Published online Aug 26, 2012. doi: 10.4252/wjsc.v4.i8.87
Revised: March 12, 2012
Accepted: March 18, 2012
Published online: August 26, 2012
AIM: To examine the imprinted Dlk1-Dio3 locus in pluripotent embryonic stem (ES) cell/fibroblast hybrid cells.
METHODS: Gtl2, Rian, and Mirg mRNA expression in mouse pluripotent ES cell/fibroblast hybrid cells was examined by real-time reverse transcription-polymerase chain reaction. Pyrosequencing and bisulfate sequencing were used to determine the DNA methylation level of the Dlk1-Dio3 locus imprinting control region.
RESULTS: The selected hybrid clones had a near-tetraploid karyotype and were highly pluripotent judging from their capacity to generate chimeric embryos and adult chimeras. Our data clearly demonstrate that Gtl2, Rian, and Mirg, which are imprinted genes within the Dlk1-Dio3 locus, are active in all examined ES cell/fibroblast hybrid clones. In spite of interclonal variability, the expression of the imprinted genes is comparable to that of ES cells and fibroblasts. Quantitative analysis of the DNA methylation status of the intergenic differentially methylated region (IG DMR) within the Dlk1-Dio3 locus by pyrosequencing and bisulfite sequencing clearly showed that the DNA methylation status of the imprinted region in the tested hybrid clones was comparable to that of both ES cells and fibroblasts.
CONCLUSION: Reprogramming process in a hybrid cell system is achieved without marked alteration of the imprinted Dlk1-Dio3 locus.
-
Citation: Battulin NR, Khabarova AA, Boyarskikh UA, Menzorov AG, Filipenko ML, Serov OL. Reprogramming somatic cells by fusion with embryonic stem cells does not cause silencing of the
Dlk1-Dio3 region in mice. World J Stem Cells 2012; 4(8): 87-93 - URL: https://www.wjgnet.com/1948-0210/full/v4/i8/87.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v4.i8.87
Cell fusion is one approach that has been used to demonstrate nuclear reprogramming of somatic cells to a pluripotent-like state. In fact, embryonic stem (ES) hybrid cells obtained by the fusion of ES cells with various somatic cell types have characteristics similar to ES cells[1-5]. The great potential of ES cell/somatic cell hybrids was confirmed by the generation of chimeric embryos[6,7] and chimeric adults[1,8,9]. In addition, reprogramming in ES cell/somatic hybrid cells occurs rapidly, generally within 5-10 d[5,10]. Such remarkable reprogramming effects could be explained by the presence of numerous reprogramming factors in ES cells compared to the limited numbers usually used in the generation of induced pluripotent stem (iPS) cells[5,11].
iPS cell derivation by forced expression of a few reprogramming factors is now considered to be a promising method of reprogramming[11-17]. Recently, several groups, using a number of different parameters, have shown that iPS cells differ from ES cells, which are considered to be the “gold standard” of pluripotency[11,18,19]. In other words, several errors can occur during the generation of iPS cells. One such “reprogramming error” is aberrant silencing of the imprinted Dlk1-Dio3 locus located on mouse chromosome 12 caused by DNA hypermethylation of key imprinting control regions[20,21]. Dysregulation of this locus leads to altered gene expression that drastically limits developmental capacity so that such iPS cells after their injection into tetraploid blastocysts do result in the birth of “all iPS-cell derived” mice but rather generate chimeras with a low contribution of the tested cells. This phenomenon was observed in 95% of mouse iPS cell lines[21]. It should be noted that methylation level of CpG-sites in the imprinted Dlk1-Dio3 locus is usually about 50% in both somatic and ES cells.
We have previously established ten stable hybrid clones, three of which are ES cell/embryonic fibroblast cell type and seven that are ES cell/adult fibroblast cell type[9]. Based on cytogenetic analysis, four of ten clones in which more than 80% of cells contained 76-80 chromosomes were selected; in other words, the hybrid cells had a near-tetraploid chromosome set. Injection of the GFP-labeled hybrid cells into blastocysts demonstrated that all four hybrid clones were able to give rise to chimeric embryos with a high contribution of GFP-labeled hybrid cell descendents. Furthermore, three clones resulted in the birth of about two dozen adult chimeras[9]. Taken together, the selected hybrid clones had highly pluripotency comparable with parental ES cells. It is important to note that cytogenetic and microsatellite analyses have demonstrated that the initial near-tetraploid karyotype of the hybrid cells remained stable during the development of the chimeras[9].
