Published online Nov 26, 2024. doi: 10.4252/wjsc.v16.i11.906
Revised: July 24, 2024
Accepted: October 12, 2024
Published online: November 26, 2024
Processing time: 306 Days and 6.2 Hours
Thin endometrium seriously affects endometrial receptivity, resulting in a significant reduction in embryo implantation, and clinical pregnancy and live birth rates, and there is no gold standard for treatment. The main pathophysio
To determine the role of hypoxic-EEC-derived exosomes in function of HucMSCs and explore the potential mechanism.
Exosomes were isolated from normal EECs (EEC-exs) and hypoxia-damaged EECs (EECD-exs), before characterization using Western blotting, nanoparticle-tracking analysis, and transmission electron microscopy. HucMSCs were cocu
MiR-21-5p and miR-214-5p were lowly expressed in EECD-ex-pretreated HucMSCs. MiR-214-5p and miR-21-5p inhibitors facilitated the migratory and differentiative potentials of HucMSCs. MiR-21-5p and miR-214-5p targeted STAT3 and protein inhibitor of activated STAT3, respectively, and negatively regulated phospho-STAT3. MiR-21-5p- and miR-214-5p-inhibitor-induced promotive effects on HucMSC function were reversed by STAT3 inhibition. MiR-21-5p and miR-214-5p overexpression repressed HucMSC migration and differentiation, while STAT3 activation reversed these effects.
Low expression of miR-21-5p/miR-214-5p in hypoxic-EEC-derived exosomes promotes migration and differentiation of HucMSCs into EECs via STAT3 signaling. Exosomal miR-214-5p/miR-21-5p may function as valuable targets for thin endometrium.
Core Tip: Thin endometrium is a primary cause of repeated implantation failure and infertility. In this study, with the help of microRNA sequencing, we investigated the role of hypoxic endometrial epithelial cell (EEC)-derived exosomes in cell function of human umbilical cord mesenchymal stem cells and explored the potential mechanism. Eventually, we found that lowly-expressed miR-21-5p and miR-214-5p in hypoxic EEC-derived exosomes promoted migration and differentiation of human umbilical cord mesenchymal stem cells into EECs via signal transducer and activator of transcription 3 signaling. Exosomal miR-214-5p and miR-21-5p may function as valuable targets for thin endometrium.
- Citation: Zhang WY, Wang HB, Deng CY. Effects of miR-214-5p and miR-21-5p in hypoxic endometrial epithelial-cell-derived exosomes on human umbilical cord mesenchymal stem cells. World J Stem Cells 2024; 16(11): 906-925
- URL: https://www.wjgnet.com/1948-0210/full/v16/i11/906.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v16.i11.906
The endometrium is a dynamic, repetitively cycling tissue that mediates blastocyst implantation[1]. Thin endometrium is considered to be a major cause of unsuccessful embryo transfer, leading to long-term infertility[2]. Possible pathological causes of thin endometrium include radiation, infection, history of uterine surgery, and Asherman syndrome[3]. Nume
Exosomes are nanosized structures released by normal and pathological cells, which are responsible for delivering bioactive cargoes such as nucleic acids, lipids, and proteins[5]. Exosomes play a role in regulating cell function in disorders of the endometrium[6]. MicroRNAs (miRNAs) are noncoding small RNAs comprised of about 22 nucleotides, which bind to target mRNAs for transcription and translation regulation[7]. Exosomes-derived miRNAs are related to various cellular processes, including cell proliferation, differentiation, and migration[8]. In ovarian endometriosis, exosomes derived from endometrial epithelial cells (EECs) carry miRNA-30c and repress cell migration and invasion[9]. Exosomes from bovine EECs ensure the development of trophoblast cells via secreting miR-218[10]. Additionally, exosomes can be used to deliver miR-218 from EECs into the uterine microenvironment to modulate immune responses[11].
Mesenchymal stem cells (MSCs) are multipotent cells possessing the capabilities of self-renewal and multidirectional differentiation[12]. Recent clinical applications use umbilical cord-derived MSCs (ucMSCs) for treating human diseases, including metabolic/endocrine-related diseases and reproductive disorders[13]. Previous studies have validated the beneficial role of ucMSCs in disorders of the endometrium. UcMSC transplantation contributes to the repair of endo
HucMSCs at passage 3 were obtained from Cellverse Bioscience Technology Co. Ltd. (Shanghai, China) and cultured in Dulbecco’s modified Eagle’s medium/nutrient mixture F-12 (DMEM/F12) (03.2001C, EallBio, Beijing, China) with 10% fetal bovine serum (FBS) (SH30406.05, HyClone, Marlborough, MA, United States). Primary human EECs were bought from Cellverse Bioscience Technology and maintained in DMEM (11965092, Gibco, Carlsbad, CA, United States) containing 10% FBS. Streptomycin (100 U/mL) and 100 mg/mL penicillin were added to the medium. Cells were cultured in normoxic (21% O2, 75% N2, and 5% CO2) conditions at 37 °C. HucMSCs at passages 4-6 were used for the next experiments, as previously described[18].
EECs were grown to about 70% confluence in normoxic air. To induce hypoxia, cells were maintained in a modular incubator chamber in a humidified hypoxic condition (5% CO2, 94% N2, and 1% O2) at 37 °C for 4 h. In the control group, EECs were only incubated in normoxic conditions.
Exosomes in culture medium of hypoxia damaged-EECs (EECD-exs) or normal EECs (EEC-exs) were extracted by ultracentrifugation. EECs were maintained for 48 h with exosome-free DMEM. To remove cell debris, the medium was centrifuged (2000 × g) at 4 °C for 20 min, prior to 10000 × g at 4 °C for 30 min. A 0.2-μm filter was used to filter the supernatants. The supernatants were ultracentrifuged at 100000 × g at 4 °C for 1 h. The precipitate was resuspended in phosphate-buffered saline, followed by ultracentrifugation at 100000 × g for 1 h. Isolated exosomes were gathered with exosome precipitation reagent.
Exosome morphology was identified by transmission electron microscopy (TEM) (HT7800; Hitachi, Tokyo, Japan). Nanoparticle-tracking analysis (NTA) was performed to analyze the concentration and size of exosomes with an analyzer (ZetaVIEW S/N 252, ParticleMetrix, Germany). Specific markers of exosomes [CD81/tetraspanin-28 (Tspan-28) and CD63] were determined by Western blotting with corresponding primary antibodies against CD81/Tspan-28 (ab109201, Abcam, Cambridge, United Kingdom) and CD63 (ab134045, Abcam).
Total RNA was isolated from cells, and an Agilent Technologies 2100 Bioanalyzer (CA, United States) was used to examine the quality of total RNA. The small RNA library was prepared by using the TruSeq small RNA library prep kit (RS-200-0036, Illumina, San Diego, CA, United States). Following multiplexing in equimolar amounts, indexed small RNA libraries were denatured and loaded for cluster generation on GAIIx flow cell lanes with cBot station and Illumina cluster generation kits. Differentially expressed miRNAs showing raw reads ≥ 5 in samples and P < 0.05 were chosen.
