Published online Aug 26, 2023. doi: 10.4252/wjsc.v15.i8.807
Peer-review started: March 28, 2023
First decision: June 25, 2023
Revised: June 29, 2023
Accepted: July 14, 2023
Article in press: July 14, 2023
Published online: August 26, 2023
Processing time: 149 Days and 21.6 Hours
Bone marrow mesenchymal stromal cells (BMSCs) are the commonly used seed cells in tissue engineering. Aryl hydrocarbon receptor (AhR) is a transcription factor involved in various cellular processes. However, the function of constitutive AhR in BMSCs remains unclear.
To investigate the role of AhR in the osteogenic and macrophage-modulating potential of mouse BMSCs (mBMSCs) and the underlying mechanism.
Immunochemistry and immunofluorescent staining were used to observe the expression of AhR in mouse bone marrow tissue and mBMSCs. The overexpression or knockdown of AhR was achieved by lentivirus-mediated plasmid. The osteogenic potential was observed by alkaline phosphatase and alizarin red staining. The mRNA and protein levels of osteogenic markers were detected by quantitative polymerase chain reaction (qPCR) and western blot. After coculture with different mBMSCs, the cluster of differentiation (CD) 86 and CD206 expressions levels in RAW 264.7 cells were analyzed by flow cytometry. To explore the underlying molecular mechanism, the interaction of AhR with signal transducer and activator of transcription 3 (STAT3) was observed by co-immunoprecipitation and phosphorylation of STAT3 was detected by western blot.
AhR expressions in mouse bone marrow tissue and isolated mBMSCs were detected. AhR overexpression enhanced the osteogenic potential of mBMSCs while AhR knockdown suppressed it. The ratio of CD86+ RAW 264.7 cells cocultured with AhR-overexpressed mBMSCs was reduced and that of CD206+ cells was increased. AhR directly interacted with STAT3. AhR overexpression increased the phosphorylation of STAT3. After inhibition of STAT3 via stattic, the promotive effects of AhR overexpression on the osteogenic differentiation and macrophage-modulating were partially counteracted.
AhR plays a beneficial role in the regenerative potential of mBMSCs partially by increasing phosphorylation of STAT3.
Core Tip: Aryl hydrocarbon receptor (AhR) was positively expressed in murine bone marrow tissue and bone marrow mesenchymal stromal cells (BMSCs). In vitro, overexpression of AhR enhanced the osteogenic potential of mouse BMSCs. Additionally, AhR-overexpressed BMSCs had an increased ability to polarize macrophages to an anti-inflammatory phenotype. While knockdown of AhR showed the opposite effects. Mechanistically, the beneficial effects of AhR were partially dependent on increased phosphorylation of signal transducer and activator of transcription 3. This study suggests that AhR might be a target for achieving optimal bone regeneration in mouse BMSCs-based tissue engineering.
- Citation: Huang J, Wang YN, Zhou Y. Constitutive aryl hydrocarbon receptor facilitates the regenerative potential of mouse bone marrow mesenchymal stromal cells. World J Stem Cells 2023; 15(8): 807-820
- URL: https://www.wjgnet.com/1948-0210/full/v15/i8/807.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v15.i8.807
Cell-based tissue engineering is an important method for the treatment of bone defects. Bone marrow mesenchymal stromal cells (BMSCs) are one of the most commonly used seed cells. The osteogenic potential of BMSCs is the premise of their applications in bone regeneration[1]. Additionally, the crosstalk between BMSCs and immune cells like macrophages has been recognized as a critical element in achieving ideal bone tissue repair[2]. Plenty studies have demonstrated that BMSCs are able to trigger a functional switch in macrophages from pro-inflammatory classically activated macrophages (M1) to anti-inflammatory alternatively activated macrophages (M2)[3]. In turn, macrophage polarization is essential for the osteogenic potency of BMSCs. M2 macrophages promote the osteogenesis of BMSCs by secreting pro-regenerative cytokines[4], while M1 macrophages suppress the process[5]. Accordingly, approaches to enhance the osteogenic potential and macrophage-modulating capacity of BMSCs, such as genetic engineering to express specific genes, are continuously being explored.
Aryl hydrocarbon receptor (AhR) is a member of the helix-loop-helix transcription factor superfamily[6]. Historically, AhR has been recognized as a nuclear receptor that responds to environmental toxic stimuli. Recently, increasing number of studies have demonstrated that AhR is an essential modulator in bone turnover[7] and immune responses[8]. AhR can be activated by chemosynthetic agonists such 6-formyl (3,2-b) carbazole (FICZ)[9]. In our previous studies, AhR signaling was suppressed in periodontitis, and FICZ alleviated the inflammatory responses by activating AhR and promoting the phosphorylation of signal transducer and activator of transcription 3 (STAT3)[10]. In another study of our group, FICZ was found to play a beneficial role in the proliferation, osteogenic potential and macrophage-modulation of rat BMSCs and primed cartilage templates[11].
Except for ligand-activated AhR, the role of constitutive unligated AhR in the osteogenic and macrophage-modulating potential of BMSCs has not been investigated. Therefore, the aim of the present study was to: (1) Establish stable AhR-overexpressing or AhR-knockdown mouse BMSCs (mBMSCs); (2) explore the osteogenic differentiation of different mBMSCs; (3) observe the phenotype of macrophages cocultured with different mBMSCs; and (4) investigate the involved molecular mechanism.
Six-week-old male C57BL/6 mice were obtained from the Hubei Research Centre of Laboratory Animals (Wuhan, China) and kept in specific pathogen free condition. All experimental protocols were approved by the Institutional Animal Care and Use Committee of School and Hospital of Stomatology, Wuhan University (No. 2020-A08).
