H, Kang J. MiR-138 promotes induced pluripotent stem cell generation through the regulation of the p53 signaling. Stem Cells. 2012;30:1645-1654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 75] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 159. | Yang CS, Li Z, Rana TM. microRNAs modulate iPS cell generation. RNA. 2011;17:1451-1460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 92] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 160. | Zardo G, Ciolfi A, Vian L, Billi M, Racanicchi S, Grignani F, Nervi C. Transcriptional targeting by microRNA-polycomb complexes: a novel route in cell fate determination. Cell Cycle. 2012;11:3543-3549. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 161. | Guo X, Liu Q, Wang G, Zhu S, Gao L, Hong W, Chen Y, Wu M, Liu H, Jiang C. microRNA-29b is a novel mediator of Sox2 function in the regulation of somatic cell reprogramming. Cell Res. 2013;23:142-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 162. | Ding S, Schultz PG. Small molecules and future regenerative medicine. Curr Top Med Chem. 2005;5:383-395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 37] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 163. | Li W, Jiang K, Ding S. Concise review: A chemical approach to control cell fate and function. Stem Cells. 2012;30:61-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 76] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 164. | Ding S, Schultz PG. A role for chemistry in stem cell biology. Nat Biotechnol. 2004;22:833-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 227] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 165. | Buehr M, Meek S, Blair K, Yang J, Ure J, Silva J, McLay R, Hall J, Ying QL, Smith A. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287-1298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 654] [Cited by in RCA: 589] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 166. | Chen G, Hou Z, Gulbranson DR, Thomson JA. Actin-myosin contractility is responsible for the reduced viability of dissociated human embryonic stem cells. Cell Stem Cell. 2010;7:240-248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 244] [Cited by in RCA: 234] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 167. | Ohgushi M, Matsumura M, Eiraku M, Murakami K, Aramaki T, Nishiyama A, Muguruma K, Nakano T, Suga H, Ueno M. Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell. 2010;7:225-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 344] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 168. | Xu Y, Zhu X, Hahm HS, Wei W, Hao E, Hayek A, Ding S. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc Natl Acad Sci USA. 2010;107:8129-8134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 282] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 169. | Tsutsui H, Valamehr B, Hindoyan A, Qiao R, Ding X, Guo S, Witte ON, Liu X, Ho CM, Wu H. An optimized small molecule inhibitor cocktail supports long-term maintenance of human embryonic stem cells. Nat Commun. 2011;2:167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 133] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 170. | Ni TT, Rellinger EJ, Mukherjee A, Xie S, Stephens L, Thorne CA, Kim K, Hu J, Lee E, Marnett L. Discovering small molecules that promote cardiomyocyte generation by modulating Wnt signaling. Chem Biol. 2011;18:1658-1668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 47] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 171. | James D, Nam HS, Seandel M, Nolan D, Janovitz T, Tomishima M, Studer L, Lee G, Lyden D, Benezra R. Expansion and maintenance of human embryonic stem cell-derived endothelial cells by TGFbeta inhibition is Id1 dependent. Nat Biotechnol. 2010;28:161-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 253] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 172. | Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275-280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3036] [Cited by in RCA: 2794] [Article Influence: 164.4] [Reference Citation Analysis (0)] |
| 173. | Zhang Y, Li W, Laurent T, Ding S. Small molecules, big roles -- the chemical manipulation of stem cell fate and somatic cell reprogramming. J Cell Sci. 2012;125:5609-5620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 126] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 174. | Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269-1275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 994] [Cited by in RCA: 954] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 175. | Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7:651-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 506] [Cited by in RCA: 520] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 176. | Shi Y, Desponts C, Do JT, Hahm HS, Schöler HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 666] [Cited by in RCA: 643] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 177. | Shi Y, Do JT, Desponts C, Hahm HS, Schöler HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525-528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 544] [Cited by in RCA: 502] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 178. | Lee J, Xia Y, Son MY, Jin G, Seol B, Kim MJ, Son MJ, Do M, Lee M, Kim D. A novel small molecule facilitates the reprogramming of human somatic cells into a pluripotent state and supports the maintenance of an undifferentiated state of human pluripotent stem cells. Angew Chem Int Ed Engl. 2012;51:12509-12513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 179. | Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 632] [Cited by in RCA: 630] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 180. | Van Zant G, Liang Y. Concise review: hematopoietic stem cell aging, life span, and transplantation. Stem Cells Transl Med. 2012;1:651-657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 181. | Young JC, Wu S, Hansteen G, Du C, Sambucetti L, Remiszewski S, O’Farrell AM, Hill B, Lavau C, Murray LJ. Inhibitors of histone deacetylases promote hematopoietic stem cell self-renewal. Cytotherapy. 2004;6:328-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 50] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 182. | Boitano AE, Wang J, Romeo R, Bouchez LC, Parker AE, Sutton SE, Walker JR, Flaveny CA, Perdew GH, Denison MS. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329:1345-1348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 884] [Cited by in RCA: 847] [Article Influence: 52.9] [Reference Citation Analysis (0)] |