HucMSCs were pretreated with EEC-exs or EECD-exs (5 μg/mL) for 24 h, which formed the EEC-ex + HucMSC or EECD-ex + HucMSC group, respectively. For cell transfection, the negative controls, miR-214-5p/miR-21-5p inhibitors, and miR-214-5p/miR-21-5p mimics were synthesized by Sangon Biotech, Shanghai, China. HucMSCs (2 × 104/well) were grown to 80% confluence in six-well plates. HucMSCs were transfected with normal control (NC) mimic, NC inhibitor, miR-214-5p/miR-21-5p inhibitors, or miR-214-5p/miR-21-5p mimics (30 nM/well) using jetPRIME (101000046, Polyplus, Illkrich, France). For inhibition or activation of signal transducer and activator of transcription 3 (STAT3), the STAT3 inhibitor Stattic (HY-13818, MedChemExpress, Junction, NJ) or agonist colivelin (HY-P1061, MedChemExpress) was added to HucMSCs.
For the wound healing assay, cells were implanted in a six-well plate. A pipette tip was used to scratch the cell monolayer on the bottom of the plate to draw a gap between cells. Images were obtained at the indicated times to observe cell mi
RIPA buffer with protease inhibitors (P0013B, Beyotime, Shanghai, China) was applied to extract proteins from the cells. Protein samples were separated on sodium-dodecyl sulfate gel electrophoresis, before transfer to polyvinylidene di
The differentiation of HucMSCs into endometrial stromal cells (ESCs) and EECs were identified by staining with an
TRIzol reagent (R1100, Solarbio, Beijing, China) was applied to isolate total RNA from cells, prior to the preparation of cDNA with total RNA (50 ng) in a reaction mixture (20 μL) using iScript cDNA Synthesis Kit (KR118, Tiangen, Beijing, China). Gene expression was quantified by performing reverse transcription-quantitative polymerase chain reaction (RT-qPCR) with SYBR Green (FP205, Tiangen) and specific primers (Table 1), and optimized on the Real-Time PCR Detection System (ABI7500, Thermo Scientific, Waltham, MA, United States). RNA expression was determined using the 2-ΔΔCT method. For each sample, the expression of miRNA and mRNA was normalized to U6 and GAPDH, respectively.
| Gene | Primer sequence (5’-3’) | |
| U6 | F | ACGATACAGAGAAGATTAGCATGG |
| R | AAATATGGAACGCTTCACGAA | |
| hsa-miR-1303 | F | CTCAACTGGTGTCGTGGAGT |
| R | TCGGCAGGTTTTAGAGACGGGGTCT | |
| RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTCTCGT | |
| hsa-miR-4521 | F | CTCAACTGGTGTCGTGGAGT |
| R | TCGGCAGGGCTAAGGAAGTCCTGTGCT | |
| RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAAACTG | |
| hsa-miR-7974 | F | CTCAACTGGTGTCGTGGAGT |
| R | TCGGCAGGAGGCTGTGATGCTCTCCT | |
| RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCGGGCTC | |
| hsa-miR-210-3p | F | CTCAACTGGTGTCGTGGAGT |
| R | TCGGCAGGCTGTGCGTGTGACAGCG | |
| RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTCAGCC | |
| hsa-miR-31-5p | F | CTCAACTGGTGTCGTGGAGT |
| R | TCGGCAGGAGGCAAGATGCTGGCATAGCTGT | |
| RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCACAGCT | |
| hsa-miR-214-5p | F | CTCAACTGGTGTCGTGGAGT |
| R | TCGGCAGGTGCCTGTCTACACTTGC | |
| RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCGCACAG | |
| hsa-miR-21-5p | F | CTCAACTGGTGTCGTGGAGT |
| R | TCGGCAGGTAGCTTATCAGACTGAT | |
| RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCGTCAAC | |
| PIAS3 | F | CTGGGCGAATTAAAGCACATGG |
| R | AAAGCGTCGTCGGTAAAGCTC | |
| STAT3 | F | ACCAGCAGTATAGCCGCTTC |
| R | GCCACAATCCGGGCAATCT | |
| GAPDH | F | ACGGATTTGGTCGTATTGGG |
| R | GGGATCTCGCTCCTGGAAG |
TargetScan and miRDB were utilized to predict the target genes of miR-214-5p or miR-21-5p. The 3’-untranslated region (UTR) sequence of PIAS3 or STAT3 containing a binding site for miR-214-5p or miR-21-5p was inserted into psiCHECK-2 vector (General Bio, Anhui, China) to generate PIAS3-WT or STAT3-WT. The 3’-UTR sequence of PIAS3 or STAT3 was mutated for construction of PIAS3-Mutant or STAT3-Mutant. Cells were seeded in a 24-well plate and incubated for 24 h. MiR-214-5p/miR-21-5p mimic and PIAS3/STAT3-WT or PIAS3/STAT3-Mutant were transfected into the cells. A dual-luciferase reporter system (RG028, Beyotime) was used to evaluate luciferase activity 24 h after transfection.
Results are described as the mean ± SD. One-way ANOVA was carried out for multiple comparisons. Student’s t test was used for the comparisons between two groups. Statistical analyses were undertaken employing GraphPad Prism 7.0. Differences were considered significant at P < 0.05.