BMSCs was isolated from 6-week-old male C57BL/6 mice via whole femur bone marrow adherent culturing. The femora were excised aseptically, cleaned of soft tissues, and passed through 3 washes with phosphate buffered saline (PBS). The ends of the bones were removed, and the marrow flushed out. The released cells were collected in two 75 cm2 flasks (Corning) containing 10 mL of 10% fetal bovine serum (Hyclone) in -minimum essential medium. Cultures were maintained in a humidified atmosphere of 95% air and 5% CO2, at 37°C. After 72 h, all medium was aspirated and replaced to remove the non-adherent cells. The medium was replaced every 2–3 d.
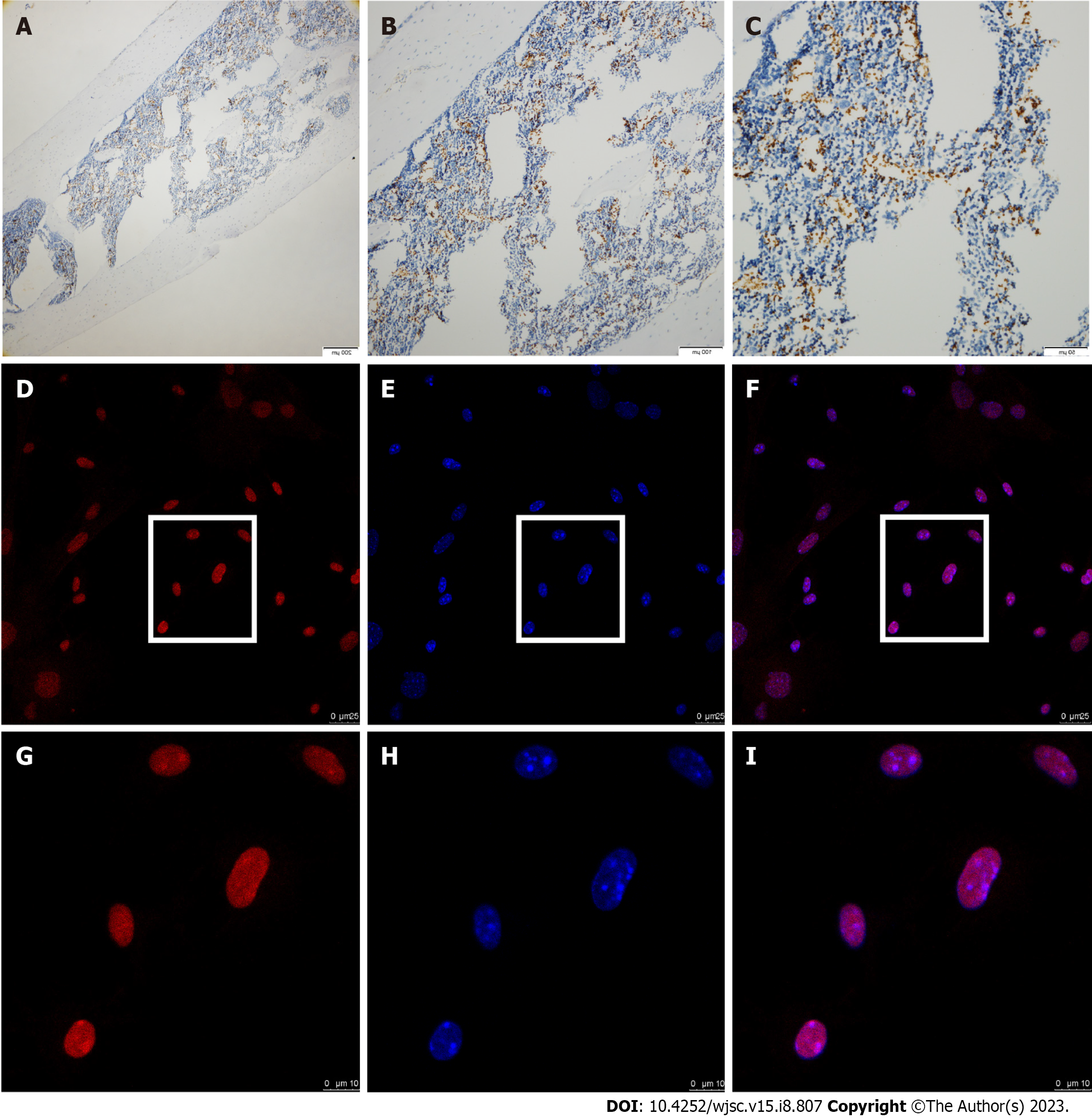
The mice femurs were collected and fixed with 4% paraformaldehyde and then decalcified in 10% ethylene diamine tetraacetic acid for 6 wk. The tissue was subsequently processed for paraffin embedding and serial 4-mm-thick sections were prepared. Then the sections were dewaxed in xylene and rehydrated through graded ethanol to water. Antigen retrieval was conducted in stomach enzyme antigen repair solution for 30 min at 37 °C. Immunostaining was performed by incubating the sections with anti-AhR (NB300-515, 1:200, Novus) at 4 °C overnight. The slides were then washed with PBS and incubated with secondary antibody (Maxim Biotechnology) for 30 min at 37 °C. Staining was visualized with 3, 3-diaminobenzidine and counterstained with hematoxylin.
BMSCs at third passage were seeded in cell dish (801002, NEST). After the cells reached 80% confluence, BMSCs were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 for 20min. Then the cells were blocked with bovine serum albumin (BSA) for 1 h. Subsequently, the cells were incubated with primary antibody against AhR (NB300-515, 1:100, Novus) at 4 °C overnight. After washing, cells were incubated with dylight 594-conjugated secondary antibody (1:200, A23420, Abbkine) for 1 h at room temperature. Then, the cells were stained with 4',6-diamidino-2-phenylindole staining solution (C1005, Beyotime) for 5 min. Finally, the stained cells were observed and photographed under confocal microscope (Leica-LCS-SP8-STED).