| 183. | North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, Weber GJ, Bowman TV, Jang IH, Grosser T. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007-1011. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 962] [Cited by in RCA: 915] [Article Influence: 48.2] [Reference Citation Analysis (0)] |
| 184. | Johnson K, Zhu S, Tremblay MS, Payette JN, Wang J, Bouchez LC, Meeusen S, Althage A, Cho CY, Wu X. A stem cell-based approach to cartilage repair. Science. 2012;336:717-721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 463] [Cited by in RCA: 605] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
| 185. | Faghihi F, Baghaban Eslaminejad M, Nekookar A, Najar M, Salekdeh GH. The effect of purmorphamine and sirolimus on osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. Biomed Pharmacother. 2013;67:31-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 186. | Kim JM, Choi JS, Kim YH, Jin SH, Lim S, Jang HJ, Kim KT, Ryu SH, Suh PG. An activator of the cAMP/PKA/CREB pathway promotes osteogenesis from human mesenchymal stem cells. J Cell Physiol. 2013;228:617-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 187. | Ouyang J, Shao J, Zou H, Lou Y, Yu Y. Hepatic differentiation of rat mesenchymal stem cells by a small molecule. ChemMedChem. 2012;7:1447-1452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 188. | Chang W, Lim S, Song BW, Lee CY, Park MS, Chung YA, Yoon C, Lee SY, Ham O, Park JH. Phorbol myristate acetate differentiates human adipose-derived mesenchymal stem cells into functional cardiogenic cells. Biochem Biophys Res Commun. 2012;424:740-746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 189. | Song H, Hwang HJ, Chang W, Song BW, Cha MJ, Kim IK, Lim S, Choi EJ, Ham O, Lee CY. Cardiomyocytes from phorbol myristate acetate-activated mesenchymal stem cells restore electromechanical function in infarcted rat hearts. Proc Natl Acad Sci USA. 2011;108:296-301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 190. | Yang HJ, Xia YY, Wang L, Liu R, Goh KJ, Ju PJ, Feng ZW. A novel role for neural cell adhesion molecule in modulating insulin signaling and adipocyte differentiation of mouse mesenchymal stem cells. J Cell Sci. 2011;124:2552-2560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 191. | Qin J, Li WQ, Zhang L, Chen F, Liang WH, Mao FF, Zhang XM, Lahn BT, Yu WH, Xiang AP. A stem cell-based tool for small molecule screening in adipogenesis. PLoS One. 2010;5:e13014. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 192. | Alexanian AR, Liu QS, Zhang Z. Enhancing the efficiency of direct reprogramming of human mesenchymal stem cells into mature neuronal-like cells with the combination of small molecule modulators of chromatin modifying enzymes, SMAD signaling and cyclic adenosine monophosphate levels. Int J Biochem Cell Biol. 2013;45:1633-1638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 193. | Wang Y, He W, Bian H, Liu C, Li S. Small molecule induction of neural-like cells from bone marrow-mesenchymal stem cells. J Cell Biochem. 2012;113:1527-1536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 194. | Kim NR, Kang SK, Ahn HH, Kwon SW, Park WS, Kim KS, Kim SS, Jung HJ, Choi SU, Ahn JH. Discovery of a new and efficient small molecule for neuronal differentiation from mesenchymal stem cell. J Med Chem. 2009;52:7931-7933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 195. | Saraswati S, Deskins DL, Holt GE, Young PP. Pyrvinium, a potent small molecule Wnt inhibitor, increases engraftment and inhibits lineage commitment of mesenchymal stem cells (MSCs). Wound Repair Regen. 2012;20:185-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 196. | Gwak J, Hwang SG, Park HS, Choi SR, Park SH, Kim H, Ha NC, Bae SJ, Han JK, Kim DE. Small molecule-based disruption of the Axin/β-catenin protein complex regulates mesenchymal stem cell differentiation. Cell Res. 2012;22:237-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 118] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 197. | Gumireddy K, Young DD, Xiong X, Hogenesch JB, Huang Q, Deiters A. Small-molecule inhibitors of microrna miR-21 function. Angew Chem Int Ed Engl. 2008;47:7482-7484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 345] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 198. | Young DD, Connelly CM, Grohmann C, Deiters A. Small molecule modifiers of microRNA miR-122 function for the treatment of hepatitis C virus infection and hepatocellular carcinoma. J Am Chem Soc. 2010;132:7976-7981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 211] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 199. | Connelly CM, Thomas M, Deiters A. High-throughput luciferase reporter assay for small-molecule inhibitors of microRNA function. J Biomol Screen. 2012;17:822-828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 200. | 100 Deiters A. Small molecule modifiers of the microRNA and RNA interference pathway. AAPS J. 2010;12:51-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 74] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 201. | Liu X, Wang S, Meng F, Wang J, Zhang Y, Dai E, Yu X, Li X, Jiang W. SM2miR: a database of the experimentally validated small molecules’ effects on microRNA expression. Bioinformatics. 2013;29:409-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 172] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 202. | Jiang W, Chen X, Liao M, Li W, Lian B, Wang L, Meng F, Liu X, Chen X, Jin Y. Identification of links between small molecules and miRNAs in human cancers based on transcriptional responses. Sci Rep. 2012;2:282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
P- Reviewers Guo ZS, Yarema KJ S- Editor Song XX L- Editor A E- Editor Wu HL