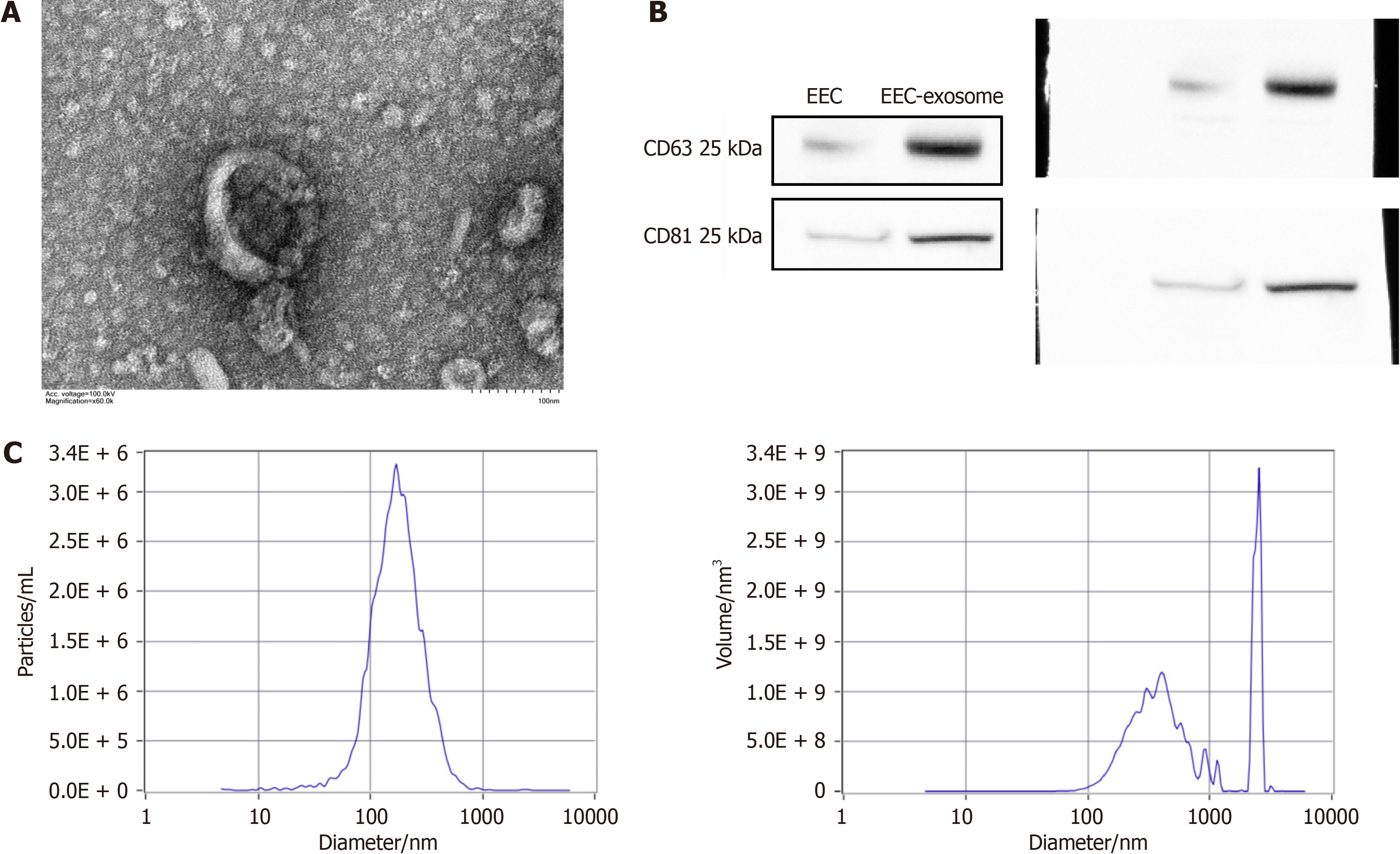
EECD-exs and EEC-exs were extracted, then TEM, NTA, and Western blotting were performed to identify the extracted exosomes. The vesicle-like exosomes (EEC-exs) were spherical on TEM imaging (Figure 1A). The isolated exosomes (EEC-exs) expressed typical positive markers (CD63 and CD81/Tspan-28) (Figure 1B). NTA data indicated that the exosomes (EEC-exs) were about 150 nm in size (Figure 1C). These results demonstrated the successful isolation of exosomes from culture medium of EECs.
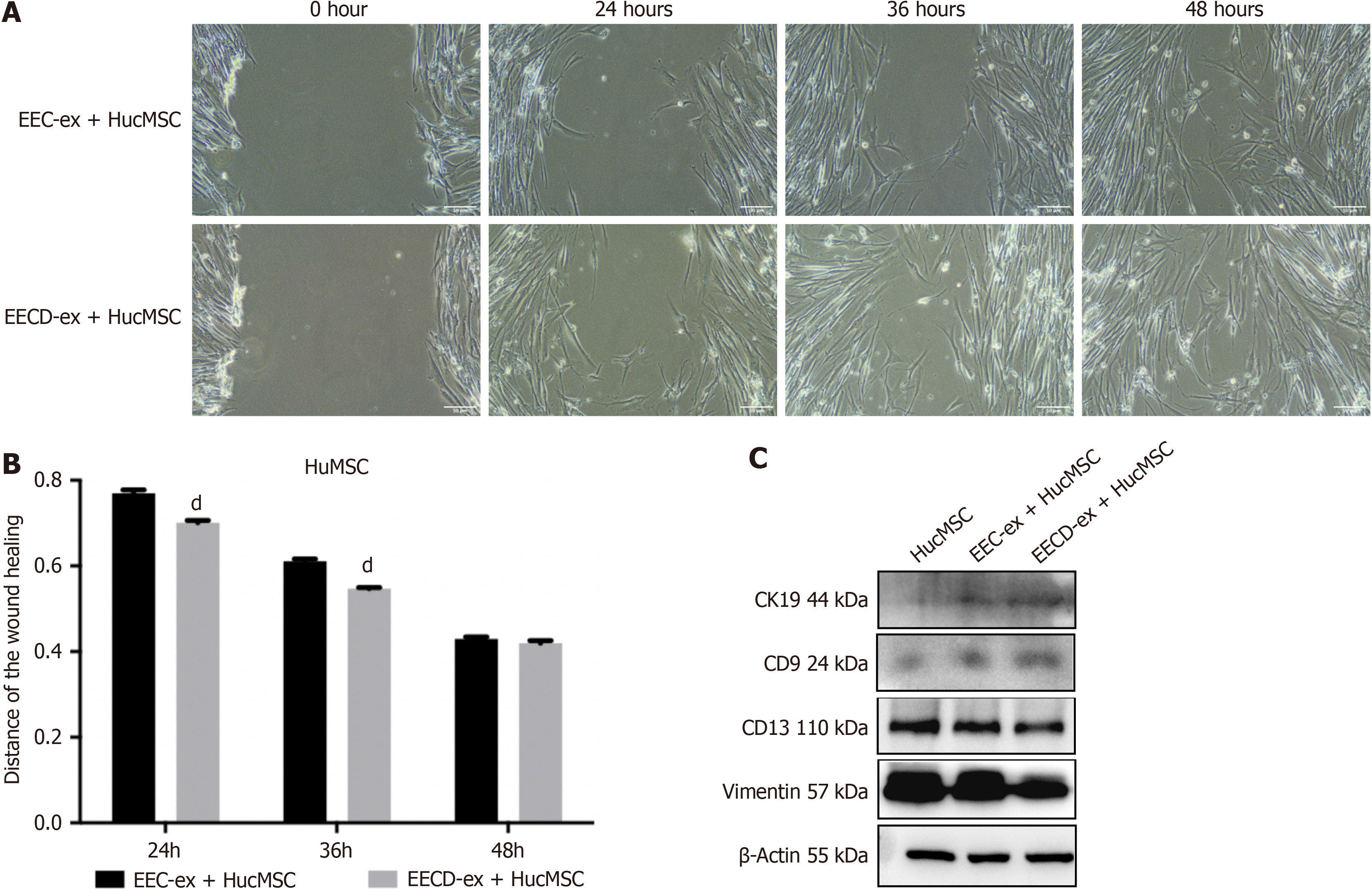
To determine the function of EECD-exs in the migration of HucMSCs, HucMSCs were cocultured with EEC-exs or EECD-exs. Compared with the EEC-ex + HucMSC group, the migration of HucMSCs was promoted after coculture with EECD-exs, exhibited by the wound healing assay (Figure 2A and B). The data indicated that EECD-exs promoted the migration of HucMSCs. EECD-ex coculture increased protein expression of the markers of epithelial cell lineage (CD9 and CK19), and decreased expression of the markers of stromal lineage (vimentin and CD13) in HucMSCs (Figure 2C), suggesting that EECD-exs promoted differentiation of HucMSCs into EECs.