Based on the published sequence of mouse AhR (NM_013464), certain short hairpin RNA (shRNAs) specifically targeting AhR were designed to knockdown their expressions in mBMSCs. The shRNA sequences are as follows: 5’-CATCGACATAACGGACGAAAT-3’ (AhR sense) and 5’-ATTTCGTCCGTTATGTCGATG–3’ (antisense). These plasmid DNAs transcribed shRNAs with loop sequences of 5’-CTCGAG–3’. In parallel, a negative control (NC) sequences were projected (sense: 5’-TTCTCCGAACGTGTCACGT-3’, antisense: 5’-ACGTGACACGTTCGGAGAA-3’), which had no homology with human proteins. The generated oligo DNA was cloned into GV493 vector (hU6-MCS-CBh-gcGFP-IRES-puromycin) (GENECHEM, Shanghai).
The coding sequences of mouse AhR (NM_013464) was cloned into GV358 vector (Ubi-MCS-3FLAG-SV40-EGFP-IRES-puromycin) (GENECHEM, Shanghai). The empty Ubi-MCS-SV40-EGFP-IRES-puromycin vector was served as NC.
The 293T packaging cell line was co-transfected with plasmid and lentiviral helper vectors (GENECHEM, Shanghai). The medium was replaced with fresh dulbecco’s modified eagle medium overnight. After another 48 h, the supernatants were collected and filtered. The viral supernatants were concentrated via ultracentrifuging at the speed of 25000 rpm for 2 h at 4°C. One day prior to infection, mBMSCs were seeded in 6-well plates at the density of 105 cells/well. Then the mBMSCs were infected with lentiviral particles containing plasmids of knockdown-NC (sh-NC), knockdown-AhR (sh-AhR), overexpression-NC (oe-NC) or overexpression-AhR (oe-AhR) via multiplicity of infection level of 20. After 12-16 h, the culture medium was refreshed. Then the cells were cultured in complete medium containing 2 mg/mL puromycin.
Total RNA from different cell samples were isolated by Trizol reagent (Takara Bio). SYBR Green Reagent (Takara Bio) was used to perform qPCR in a 7500 Fast Real-Time PCR system (Applied Biosystem). The primer sequences used in the study was showed in Table 1. The relative expression levels were calculated using the 2-DDCt method. Three biological replicates were conducted.
| Gene | Sequences |
| GAPDH | Forward: TGGAAAGCTGTGGCGTGAT |
| Reverse: GTCATCATACTTGGCAGGTTTCT | |
| AhR | Forward: GGCTTTCAGCAGTCTGATGTC |
| Reverse: CATGAAAGAAGCGTTCTCTGG | |
| ALPL | Forward: GGGCGTCTCCACAGTAACCG |
| Reverse: ACTCCCACTGTGCCCTCGTT | |
| RUNX2 | Forward: GAGTCAGATTACAGATCCCA |
| Reverse: TGGCTCTTCTTACTGAGAGA |
Total proteins were extracted from cells with radio immunoprecipitation assay buffer supplemented with 1:100 proteinase and phosphatase inhibitors. The proteins were separated by 10% sodium dodecyl sulphate (SDS)
For osteogenic induction, mBMSCs at passage 3 from different groups (sh-NC, sh-AhR, oe-NC or oe-AhR) were cultured in medium supplemented with 50 mg/mL ascorbic acid, 10 mmol/L b-glycerophosphate and 10-8 mol/L dexamethasone (Sigma). Then, cells were fixed in 4% paraformaldehyde for 10 min and stained using alkaline phosphatase (ALP) Color Development Kit (C3206, Beyotime) following the manufacturer’s instructions. For alizarin red staining (ARS), cells were fixed and stained with alizarin red solution (Cyagen) for 10 min. Three biological replicates were conducted.
For direct coculture, mBMSCs from all groups (sh-NC, sh-AhR, oe-NC or oe-AhR) were plated in monoculture at the density of 5 105 cells/well at 6-well plate. After the adherence of mBMSCs, the macrophage linage RAW264.7 cells (ScienceCell, Shanghai) (1 105 cells/well) were seeded into the wells. The cells were harvested after 24 h of coculture.
For indirect coculture, conditional medium from the culture of sh-NC, sh-AhR, oe-NC or oe-AhR mBMSCs were collected. The conditioned medium was harvested and centrifuged for 10 min at 1000 rpm and then frozen at -20 °C until used. The RAW 264.7 was treated with 1:2 fresh medium and conditioned medium for 24 h.
The macrophage surface markers of RAW264.7 cells in the coculture system was detected by flow cytometry. Three biological replicates were conducted.
For direct coculture, the mBMSCs and RAW264.7 cells were collected from 6-well plates after coculture for 24 h and washed three times with PBS. The cell suspensions were divided into 1.5 mL Eppendorf micro test tubes (EP tubes) and incubated with blocking 3% BSA. Then the cell suspensions were incubated with allophycocyanin anti-F4/80 (157305, BioLegend), combined with fluorescein isothiocyanate (FITC) anti-cluster of differentiation (CD) 86 (105005, BioLegend) or PerCP/Cy5.5 anti-CD206 (141715, BioLegend) at 4°C for 1 h.