This study examined the imprinted Dlk1-Dio3 locus in pluripotent ES cell/fibroblast hybrid clones. The aim was to determine whether alterations of the Dlk1-Dio3 locus observed in iPS cells are common in other reprogramming systems, particularly cell fusion, or whether the alternations are caused by the lack of some reprogramming factors used in generating iPS cells.
We used the following cell lines in this study: (1) the murine ES cell line E14Tg2aSc4TP6.3 (tauGFP)[22], in which the hypoxanthine phosphoribosyl transferase gene has been deleted, the pTP6 transgene contains a tau-tagged green fluorescent protein (GFP) and the puromycin resistance (Puro) gene. Culture conditions of this ES cell line were described previously in detail[9]; (2) the MA01 ES cell line, prepared from a blastocyst derived from the 129/Ola x BALB mouse using a previously published protocol[23]. MA01 cells had morphology and growth characteristics typical of ES cells, were positive for Oct4 and Nanog expression, and had a diploid karyotype without visible chromosomal rearrangements. MA01 cells had undergone five to eleven passages; (3) mouse embryonic fibroblast (MEF) cultures from 13.5 dpc embryos derived from DD/c mice, prepared and cultures as described previously[9]; and (4) a set of hybrid cell clones: tef4, taf2, taf5, and tef9 produced by fusing diploid tauGFP ES cells and diploid embryonic (series tef) or adult (series taf) fibroblasts. As mentioned above, these hybrid clones had near-tetraploid chromosome complements that indicated on absence of marked segregation of parental chromosomes. The ES cells and hybrid clones were cultured without feeder in Glasgow Modified Eagle’s Medium MEM (Gibco/BRL-Life Technologies, UK) containing 10% fetal bovine serum (FBS; PAA, Austria), 1% non-essential amino acids (Gibco/BRL-Life Technologies), 10-4 mol/L β-mercaptoethanol, 100 μg/mL penicillin and streptomycin, and 103 U/mL murine leukemia inhibitory factor (mLIF; Chemicon, UK). The cells were cultured in plastic dishes coated with 0.1% gelatin (Fluka, Germany) at 37 °C in a 5% CO2 atmosphere with high humidity. The medium was changed every 2-3 d. Preparation of the hybrid cell clones was previously described[9].
To distinguish the parental chromosomes in hybrid cells, a set of microsatellite markers that allow unambiguous marking of each parental chromosome was used. Primers and polymerase chain reaction (PCR) conditions for the microsatellites were previously described[9].
Genomic DNA extraction and bisulfite mutagenesis sequencing analysis were conducted using the EZ DNA Methylation-Direct kit (Zymo Research, Orange, CA, USA), as described previously[24]. The genomic DNA was eluted with 10 μL of elution buffer and used for two successive rounds of PCR with nested primer pairs (inner and outer), which were specific for the bottom strand of the mutagenized DNA (first-round primer set: IGDMR_out_for 5'-AAGGTATATTATGTTAGTGTTAGGAAGGATTGTGA-3' and IGDMR_out_rev 5'-CAAAACATTCTCCATTAACAAAATAATACAACCCT-3'; second-round primer set: IGDMR_in_for 5'-TGTGGTTTGTTATGGGTAAGTTTTATGGTTTATTG-3' and IGDMR_in_rev 5'-AATACAACCCTTCCCTCACTCCAAAAATTAAAAAA-3'). The conditions for the first round of PCR were as follows: initial denaturation at 94 °C for 3 min, followed by 30 cycles of 94 °C for 15 s, 58 °C for 15 s, 72 °C for 30 s, and a final extension at 72 °C for 3 min. The first round PCR product was purified using an affinity column to remove the outer primers. The PCR products were eluted in 10 μL of water and used for the second PCR round: initial denaturation at 94 °C for 3 min, 30 cycles of 94 °C for 15 s, 58 °C for 15 s, and 72 °C for 20 s, and a final extension at 72 °C for 3 min. All PCRs were performed with Taq DNA polymerase. The PCR products were run on a 2% agarose gel to verify the amplification of specific bands, which were then excised from the gel and purified with the MinElute Gel Extraction Kit (Qiagen, Hilden, Germany). The purified PCR products were subcloned using the pGEMT Easy kit (Promega, Madison, WI, USA), and then individual clones were sequenced. Sequencing of individually cloned products was performed using the M13 reverse and forward primers. Clones were accepted only with ≥ 90% cytosine conversion. Resultant sequences were aligned to the DNA sequences of the selected loci of the gene of interest using the online quantification tool for methylation analysis (http://quma.cdb.riken.jp/top/index.html).