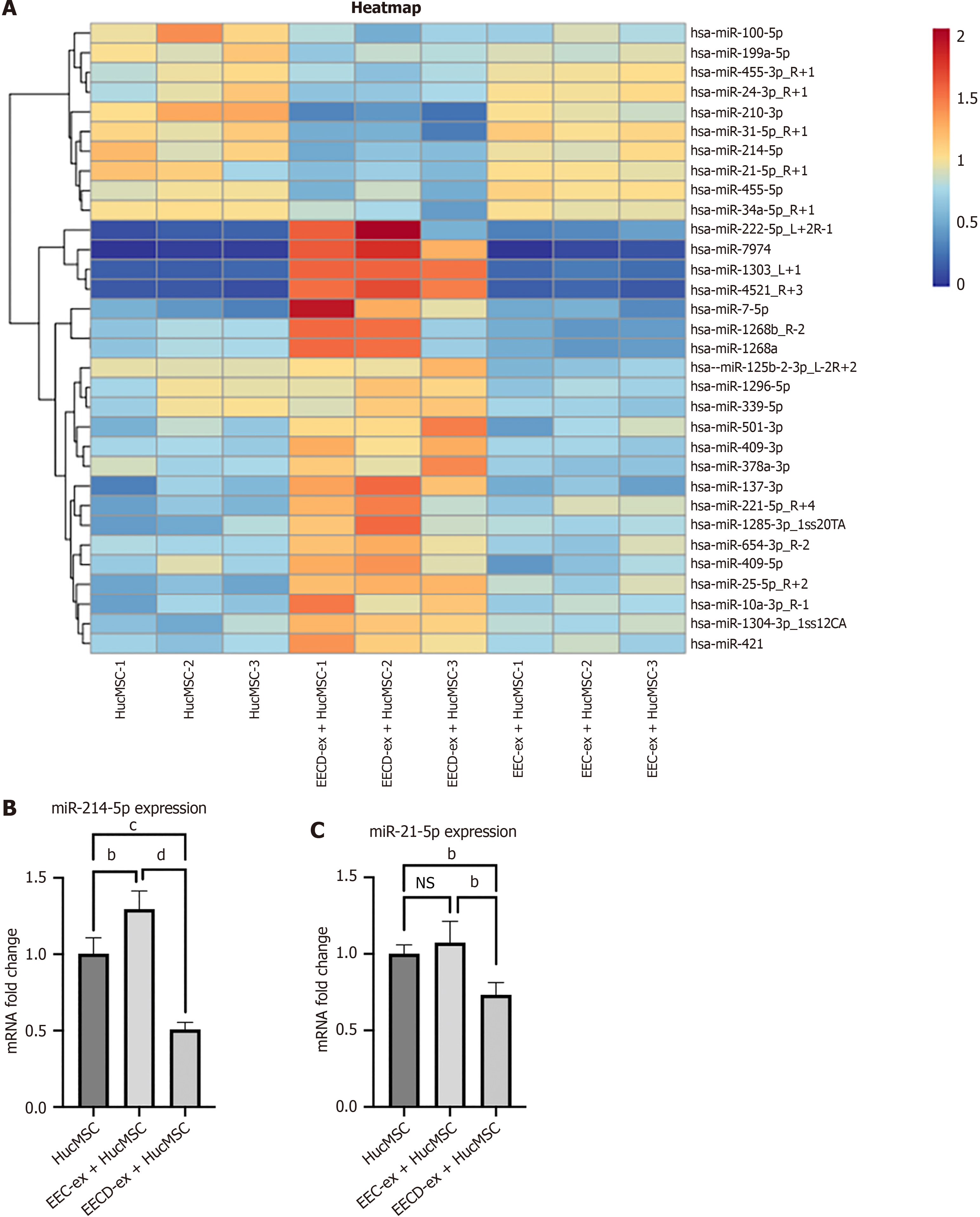
To determine differentially expressed miRNAs in exosomes cocultured with HucMSCs, miRNA sequencing analysis was performed. Heat maps showed the miRNA profiles that were differentially expressed in the EECD-ex + HucMSC group compared with the EEC-ex + HucMSC group (Figure 3A). RT-qPCR showed low expression of miR-214-5p and miR-21-5p in the EECD-ex + HucMSC group, which was consistent with the data from miRNA sequencing (Figure 3B and C). RT-qPCR showed that expression of miR-1303, miR-4521, miR-7974, miR-210-3p, and miR-31-5p was inconsistent with the data from miRNA sequencing (Supplementary Figure 1). We propose that EECD-exs might deliver miRNA into Huc
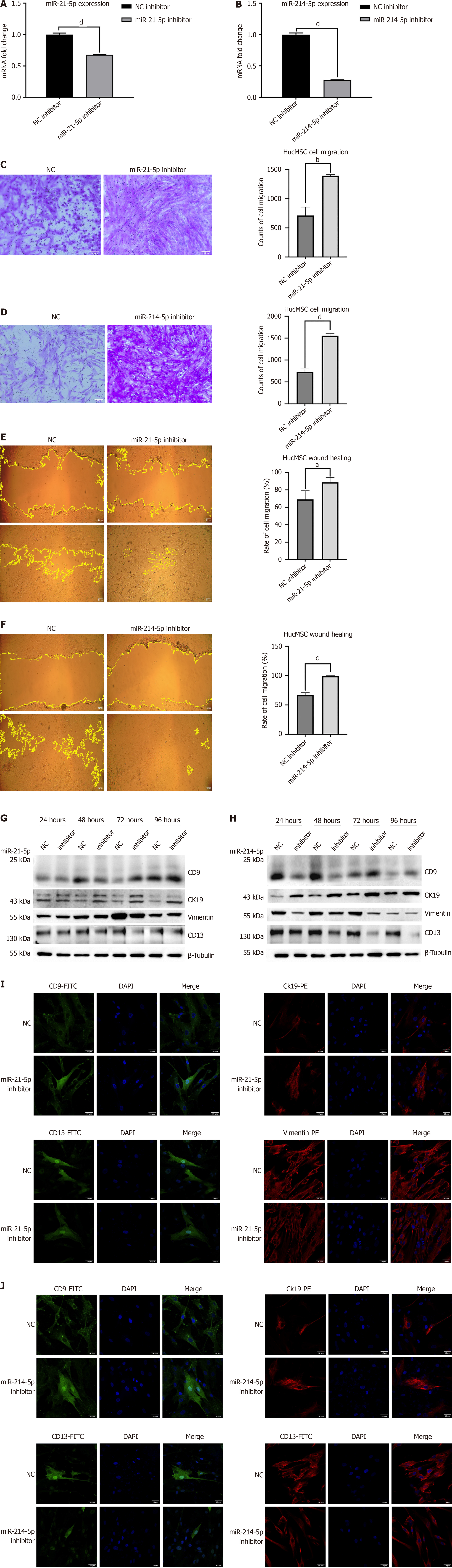
To explore whether miR-21-5p or miR-214-5p is implicated in regulating HucMSC function, miR-21-5p or miR-214-5p inhibitor was transfected into HucMSCs. The successful inhibitor of miR-21-5p or miR-214-5p was validated by RT-qPCR (Figure 4A and B). Transwell assay showed that HucMSC migration was enhanced by miR-21-5p or miR-214-5p inhibitor (Figure 4C and D). Similar results were obtained by wound healing assay (Figure 4E and F). MiR-21-5p or miR-214-5p inhibitor led to elevated expression of CD9 and CK19 and decreased expression of vimentin and CD13 in HucMSCs (Figure 4G and H). Immunofluorescence staining also showed that CD9 and CK19 were increased while CD13 and vimentin were decreased in HucMSCs after miR-21-5p or miR-214-5p silencing (Figure 4I and J). Compared with the NC inhibitor control, after transfection with miR-21-5p or miR-214-5p inhibitor, the alizarin red staining of HucMSCs after induction of osteogenic differentiation was significantly weakened, and the alginate-like polymers level significantly decreased (Supplementary Figure 2A-C and E-G). The five key regulatory genes related to osteogenic differentiation, alginate-like polymers, osteoprotegerin, bone morphogenetic protein 2, osteocalcin, and RunX2, were