For indirect coculture, the RAW264.7 cells were collected and washed three times with PBS. The cell suspensions were divided into 1.5 mL EP tubes and incubated with blocking 2% BSA. Then the cell suspensions were successively incubated with CD86 (13395-1-AP, Proteintech) or CD206 (18704-1-AP, Proteintech), and FITC conjugated immunoglobulin G (BA1105, Boster).
The above cells were washed three times after incubation and suspended in 500 mL containing 3% fetal bovine serum and then detected by Beckman Coulter CytoFLEX S and analyzed by CytoExpert.
The AhR and p65 protein-protein interaction was detected by co-immunoprecipitation (Co-IP) kit (P2179S, Beyotime) following manufacturer’s instructions. The mBMSCs protein sample was extracted from cells with lysis buffer supplemented with protease inhibitor cocktail. The AhR antibody (NB300-515, 1:100, Novus) was incubated with protein A+G beads at room temperature for 2 h and then washed three times using Tris buffered saline. Then the beads-antibody complex was immunoprecipitated with mBMSCs protein sample at 4°C overnight. After washed three times using lysis buffer, the beads-antibody-antigen complex was eluted SDS-polyacrylamide gel electrophoresis sample loading buffer. After magnetic separation, the expression of STAT3 in the supernatant was detected using STAT3 primary antibody (9139, 1:1000, CST) via western blot. Three biological replicates were conducted.
To testify whether the effects of AhR in osteogenic differentiation and macrophage-modulating was partially STAT3-dependent or not, a specific STAT3 inhibitor stattic (Selleck) was introduced. In osteogenic induction, oe-NC and oe-AhR mBMSCs were cultured in osteogenic medium supplemented with or without 2 mmol/L stattic. For indirect coculture system of mBMSCs and RAW 264.7 cells, the mixed fresh and conditioned medium from mBMSCs was supplemented with or without 2 mmol/L stattic. Three biological replicates were conducted for each treatment.
All data were expressed as the mean ± SD. For comparison between two groups, statistical differences were evaluated by a two-tailed Student’s t test. For multiple comparisons, a one-way analysis of variance (ANOVA) followed by Tukey’s test were conducted. P-value < 0.05 was considered statistically significant.
The immunohistochemistry (IHC) staining showed that AhR was expressed in parts of mouse femur bone marrow (Figure 1A-C). In isolated mBMSCs derived from femurs, AhR expression was positively observed via immunofluorescence staining (Figure 1D-I), which was consistent with the IHC results.
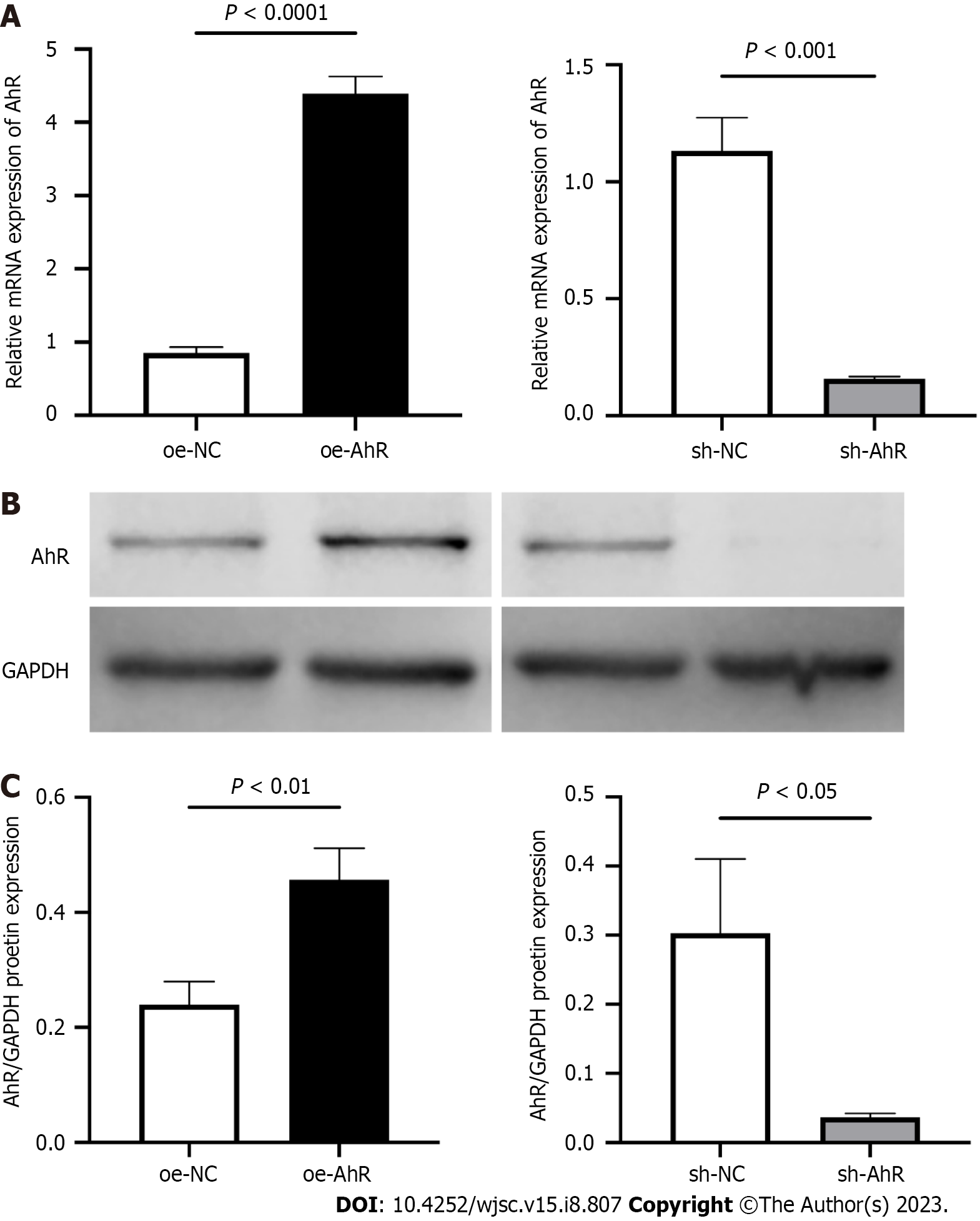
At 48 h after infection with oe-AhR or sh-AhR and their NC lentivirus (oe-NC and sh-NC), the efficiencies of AhR overexpression or knockdown were confirmed by qPCR and western blot. The qPCR data (Figure 2A) showed that the relative AhR mRNA expression level of the oe-NC and oe-AhR groups were 0.85 ± 0.08 and 4.39 ± 0.23 respectively, and those of the sh-NC and sh-AhR groups were 1.13 ± 0.14 and 0.16 ± 0.01, respectively. The western blot results (Figure 2B and C) demonstrated that the relative AhR/GAPDH protein levels of the oe-NC and oe-AhR groups were (0.29 ± 0.05) and (0.45 ± 0.07), and those of the sh-NC and sh-AhR groups were 0.31 ± 0.11 and 0.04 ± 0.01 respectively. The data indicated that AhR was significantly overexpressed or knocked down in oe-AhR or sh-AhR mBMSCs.