Amplicons for pyrosequencing reactions were generated in a 50 μl reaction volume with 300 nmol/L forward and reverse PCR primers (5'-GTTATGGATTGGTGTTAAGGT-3', biotin-5'-TACAACCCTTCCCTCACTC-3'), 10 mmol/L Tris-HCl (рН 8.9), 55 mmol/L KCl, 2.5 mmol/L MgCl2, 0.05% Tween-20, 0.2 mmol/L dNTP, 0.5 U of Hot start Taq DNA polymerase (Biosan, Novosibirsk, Russia), and 2 μL of purified first round PCR products, as described for bisulfite DNA methylation analysis. Single-stranded biotinylated PCR products were prepared for sequencing using the Vacuum Prep Tool and Streptavidin Sepharose™ HP according to the manufactur-er’s instructions. Pyrosequencing reactions were performed using the PSQ 96 SNP Reagent Kit (Pyrosequencing, Uppsala, Sweden). The degree of methylation at each CpG site was determined by Pyro Q-CpG Software (Biotage, Uppsala, Sweden). The methylation index for each sample was calculated as the mean value of methC percentage for all 12 CpGs examined. It is known that differences in sequence between methylated and unmethylated samples after bisulfite treatment can influence amplification efficiency. We constructed a calibration curve to verify that the pyrosequencing assay provided unbiased quantification. Unmethylated and 100% methylated DNA samples after bisulfite treatment were quantified and mixed at different ratios. The samples were PCR-amplified and subjected to pyrosequencing. Actual and expected methylation percentages are expected to be linearly correlated with an r2≥ 0.8.
Total RNA from cultured cells was isolated using the SV Total RNA Isolation System (Promega, Madison, WI, USA) following the manufacturer’s instructions. After DNaseI treatment, 0.25 μg of total RNA was reverse transcribed using the oligo-dT primers and ImProm-II Reverse Transcription System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Amplification reactions were run in triplicate on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Bedford, MA, USA). First, primer efficiency was validated with a standard curve of serial dilution points of a scraped section cDNA pool and a no-template control (NTC). qPCR amplification mixture was a total volume of 25 μL, which contained 5 μL single strand cDNA template diluted 20 times after reverse-transcription, 10 μL 2.5 × SYBR Green I qPCR Master Mix (Syntol, Moscow, Russia), and 1.2 μL forward and reverse primers (5 μmol/L) so that the final primer concentration was 300 nmol/L. The sequence of the primers used in the study were as follows: Ppia (reference gene) 5'-CGCGTCTCCTTCGAGCTGTTTG-3' and 5'-TGTAAAGTCACCACCCTGGCACAT-3'; Rian 5'-TCGAGACACAAGAGGACTGC-3' and 5'-ATTGGAAGTCTGAGCC-3'; Mirg 5'-TTGACTCCAGAAGATGCTCC-3' and 5'-CCTCAGGTTCCTAAGCAAGG-3'; Gtl2 5'-TTGCACATTTCCTGTGGGAC-3' and 5'-AAGCACCATGAGCCACTAGG-3'; Oct4 5'-TAGGTGAGCCGTCTTTCCAC-3' and 5'-GCTTAGCCAGGTTCGAGGAT-3'; Nanog 5'-TTGCTTACAAGGGTCTGCTACT-3' and 5'-ACTGGTAGAAGAATCAGGGCT-3'. Cycle conditions were as follows: after an initial 5 min denaturation at 95 °C, the samples were amplified using 35 cycles at 95 °C for 15 s, 58 °C for 15 s, and 72 °C for 20 s. After optimization of qPCR systems, we used a relative expression software tool (REST 2009 V2.0.13©, Qiagen) to compare gene expression between tauGFP ES cells, MEFs, and ES cell/fibroblast hybrid clones.
As mentioned above, four highly pluripotent hybrid clones: tef4, taf2, tef9, and taf5 had near-tetraploid chromosome sets. To identify parental chromosome origin in hybrid clones, we analyzed a set of microsatellites marking all parental chromosomes except chromosomes 11 and X (Table 1). This analysis showed that all four hybrid clones retain chromosomes from both parental cell types. The Dlk1-Dio3 imprinted locus is localized on chromosome 12, which was present in all analyzed hybrid clones. Also, microsatellite and repeated cytogenetic analyses at different passages from 8 to 14 demonstrated that chromosome numbers and microsatellite markers remained stable during that period.