significantly decreased at both the gene transcription (RT-qPCR) (Supplementary Figure 2D and H) and protein expression (Western blotting) (Supplementary Figure 2I-L) levels. Oil Red O staining of the cells after induction of adipogenic differentiation showed that, compared with the transfection of the NC inhibitor control sequence, after transfection with miR-21-5p or miR-214-5p inhibitor, the size and proportion of lipid droplets in HucMSCs after induction of adipogenic differentiation were significantly decreased (Supplementary Figure 3A-D). The key regulatory genes of adipogenic differentiation, CCAAT/enhancer binding protein beta-α and peroxisome proliferator-activated receptor-γ, were significantly decreased at both the gene transcription (RT-qPCR) and protein expression (Western blotting) (Supplementary Figure 3E-J) levels. The above outcomes indicated that miR-21-5p and miR-214-5p inhibitors promoted HucMSC migration and differentiation into EECs.
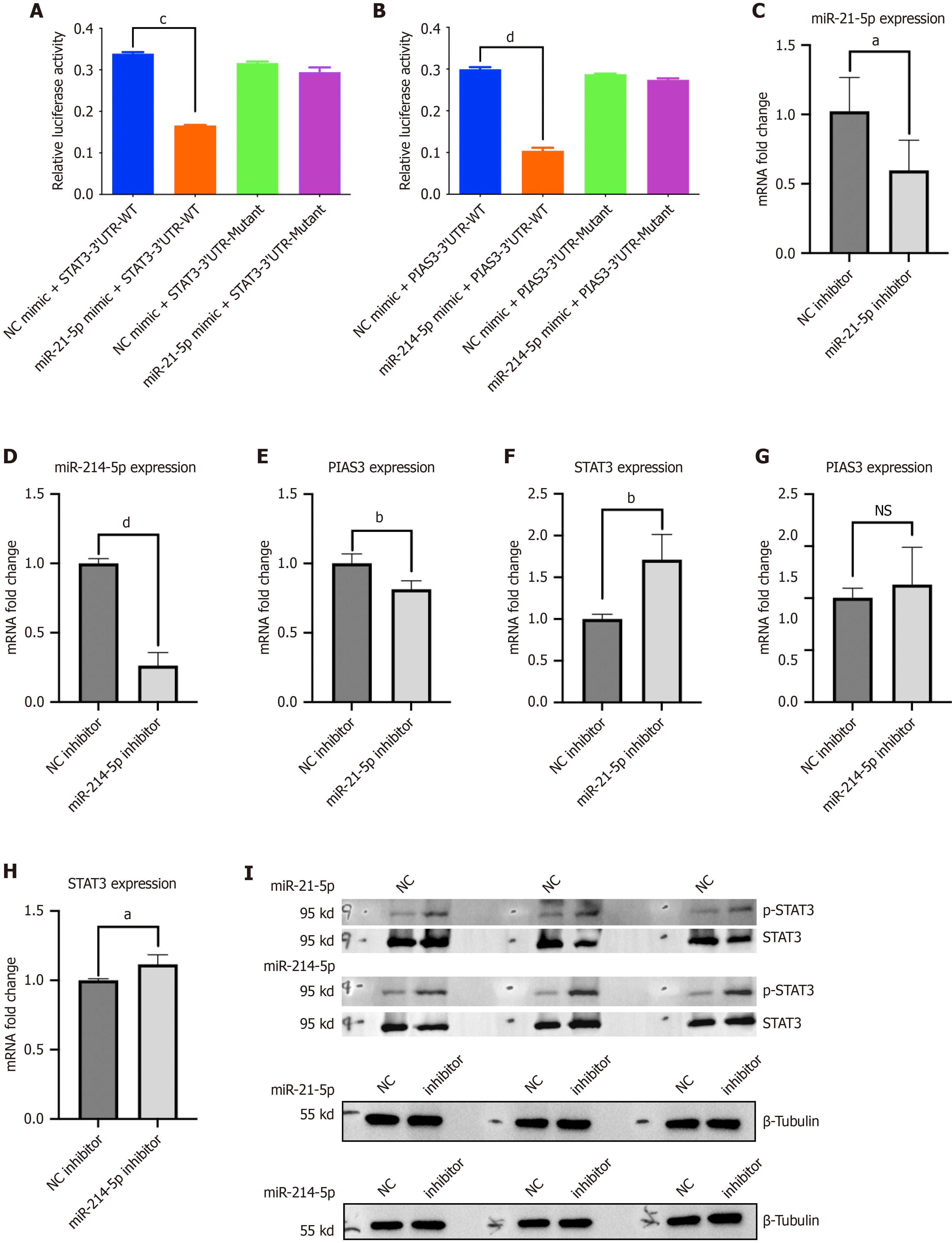
miR-214-5p was uncovered to target PIAS3-3’-UTR and miR-21-5p targets STAT3 3’-UTR, while the binding of miRNAs to mRNA 3’-UTR loosened after 3’-UTR mutation, which was verified by dual-luciferase reporter assay (Figure 5A and B). Subsequently, the silencing efficiency of miR-214-5p and miR-21-5p inhibitors was assessed through RT-PCR. Compared to the NC inhibitor group, the RNA expression level of miR-214-5p in the miR-214-5p inhibitor group was markedly reduced. Similarly, the RNA expression level of miR-21-5p in the miR-21-5p inhibitor group was significantly diminished relative to the NC inhibitor group (Figure 5C and D). MiR-21-5p inhibitor upregulated STAT3 mRNA level but downregulated PIAS3 mRNA level in HucMSCs (Figure 5E and F). MiR-214-5p inhibitor upregulated STAT3 and PIAS3 mRNA levels in HucMSCs (Figure 5G and H). Additionally, phosphorylated (p)-STAT3 protein expression was elevated after miR-214-5p or miR-21-5p silencing in HucMSCs (Figure 5I).