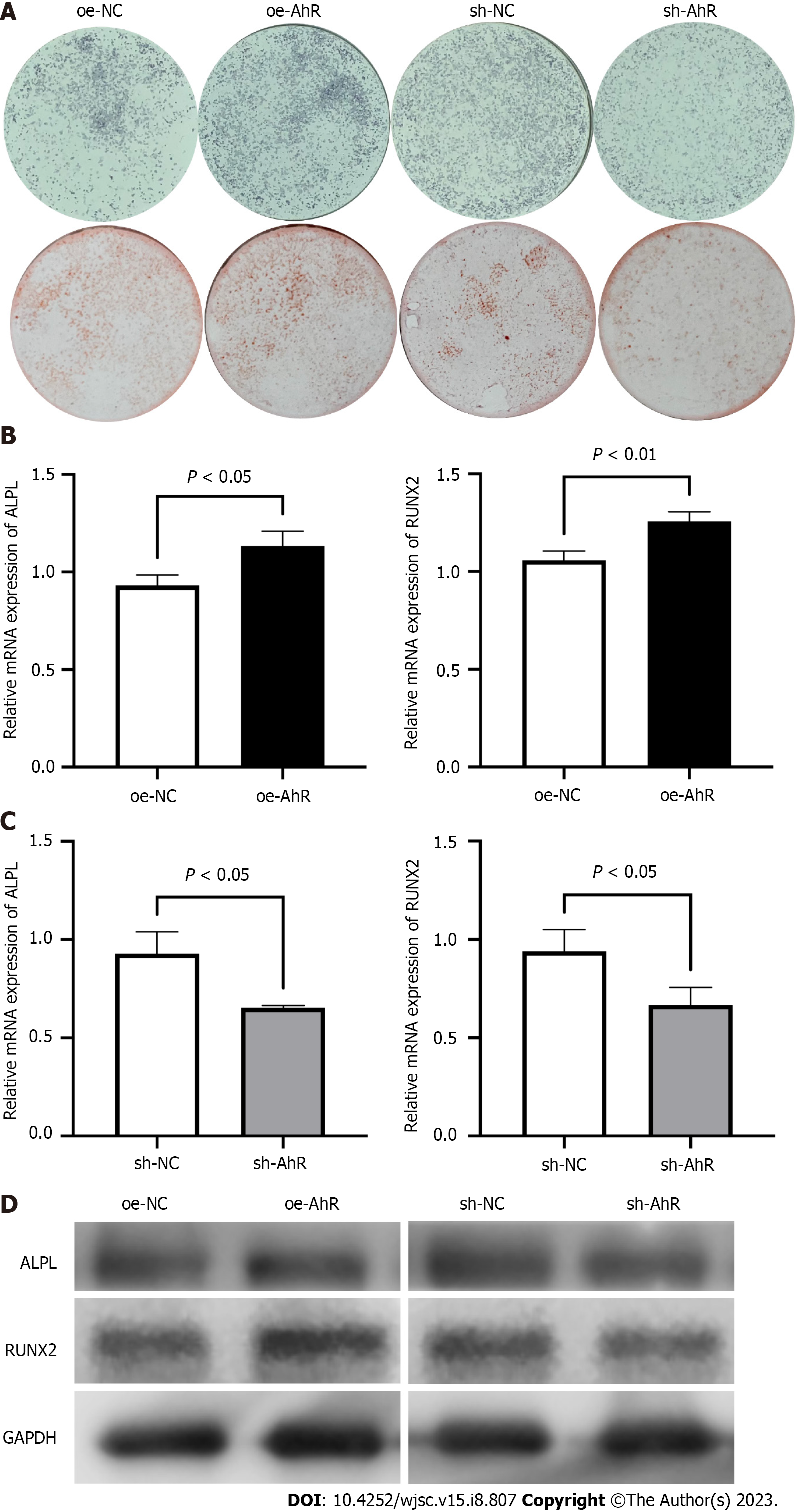
At the 7th day of osteogenic induction, ALP staining showed that AhR overexpression resulted in more positive nodules than the NC (Figure 3A). Consistently, after 14 d of osteogenic induction, the visualization of calcium deposits and mineralized nodules by ARS also demonstrated that AhR overexpression promoted the osteogenic potential of mBMSCs (Figure 3A). In contrast, AhR knockdown suppressed the nodules formation and calcium deposition, as showed by ALP staining and ARS, compared to the NC (Figure 3A).