| Microsatellite marker1 | Hybrid cell clones | |||||||
| tef4 | taf2 | tef9 | taf5 | |||||
| 129 | DD | 129 | DD | 129 | DD | 129 | DD | |
| D1Mit200 | + | + | + | + | + | + | + | + |
| D2Mit9 | + | + | + | + | + | + | + | + |
| D3Mit257 | + | + | + | + | + | + | + | + |
| D4Mit11 | + | + | + | + | + | + | + | + |
| D5Mit346 | + | + | + | + | + | + | + | + |
| D6Mit201 | + | + | + | + | + | + | + | + |
| D7Mit309 | + | + | + | + | + | + | + | + |
| D8Mit155 | + | + | + | + | + | + | + | + |
| D9Mit181 | + | + | + | + | + | + | + | + |
| D10Mit109 | + | + | + | + | + | + | + | + |
| D12Mit270 | + | + | + | + | + | + | + | + |
| D13Mit78 | + | + | + | + | + | + | + | + |
| D14Mit38 | + | + | + | + | + | + | + | + |
| D15Mit14 | + | + | + | + | + | + | + | + |
| D16Mit4 | + | + | + | + | + | + | + | + |
| D17Mit36 | + | + | + | + | + | + | + | + |
| D18Mit36 | + | + | + | + | + | + | + | + |
| D19Mit10 | + | + | + | + | + | + | + | + |
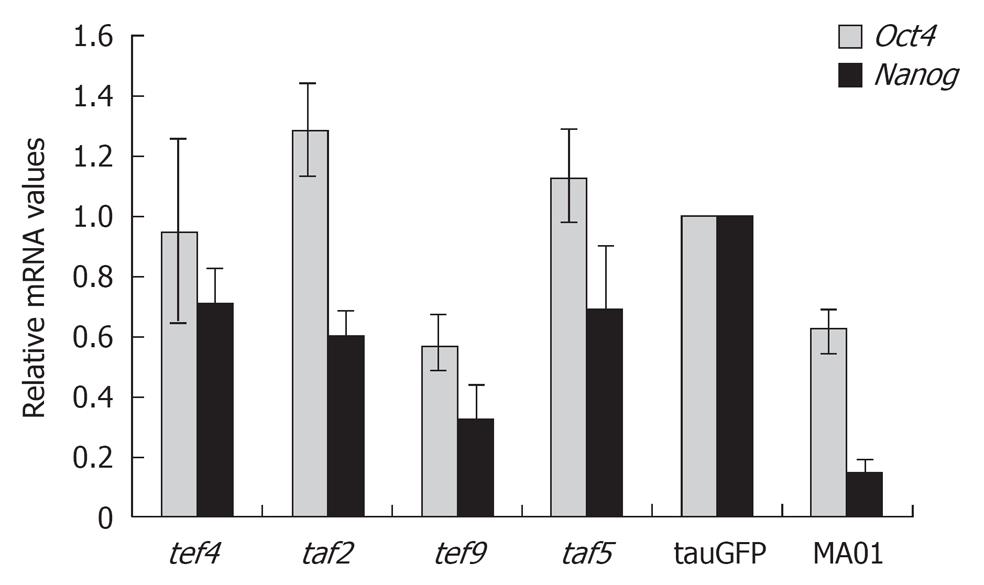
The pluripotent state of ES cells is supported by the activity of a small number of key genes such as Oct4, Nanog and Sox2. Therefore, we performed additional quantitative analysis of Oct4 and Nanog expression in hybrid cells, as shown in Figure 1. The ES cell lines tauGFP and MA01 were used as controls. As shown in Figure 1, Oct4 and Nanog expression in the tef4, taf2, and taf5 hybrid cells was similar to that in tauGFP ES cells and even somewhat higher than in MA01 ES cells. Expression of both genes was lower in tef9 cells than in tauGFP ES cells but was comparable with that of MA01 ES cells. Interestingly, expression of Oct4 and Nanog genes was higher in tauGFP ES cells (passed more than 40 passages) than MA01 cells analyzed at the eleventh passage (Figure 1).