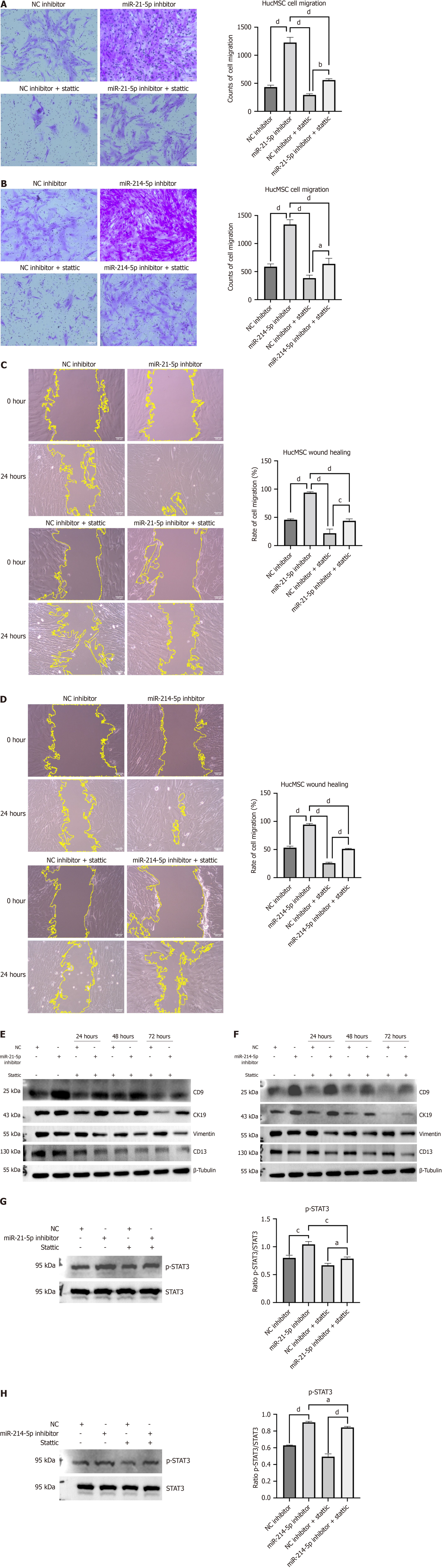
We explored whether miR-214-5p or miR-21-5p mediated the functional alterations of HucMSCs via regulation of STAT3. HucMSC transfected with miR-21-5p or miR-214-5p inhibitor were treated with the STAT3 inhibitor Stattic. Transwell assay showed that miR-214-5p or miR-21-5p inhibitor enhanced HucMSC migration, which was repressed after addition of the STAT3 inhibitor (Figure 6A and B). Wound healing assay also exhibited consistent results with those from the Transwell assay (Figure 6C and D). Compared with the NC inhibitor group, CD9 and CK19 were increased while vi
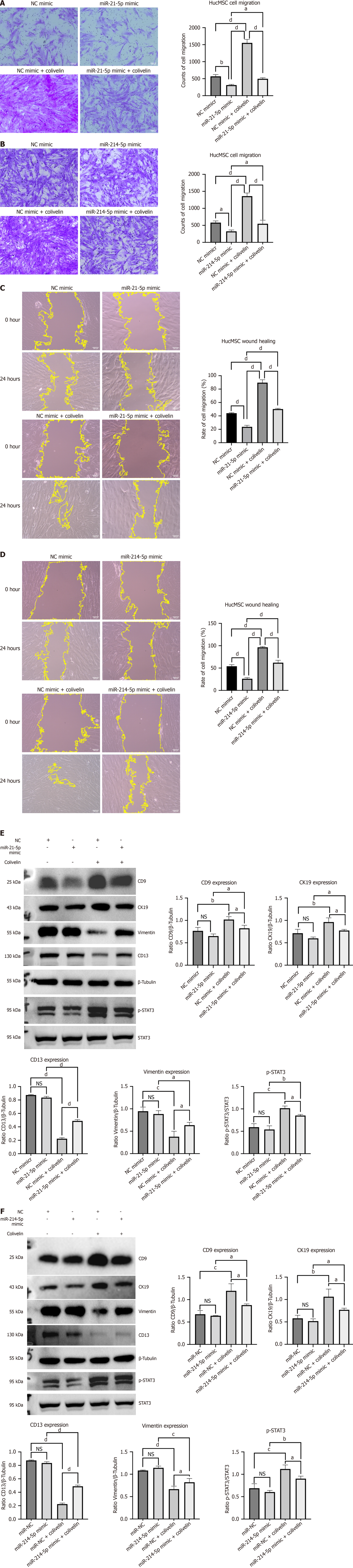
Transwell assay showed that miR-214-5p or miR-21-3p overexpression inhibited HucMSC migration compared with the NC mimic group, whereas addition of the STAT3 activator colivelin enhanced the migration of HucMSCs (Figure 7A and B). Wound healing assay showed similar results to the Transwell assay (Figure 7C and D). Compared with the NC mimic group, no obvious alteration was observed in the expression of CD9, CK19, vimentin, CD13, and p-STAT3 in HucMSCs after miR-21-5p or miR-214-5p overexpression. In contrast, CD9 and CK19 were increased, vimentin and CD13 were decreased, and p-STAT3 was increased in HucMSC treated with the STAT3 activator. Compared with the miR-21-5p or miR-214-5p mimic group, CD9 and CK19 increased, vimentin and CD13 decreased, and p-STAT3 increased in the miR-21-5p mimic + STAT3 activator or miR-214-5p mimic + STAT3 activator group (Figure 7E and F). Activation of STAT3 reversed the effects induced by miR-214-5p or miR-21-5p overexpression on HucMSC function.
The management of thin endometrium for patients undergoing assisted reproduction is a common challenge[3]. A previous study has demonstrated that HucMSC-derived exosomes repair EEC damage caused by hypoxia[18]. Here, we found that exosomes derived from hypoxic EECs promoted HucMSC migration and differentiation into EECs. MiR-21-5p and miR-214-5p were lowly expressed in EECD-ex-pretreated HucMSCs. MiR-21-5p or miR-214-5p silencing promoted HucMSC migration and EEC differentiation via upregulating p-STAT3.