At the 7th day of osteogenic induction, the mRNA levels of the osteogenic markers ALP, ALPL and RUNX2 in oe-AhR mBMSCs were significantly higher than those in oe-NC mBMSCs (Figure 3B), while those in sh-AhR cells were lower than those in sh-NC cells (Figure 3C). Then, the protein samples were harvested after 7 d of osteogenic induction and were subjected to western blotting. The bands showed increased protein expressions of ALPL and RUNX2 in oe-AhR mBMSCs compared to oe-NC mBMSCs, while sh-AhR inhibited their expressions compared to sh-NC (Figure 3D), which was basically consistent with the mRNA results.
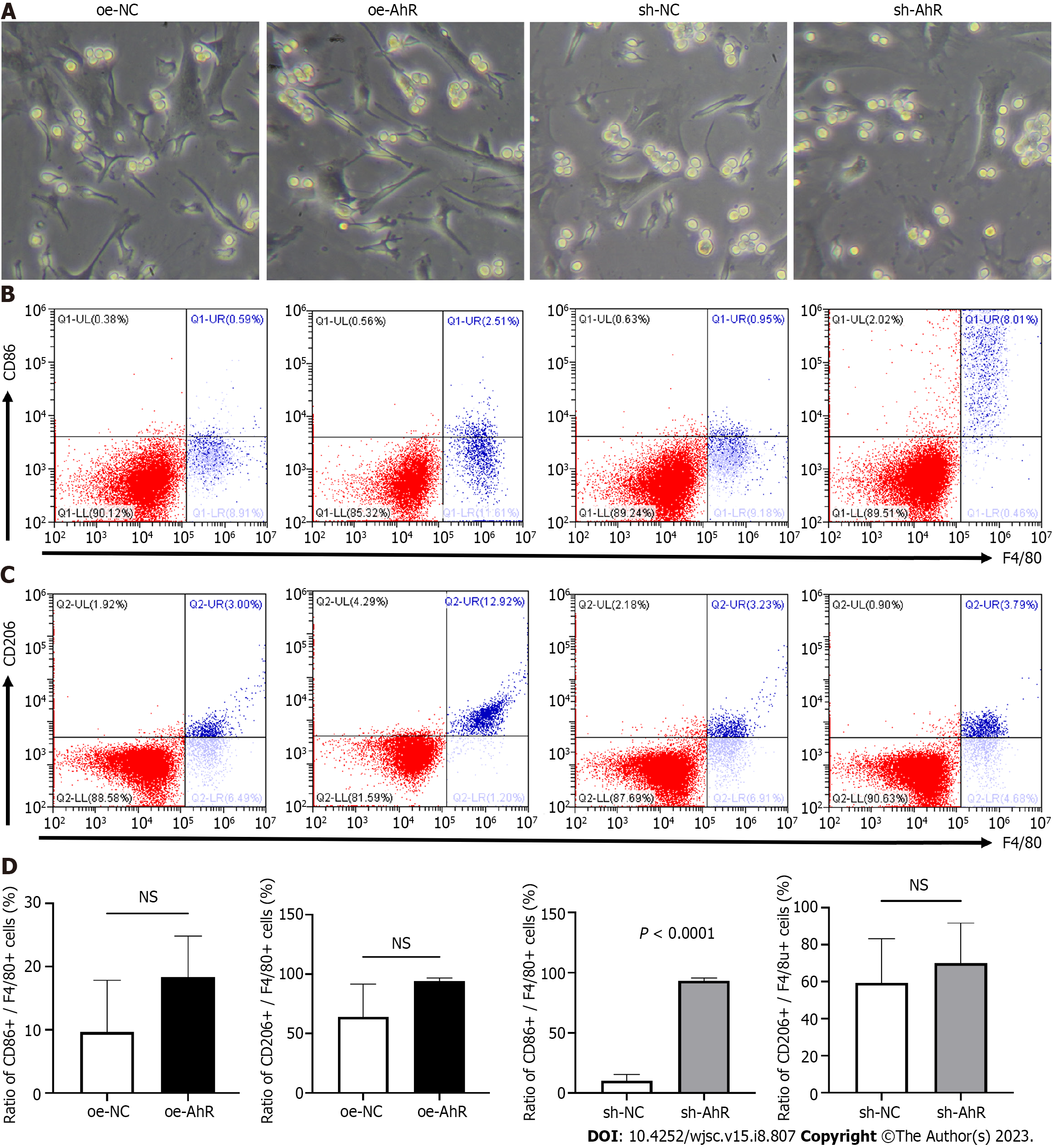
Brightfield images of direct coculture were captured prior to analysis (Figure 4A), and the morphologies of the RAW 264.7 cells and different groups of mBMSCs were similar. In flow cytometry analysis of direct coculture samples, the ratio of M1-like macrophages among RAW 264.7 cells was calculated as CD86 + (Q1-UR)/F4/80 + (Q1-UR + Q1-LR) (Figure 4B). Similarly, the ratio of M2-like macrophages among RAW 264.7 cells was calculated as CD206 + (Q2-UR)/F4/80 + (Q2-UR + Q2-LR) (Figure 4C). Quantitative analysis (Figure 4D) demonstrated that the ratio of CD86+ cells and CD206+ cells was not significantly different between RAW 264.7 cells cocultured with oe-AhR and those cocultured with oe-NC mBMSCs. While the ratio of CD86+ cells among RAW 264.7 cocultured with sh-AhR mBMSCs was significantly higher than that among RAW 264.7 cocultured with sh-NC mBMSCs.