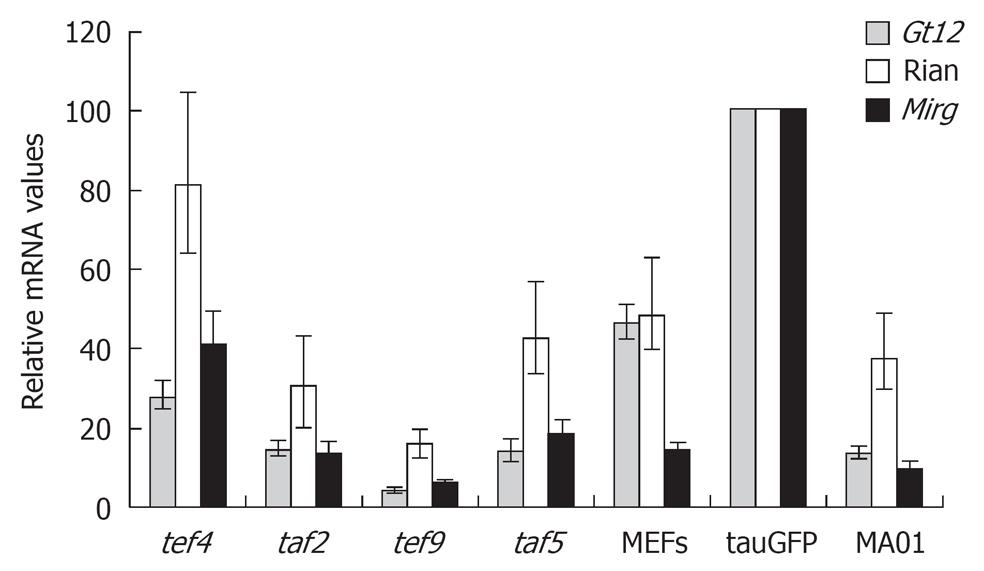
The main focus of our study was to determine whether alterations in Dlk1-Dio3 locus imprinting occurred in the cell fusion reprogramming system that were previously described in iPS cells. Quantitative estimation of Gtl2, Rian, and Mirg gene expression located within the imprinted Dlk1-Dio3 gene cluster in hybrid cell clones, tef4, taf2, tef9 and taf5, is shown in Figure 2. Among hybrid clones, expression of Gtl2, Rian and Mirg genes was highest in the tef4 clone, while the tef9 clone had the lowest expression. Levels of Gtl2, Rian, and Mirg expression in tef4, taf5 and taf2 hybrid clones (but not in tef9) were comparable with those found in MA01 and MEFs, but somewhat lower than in tauGFP ES cells (Figure 2). It should be noted that expression of these three genes was higher in tauGFP ES cells than in MA01 cells (Figure 2).
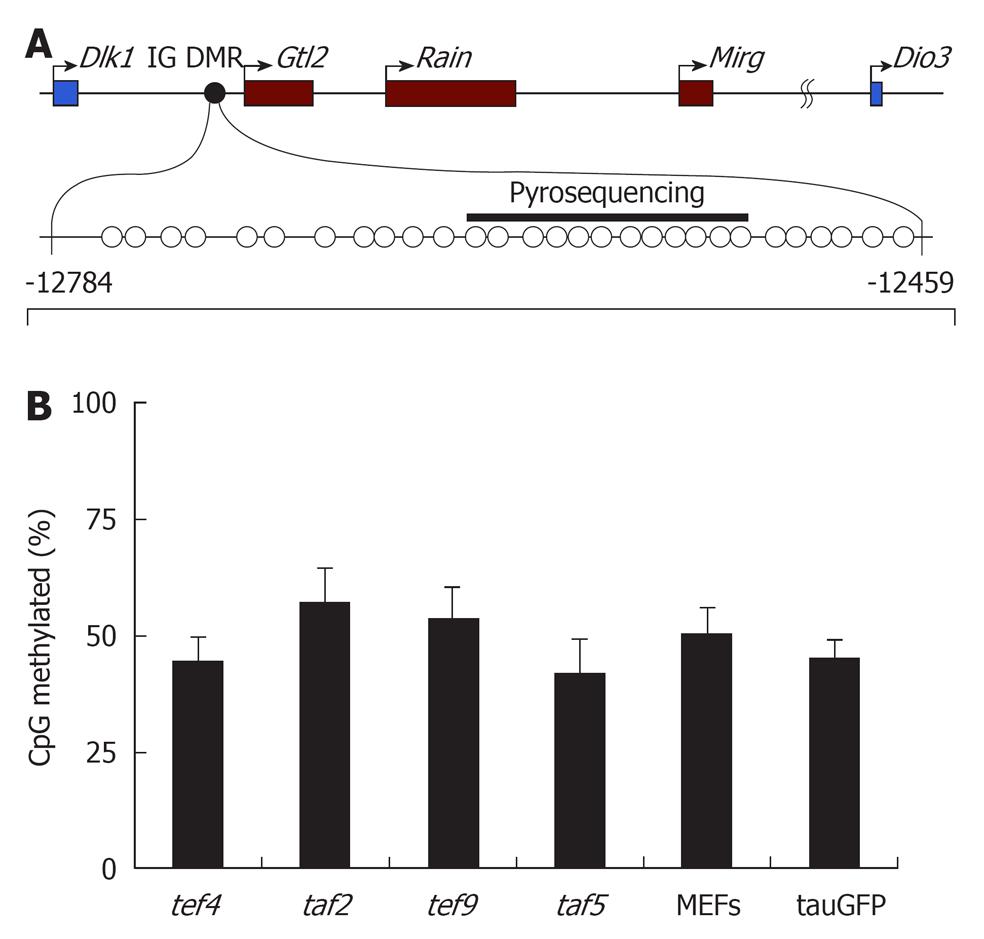
Imprinting of the Dlk1-Dio3 cluster is regulated by differentially methylated regions (DMRs) that become epigenetically modified in the germ line. One of these regions, the intergenic DMR (IG DMR), is located between the Dlk1 and Glt2 genes and is considered to be a master regulator of the entire locus (Figure 3A)[25]. To determine whether DNA methylation of this region is altered during reprogramming, we examined the methylation status of IG DMR in four hybrid cell clones, in parental tauGFP ES cells, and in MEFs (Figure 3B). Pyrosequencing analysis revealed that the level of DNA methylation of IG DMR in the parental tauGFP ES cells was about 50%, which is expected for germline-imprinted regions. Similar methylation profiles were observed in all four hybrid clones (Figure 3B).