EEC-derived exosomes suppress cell migration and invasion in ovarian endometriosis[9]. Exosomes from bovine EECs ensured trophoblast cell development[10]. Exosomes released from ectopic endometrial cells induce the migration of eutopic endometrial cells[20]. In the present study, coculture with the exosomes derived from hypoxic EECs promoted the migration of HucMSCs. A study has uncovered the capacity of MSCs derived from human umbilical cord Wharton’s jelly to differentiate into EEC-like cells in specific microenvironments[19]. We found that EECD-ex coculture promoted HucMSC differentiation into EECs, as revealed by the elevated CD9 and CK19 and reduced vimentin and CD13. The findings indicated that exosomes secreted from EECs acted as drivers of HucMSC migration and differentiation. We propose that EECD-exs might carry genetic information into HucMSCs, which promotes HucMSC migration and differentiation into EECs.
MiR-214-3p is reduced in endometrial cancer cells and tissues[21], and ectopic lesions and stromal cells in endometriosis[22]. MiR-214 level is reduced in ectopic ESCs[23]. Patients with intrauterine adhesion have a decrease in miR-21-5p in the endometrium[24]. In the present study, differentially expressed miRNAs were screened by miRNA sequencing analysis, and confirmed by RT-qPCR. We observed a low level of miR-214-5p and miR-21-5p in EECD-ex-cocultured HucMSCs. MiR-21-5p is implicated in modulating EEC proliferation and migration[25], and miR-21-5p[26,27] and miR-214-5p[28,29] are involved in regulation of osteogenic differentiation in mesenchymal stromal cells. Importantly, miR-21-5p or miR-214-5p silencing promoted HucMSC migration and differentiation into EECs, whereas miR-214-5p or miR-21-5p overexpression had opposite effects. The present study is believed to be the first to reveal the biological function of miR-214-5p and miR-21-5p in regulating differentiation of HucMSCs into EECs.
Hypoxia in endometriotic lesions induces aberrant activation of the STAT3 signaling pathway[30]. In humans, p-STAT3 is highly expressed in the endometrium from endometriosis patients and in adenomyosis lesions[31,32]. PIAS3 is a negative regulator of STAT3. PIAS3 represses STAT3 transcriptional activity by binding to the STAT3 DNA-binding domain[33]. Attenuation of PIAS3 increases p-STAT3 levels, causing aberrant STAT3 activation in endometriosis[34]. miRDB and TargetScan predicted that PIAS3 and STAT3 are the latent targets of miR-214-5p and miR-21-5p, respectively. A previous study has verified the binding of miR-21-5p to STAT3, and that miR-21-5p negatively regulates STAT3 expression[35,36]. We also verified the direct relationship between STAT3 and miR-21-5p. The direct interaction between PIAS3 and miR-214-5p was also validated. MiR-214-5p and miR-21-5p inhibitors downregulated PIAS3 mRNA and upregulated STAT3 mRNA and p-STAT3 protein in HucMSCs. A previous study has revealed that STAT3 is activated in myofibroblast differentiation of endometrial MSCs[37]. Importantly, inhibition of STAT3 reversed the effects induced by miR-21-5p or miR-214-5p inhibitor on HucMSC function. Functional assays using STAT3 activator and miR-21-5p/miR-214-5p mimics also displayed consistent outcomes. We propose that miR-21-5p or miR-214-5p inactivates the STAT3 signaling pathway to regulate the function of HucMSCs.
This research demonstrated that lowly expressed miR-214-5p and miR-21-5p in hypoxic-EEC-derived exosomes might activate the STAT3 signaling pathway to promote HucMSC migration and differentiation into EECs. Therapeutics to target and regulate miR-214-5p and miR-21-5p expression may be helpful in treating thin endometrium.
| 1. | Kliman HJ, Frankfurter D. Clinical approach to recurrent implantation failure: evidence-based evaluation of the endometrium. Fertil Steril. 2019;111:618-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 2. | Lin J, Wang Z, Huang J, Tang S, Saiding Q, Zhu Q, Cui W. Microenvironment-Protected Exosome-Hydrogel for Facilitating Endometrial Regeneration, Fertility Restoration, and Live Birth of Offspring. Small. 2021;17:e2007235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 106] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 3. | Liu KE, Hartman M, Hartman A. Management of thin endometrium in assisted reproduction: a clinical practice guideline from the Canadian Fertility and Andrology Society. Reprod Biomed Online. 2019;39:49-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 4. | Gharibeh N, Aghebati-Maleki L, Madani J, Pourakbari R, Yousefi M, Ahmadian Heris J. Cell-based therapy in thin endometrium and Asherman syndrome. Stem Cell Res Ther. 2022;13:33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 56] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 5. | Kowalczyk A, Wrzecińska M, Czerniawska-Piątkowska E, Kupczyński R. Exosomes - Spectacular role in reproduction. Biomed Pharmacother. 2022;148:112752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 57] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 6. | Mishra A, Ashary N, Sharma R, Modi D. Extracellular vesicles in embryo implantation and disorders of the endometrium. Am J Reprod Immunol. 2021;85:e13360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 7. | Bjorkman S, Taylor HS. MicroRNAs in endometriosis: biological function and emerging biomarker candidates†. Biol Reprod. 2019;100:1135-1146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 8. | Ghafouri-Fard S, Niazi V, Taheri M. Role of miRNAs in conveying message of stem cells via extracellular vesicles. Exp Mol Pathol. 2020;117:104569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 9. | Zhang M, Wang X, Xia X, Fang X, Zhang T, Huang F. Endometrial epithelial cells-derived exosomes deliver microRNA-30c to block the BCL9/Wnt/CD44 signaling and inhibit cell invasion and migration in ovarian endometriosis. Cell Death Discov. 2022;8:151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 10. | Wang X, Li Q, Xie T, Yuan M, Sheng X, Qi X, Xing K, Liu F, Guo Y, Xiao L, Ni H. Exosomes from bovine endometrial epithelial cells ensure trophoblast cell development by miR-218 targeting secreted frizzled related protein 2. J Cell Physiol. 2021;236:4565-4579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 11. | Wang X, Yao X, Xie T, Chang Z, Guo Y, Ni H. Exosome-derived uterine miR-218 isolated from cows with endometritis regulates the release of cytokines and chemokines. Microb Biotechnol. 2020;13:1103-1117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 12. | Xie Q, Liu R, Jiang J, Peng J, Yang C, Zhang W, Wang S, Song J. What is the impact of human umbilical cord mesenchymal stem cell transplantation on clinical treatment? Stem Cell Res Ther. 2020;11:519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 186] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 13. | Hoang DM, Pham PT, Bach TQ, Ngo ATL, Nguyen QT, Phan TTK, Nguyen GH, Le PTT, Hoang VT, Forsyth NR, Heke M, Nguyen LT. Stem cell-based therapy for human diseases. Signal Transduct Target Ther. 2022;7:272. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 349] [Cited by in RCA: 633] [Article Influence: 158.3] [Reference Citation Analysis (0)] |