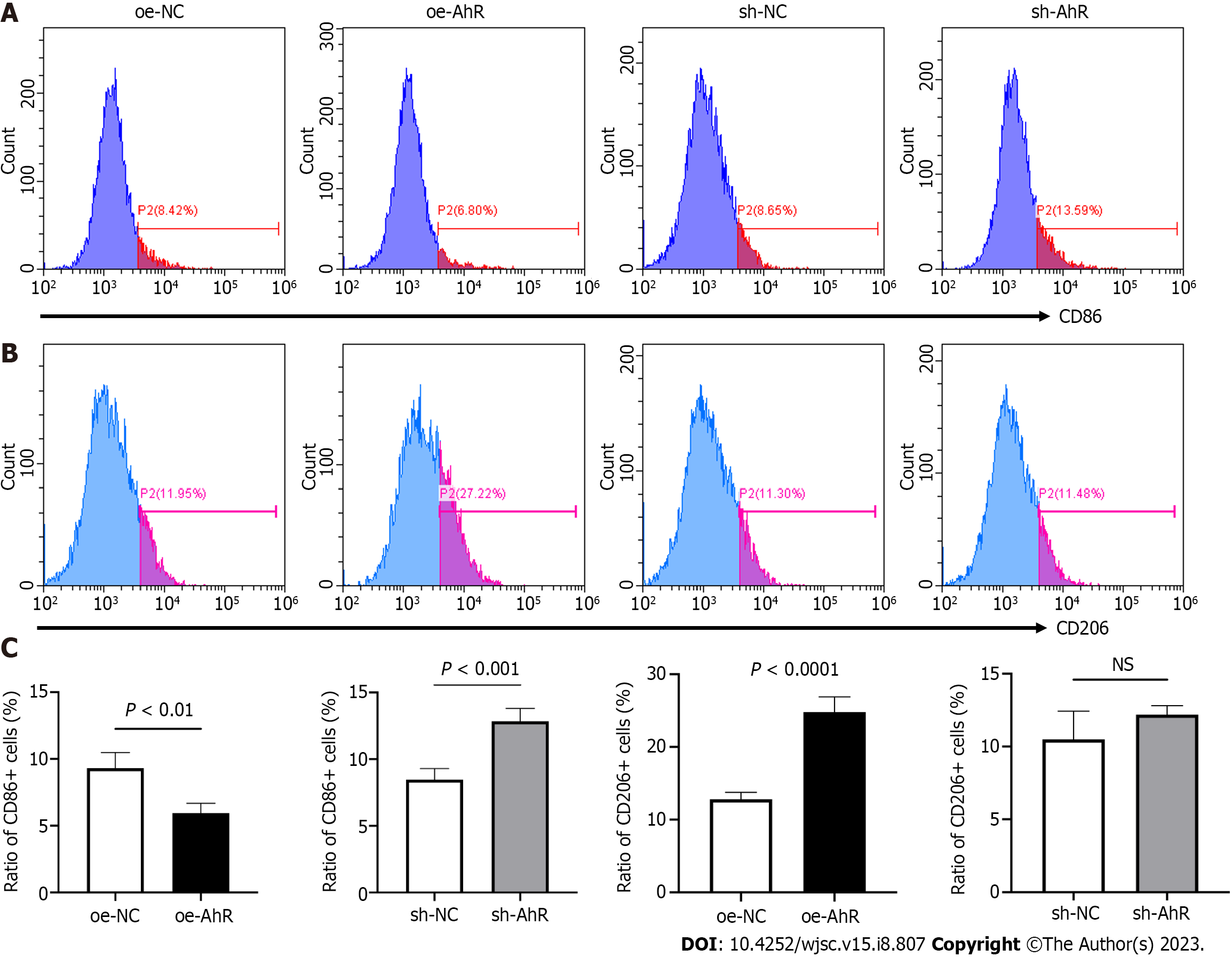
In the indirect coculture system, RAW 264.7 cells were cultured with conditioned medium from different mBMSCs. The CD86 and CD206 expressions were demonstrated using histograms (Figure 5A and B). Quantitative analysis (Figure 5C) showed that the ratio of CD86+ cells was significantly lower and the ratio of CD206+ cells was significantly higher in the oe-AhR group than oe-NC. The ratio of CD86+ cells in the sh-AhR group was significantly higher than that in sh-NC.
The above data suggested that knockdown of AhR in mBMSCs tends to drive macrophages toward the M1-like phenotype and inhibit M2-like polarization. Overexpression of AhR in mBMSCs showed the opposite macrophage-modulating effect.