These data were supported by bisulfite sequencing analysis of the IG DMR (Figure 3). As shown in Figure 3, the percentage of CpG-site methylation of IG DMR in tef4, taf2, tef9, and taf5 hybrid clones was similar to that of tauGFP ES cells and MEFs. Approximately half of the alleles at the IG DMR were hypermethylated, whereas the other half were unmethylated (Figure 4). Both approaches suggest that the hybrid clone methylation pattern of IG DMR remains stable compared to parental cells, tauGFP ES cells, and MEFs.
To determine whether Dlk1-Dio3 locus status is altered in hybrid clones we analyzed expression of several genes within that locus. It should be noted, that hybrid clones used in this study are intraspecific and have near-tetraploid karyotype. Thus it was not possible to perform a detailed analysis of the activity of each of the four alleles. Instead we used indirect methods such as quantitative evaluation of imprinted gene allele expression and assessment of the methylation status of the IG DMR which is a master regulator of the entire locus.
Our data suggest that the genes encoding Gtl2, Rian, and Mirg within the Dlk1-Dio3 cluster are active in all examined ES cell/fibroblast hybrid clones. Regardless of interclonal variability, expression of Gtl2, Rian, and Mirg in the ES cell/fibroblast hybrid clones is comparable to that in MA01 ES cells, but somewhat lower than in tauGFP ES cells. Interclonal variability is not specific to the imprinted genes because it was also observed in Oct4 and Nanog gene expression. We cannot exclude the possibility of aberrant silencing of some of the four alleles in hybrid clones. On the other hand, our data allow us to suggest that at least one allele remains active as there was no dramatic decrease of the expression level of Gtl2, Rian, and Mirg genes in iPS cells.
Stadtfeld et al[21] found that hypermethylation at the DMRs occurs within the Dlk1-Dio3 cluster in all iPS cell clones with a silent Gtl2 gene[21]. These findings prompted us to perform quantitative analysis of DNA methylation status of the IG DMR region within the Dlk1-Dio3 locus. Both quantitative methylation analyses by pyrosequencing and bisulfite sequencing clearly showed that the DNA methylation status of the imprinted region in tef4, taf2, tef9, and taf5 hybrid clones was comparable to that of tauGFP ES cells and MEFs. Since tauGFP cells and MEFs served as parental cell partners in the generation of the hybrid cells used in our study, we conclude that methylation status of the IG DMR region within the Dlk1-Dio3 locus was not altered.
Two cell lines, tauGFP and MA01 cells, were used as controls in the analysis of gene expression by qPCR. Expression of the Oct4 and Nanog genes and the imprinted Gtl2, Rian, and Mirg genes was higher in the tauGFP ES cells than other ES cells or the MA01 cell line. However, these lines showed no differences in expression levels of the Gapdh housekeeping gene (data not shown). The cause of these differences is unclear. However, long-term in vitro cultivation of tauGFP cells (over 40 passages) could lead to the appearance of undefined characteristic(s) in vitro conditions, whereas MA01 cells have only been passaged five to eleven times following isolation from blastocysts. In general, the findings agree with observation of other researchers that no abnormalities at the Dlk1-Dio3 cluster are evident in ES cells derived from normal blastocysts or even blastocysts developed after somatic cell nuclear transfer[20,21].