| 14. | Zhang L, Li Y, Guan CY, Tian S, Lv XD, Li JH, Ma X, Xia HF. Therapeutic effect of human umbilical cord-derived mesenchymal stem cells on injured rat endometrium during its chronic phase. Stem Cell Res Ther. 2018;9:36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 152] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 15. | Zhang L, Li Y, Dong YC, Guan CY, Tian S, Lv XD, Li JH, Su X, Xia HF, Ma X. Transplantation of umbilical cord-derived mesenchymal stem cells promotes the recovery of thin endometrium in rats. Sci Rep. 2022;12:412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 16. | Zhou S, Lei Y, Wang P, Chen J, Zeng L, Qu T, Maldonado M, Huang J, Han T, Wen Z, Tian E, Meng X, Zhong Y, Gu J. Human Umbilical Cord Mesenchymal Stem Cells Encapsulated with Pluronic F-127 Enhance the Regeneration and Angiogenesis of Thin Endometrium in Rat via Local IL-1β Stimulation. Stem Cells Int. 2022;2022:7819234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 17. | Saad-Naguib MH, Kenfack Y, Sherman LS, Chafitz OB, Morelli SS. Impaired receptivity of thin endometrium: therapeutic potential of mesenchymal stem cells. Front Endocrinol (Lausanne). 2023;14:1268990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 18. | Liang L, Wang L, Zhou S, Li J, Meng L, Zhang H, Cui C, Zhang C. Exosomes derived from human umbilical cord mesenchymal stem cells repair injured endometrial epithelial cells. J Assist Reprod Genet. 2020;37:395-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 19. | Shi Q, Gao J, Jiang Y, Sun B, Lu W, Su M, Xu Y, Yang X, Zhang Y. Differentiation of human umbilical cord Wharton's jelly-derived mesenchymal stem cells into endometrial cells. Stem Cell Res Ther. 2017;8:246. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 46] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 20. | Abudula M, Fan X, Zhang J, Li J, Zhou X, Chen Y. Ectopic Endometrial Cell-Derived Exosomal Moesin Induces Eutopic Endometrial Cell Migration, Enhances Angiogenesis and Cytosolic Inflammation in Lesions Contributes to Endometriosis Progression. Front Cell Dev Biol. 2022;10:824075. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 21. | Fang YY, Tan MR, Zhou J, Liang L, Liu XY, Zhao K, Bao EC. miR-214-3p inhibits epithelial-to-mesenchymal transition and metastasis of endometrial cancer cells by targeting TWIST1. Onco Targets Ther. 2019;12:9449-9458. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 22. | Zhang Y, Chang X, Wu D, Deng M, Miao J, Jin Z. Down-regulation of Exosomal miR-214-3p Targeting CCN2 Contributes to Endometriosis Fibrosis and the Role of Exosomes in the Horizontal Transfer of miR-214-3p. Reprod Sci. 2021;28:715-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 23. | Wu D, Lu P, Mi X, Miao J. Exosomal miR-214 from endometrial stromal cells inhibits endometriosis fibrosis. Mol Hum Reprod. 2018;24:357-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 24. | Song M, Zhao G, Sun H, Yao S, Zhou Z, Jiang P, Wu Q, Zhu H, Wang H, Dai C, Wang J, Li R, Cao Y, Lv H, Liu D, Dai J, Zhou Y, Hu Y. circPTPN12/miR-21-5 p/∆Np63α pathway contributes to human endometrial fibrosis. Elife. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 25. | Hua R, Zhang X, Li W, Lian W, Liu Q, Gao D, Wang Y, Lei M. Ssc-miR-21-5p regulates endometrial epithelial cell proliferation, apoptosis and migration via the PDCD4/AKT pathway. J Cell Sci. 2020;133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 26. | Valenti MT, Deiana M, Cheri S, Dotta M, Zamboni F, Gabbiani D, Schena F, Dalle Carbonare L, Mottes M. Physical Exercise Modulates miR-21-5p, miR-129-5p, miR-378-5p, and miR-188-5p Expression in Progenitor Cells Promoting Osteogenesis. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 27. | Sikora M, Śmieszek A, Pielok A, Marycz K. MiR-21-5p regulates the dynamic of mitochondria network and rejuvenates the senile phenotype of bone marrow stromal cells (BMSCs) isolated from osteoporotic SAM/P6 mice. Stem Cell Res Ther. 2023;14:54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 28. | He Q, Li R, Hu B, Li X, Wu Y, Sun P, Jia Y, Guo Y. Stromal cell-derived factor-1 promotes osteoblastic differentiation of human bone marrow mesenchymal stem cells via the lncRNA-H19/miR-214-5p/BMP2 axis. J Gene Med. 2021;23:e3366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 29. | Li L, Fang J, Liu Y, Xiao L. LncRNA LOC100506178 promotes osteogenic differentiation via regulating miR-214-5p-BMP2 axis in human bone marrow mesenchymal stem cells. PeerJ. 2020;8:e8909. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 30. | Hsiao KY, Chang N, Tsai JL, Lin SC, Tsai SJ, Wu MH. Hypoxia-inhibited DUSP2 expression promotes IL-6/STAT3 signaling in endometriosis. Am J Reprod Immunol. 2017;78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 31. | Hiraoka T, Hirota Y, Aikawa S, Iida R, Ishizawa C, Kaku T, Hirata T, Fukui Y, Akaeda S, Matsuo M, Shimizu-Hirota R, Takeda N, Osuga Y. Constant Activation of STAT3 Contributes to the Development of Adenomyosis in Females. Endocrinology. 2022;163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 32. | Kim BG, Yoo JY, Kim TH, Shin JH, Langenheim JF, Ferguson SD, Fazleabas AT, Young SL, Lessey BA, Jeong JW. Aberrant activation of signal transducer and activator of transcription-3 (STAT3) signaling in endometriosis. Hum Reprod. 2015;30:1069-1078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 93] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 33. | Chung CD, Liao J, Liu B, Rao X, Jay P, Berta P, Shuai K. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278:1803-1805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 735] [Cited by in RCA: 751] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 34. | Yoo JY, Jeong JW, Fazleabas AT, Tayade C, Young SL, Lessey BA. Protein Inhibitor of Activated STAT3 (PIAS3) Is Down-Regulated in Eutopic Endometrium of Women with Endometriosis. Biol Reprod. 2016;95:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 35. | Luo H, Wei J, Wu S, Zheng Q, Zhang N, Chen P. Exploring CircRNA N6-methyladenosine in human rheumatoid arthritis: Hyper-methylated hsa_circ_0007259 as a potential biomarker and its involvement in the hsa_circ_0007259/hsa_miR-21-5p/STAT3 axis. Int Immunopharmacol. 2023;124:110938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 36. | Huo J, Liu T, Li F, Song X, Hou X. MicroRNA215p protects melanocytes via targeting STAT3 and modulating Treg/Teff balance to alleviate vitiligo. Mol Med Rep. 2021;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 37. | Zhang Z, Wang J, Chen Y, Suo L, Chen H, Zhu L, Wan G, Han X. Activin a promotes myofibroblast differentiation of endometrial mesenchymal stem cells via STAT3-dependent Smad/CTGF pathway. Cell Commun Signal. 2019;17:45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 71] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/