As mentioned above, the examined hybrid clones tef4, taf2, tef9 and taf5 had robust levels of pluripotency judging from the generation of embryonic and adult chimeras[9]. However, adult chimera yield varied significantly between the clones. For example, injection of tef4 cells into C57BL blastocysts generated 15 adult chimeras with seven of them showing more than 50% visible color chimerism. Injection of taf2 and tef9 cells resulted in the birth of six and one adult chimeras, respectively, with moderate color chimerism. However, it is important to note that the development of tetraploid mice has generally not been observed beyond mid-gestation[26]. We have not excluded the possibility that high levels of chimerism based on excessive contribution of near-tetraploid hybrid cells could potentially halt embryonic development before birth, thereby decreasing the rate of chimera births. Of note, 30% to 65% of chimeric embryos with high contribution from tef4 and taf5 died between embryonic days 11-13[9]. Nevertheless, capacity of the tested hybrid clones to produce adult chimeras and contribute to coat color can tentatively be ranked as tef4 > taf2 > tef9. From data in Figure 3 it follows that expression levels of the maternally imprinted Gtl2, Rian, and Mirg genes can tentatively be ranked as tef4 > taf2 > tef9. Comparison of adult chimera yield and expression levels of the maternally imprinted genes suggests that there is a positive correlation. Interestingly, Liu et al[20] observed a similar phenomenon when comparing two partially pluripotent iPS cell lines with different levels of pluripotency: one cell line was germline transmittable whereas the other was not. Expression levels of all Dlk1-Dio3 miRNAs were consistently higher in the germline-transmittable cells than in the non-transmittable cells. Thus, a high level of pluripotency could be associated with precise regulation but not simply with the “switch on” state of the locus.
Our data suggest that, in contrast to iPS cells, the reprogramming process in hybrid cell systems is achieved without marked alteration of the major imprinted Dlk1-Dio3 locus, its methylation status, and Gtl2, Rian and Mirg gene expression.
Currently there are several methods of somatic cell genome reprogramming to a pluripotent state. The most promising for practical usage is induced pluripotent stem (iPS) cell derivation. The drawback of this approach is the possibility of reprogramming errors. In was shown that aberrant silencing of the Dlk1-Dio3 locus is observed in most iPS cell lines and can cause restriction of iPS cell differentiation potential. The mechanisms of this abnormality are unknown.
Cell fusion is another approach for somatic cell genome reprogramming. It was reported that cell fusion mediated reprogramming is fast, effective and relatively correct. Thus, this model is convenient for studying the basic principles of reprogramming. In this research article the authors have examined whether there is iPS cell characteristic aberrant silencing of the Dlk1-Dio3 locus in a cell fusion reprogramming system.
This report is the first study of the aberrant Dlk1-Dio3 locus reprogramming in hybrid cells. The qPCR analysis of Dlk1-Dio3 locus gene expression and its DNA methylation level in hybrids cell allowed the conclusion that, in contrast to iPS cells the reprogramming process in a hybrid cell system is achieved without marked alteration of the imprinted Dlk1-Dio3 locus.
The understanding of the reprogramming mechanisms will provide clues to development of improved protocols for the derivation of reprogrammed cells.
Reprogramming: a process of somatic cell transition from one differentiation state to another, including pluripotency; Dlk1-Dio3 locus-a large region located on mouse chromosome 12. It has important functions in the regulation of the development. Aberrant expression of its genes results in the termination of prenatal development.
In general, the manuscript is interesting and well written and the authors provide convincing evidences suggesting that the reprogramming process in a hybrid cells did not alter the imprinted Dlk1-Dio3 locus (its methylation status, or the expression of Gtl2, Rian and Mirg genes).
| 1. | Matveeva NM, Shilov AG, Kaftanovskaya EM, Maximovsky LP, Zhelezova AI, Golubitsa AN, Bayborodin SI, Fokina MM, Serov OL. In vitro and in vivo study of pluripotency in intraspecific hybrid cells obtained by fusion of murine embryonic stem cells with splenocytes. Mol Reprod Dev. 1998;50:128-138. [PubMed] |
| 2. | Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol. 2001;11:1553-1558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 662] [Cited by in RCA: 590] [Article Influence: 23.6] [Reference Citation Analysis (1)] |
| 3. | Cowan CA, Atienza J, Melton DA, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369-1373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 689] [Cited by in RCA: 596] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 4. | Ambrosi DJ, Tanasijevic B, Kaur A, Obergfell C, O'Neill RJ, Krueger W, Rasmussen TP. Genome-wide reprogramming in hybrids of somatic cells and embryonic stem cells. Stem Cells. 2007;25:1104-1113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 47] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 5. | Hasegawa K, Zhang P, Wei Z, Pomeroy JE, Lu W, Pera MF. Comparison of reprogramming efficiency between transduction of reprogramming factors, cell-cell fusion, and cytoplast fusion. Stem Cells. 2010;28:1338-1348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 6. | Ying QL, Nichols J, Evans EP, Smith AG. Changing potency by spontaneous fusion. Nature. 2002;416:545-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1123] [Cited by in RCA: 1037] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
| 7. | Pells S, Di Domenico AI, Gallagher EJ, McWhir J. Multipotentiality of neuronal cells after spontaneous fusion with embryonic stem cells and nuclear reprogramming in vitro. Cloning Stem Cells. 2002;4:331-338. [PubMed] |
| 8. | Vasilkova AA, Kizilova HA, Puzakov MV, Shilov AG, Zhelezova AI, Golubitsa AN, Battulin NR, Vedernikov VE, Menzorov AG, Matveeva NM. Dominant manifestation of pluripotency in embryonic stem cell hybrids with various numbers of somatic chromosomes. Mol Reprod Dev. 2007;74:941-951. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 16] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 9. | Kruglova AA, Kizilova EA, Zhelezova AI, Gridina MM, Golubitsa AN, Serov OL. Embryonic stem cell/fibroblast hybrid cells with near-tetraploid karyotype provide high yield of chimeras. Cell Tissue Res. 2008;334:371-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 10. | Gridina MM, Serov OL. Bidirectional reprogramming of mouse embryonic stem cell/fibroblast hybrid cells is initiated at the heterokaryon stage. Cell Tissue Res. 2010;342:377-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 11. | Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508-525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 582] [Cited by in RCA: 515] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 12. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17989] [Cited by in RCA: 18606] [Article Influence: 930.3] [Reference Citation Analysis (1)] |
| 13. | Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2048] [Cited by in RCA: 1909] [Article Influence: 100.5] [Reference Citation Analysis (0)] |
| 14. | Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1459] [Cited by in RCA: 1336] [Article Influence: 74.2] [Reference Citation Analysis (0)] |
| 15. | Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G, Kupriyanov S, Baldwin KK. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 320] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 16. | Zhao XY, Li W, Lv Z, Liu L, Tong M, Hai T, Hao J, Guo CL, Ma QW, Wang L. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 603] [Cited by in RCA: 556] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 17. | Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704-712. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 602] [Cited by in RCA: 536] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 18. | Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, Huo H, Miller JD, Hartung O, Rho J, Ince TA. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27:1033-1037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 400] [Cited by in RCA: 380] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 19. | Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285-290. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1842] [Cited by in RCA: 1723] [Article Influence: 107.7] [Reference Citation Analysis (15)] |
| 20. | Liu L, Luo GZ, Yang W, Zhao X, Zheng Q, Lv Z, Li W, Wu HJ, Wang L, Wang XJ. Activation of the imprinted Dlk1-Dio3 region correlates with pluripotency levels of mouse stem cells. J Biol Chem. 2010;285:19483-19490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 224] [Cited by in RCA: 226] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 21. | Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 655] [Cited by in RCA: 615] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 22. | Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003;35:97-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 394] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 23. | Eakin GS, Behringer RR. Tetraploid development in the mouse. Dev Dyn. 2003;228:751-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 64] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 24. | Battulin NR, Pristyazhnyuk IE, Matveeva NM, Fishman VS, Vasilkova AA, Serov OL. Allelic expression and DNA methylation profiles of promoters at the parental Oct4 and Nanog genes in Mus musculus ES cell/Mus caroli splenocyte hybrid cells. Cell Tissue Res. 2009;337:439-448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 25. | Pratt T, Sharp L, Nichols J, Price DJ, Mason JO. Embryonic stem cells and transgenic mice ubiquitously expressing a tau-tagged green fluorescent protein. Dev Biol. 2000;228:19-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 101] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 26. | Bryja V, Bonilla S, Cajánek L, Parish CL, Schwartz CM, Luo Y, Rao MS, Arenas E. An efficient method for the derivation of mouse embryonic stem cells. Stem Cells. 2006;24:844-849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 61] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
Peer reviewer: Luis Politi, PhD, Principal Investigator and Professor of Cell Biology and Neurobiology, National Research Council of Argentina and Universidad Nacional del Sur, Camino La Carrindanga Km 7, 8000 Bahía Blanca, Argentina
S- Editor Wang JL L- Editor Hughes D E- Editor Zheng XM