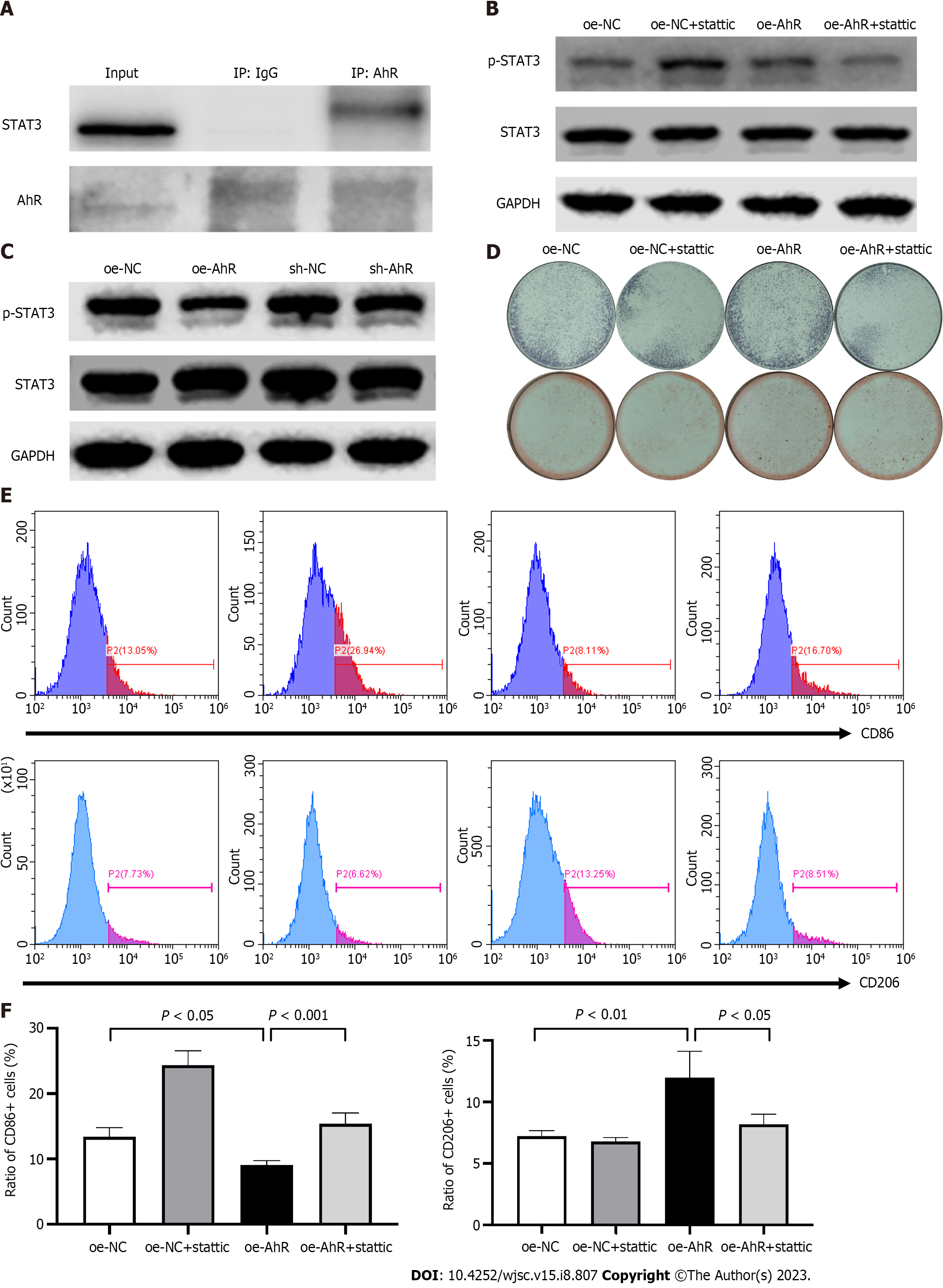
First, the AhR and STAT3 protein-protein interaction was detected via Co-IP (Figure 6A). Then, the effect of AhR overexpression or knockdown on the phosphorylation of STAT3 was explored by western blotting (Figure 6B). The results showed that oe-AhR promoted the phosphorylation of STAT3 while sh-AhR suppressed it. To test whether the effects of AhR on osteogenic differentiation and macrophage-modulating were partially STAT3-dependent or not, a specific STAT3 inhibitor, stattic, was introduced. The western blotting lanes testified that 2 mmol/L stattic partially alleviated the AhR overexpression-mediated increase in STAT3 phosphorylation (Figure 6C). ALP staining at the 7th day of osteogenic induction and ARS at the 15th day indicated that 2 mmol/L stattic partially reversed the elevated osteogenic potential mediated by AhR overexpression (Figure 6D). Regarding macrophage modulation, the flow cytometry and quantitative analysis manifested that 2 mmol/L stattic partially reversed the inhibition of CD86 expression and promotion of CD206 in RAW 264.7 cells cultured with conditioned medium from oe-AhR mBMSCs (Figure 6E and F). The above data suggested that AhR promoted the osteogenic and macrophage-modulating potentials of mBMSCs partially by interacting with STAT3 and increasing the phosphorylation of STAT3.
Based on the results, and within the limitations of the present study, it could be concluded that: (1) Endogenous unligated AhR promoted osteogenic potential of mBMSCs; (2) endogenous unligated AhR played a positive role in polarizing macrophages towards the M2-like phenotype of mBMSCs; and (3) the function of AhR was partially dependent on interacting with STAT3 and increasing the phosphorylation of STAT3. The present study offered new insights into the role of AhR and the involved molecular mechanism in the regenerative potential of mBMSCs, which might be a target for achieving optimal bone regeneration in mBMSCs-based tissue engineering.
The nuclear receptor AhR is positively expressed in bone tissue, including osteoblasts and osteoclasts, and the AhR signaling pathway plays a vital role in bone homeostasis[12]. AhR was previously considered as an environment xenobiotic sensor and mediates oxidative stress[13]. In recent decades, the roles of AhR as a transcription factor in regulating various biological processes have been revealed. Plenty studies have focused on the functions of ligand-activated AhR in biological processes. However, the effects of AhR might vary due to interactions with different ligands. The first identified AhR ligand was a dioxin-like compound (DLC), 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), which mediates toxic effects after binding to AhR. Then, various kinds of non-dioxin-like endogenous ligands were identified, such as FICZ, 2-(1′-H–indole–3-carbonyl) thiazole–4-carboxylic acid methyl ester, and tryptophan metabolites such as kynurenine, which demonstrated functional diversity[9].
Regarding osteogenic potential, AhR activated by DLC ligands such as TCDD is basically harmful[14], but the effects of endogenous ligands such as FICZ are controversial. In a recent study, two different AhR ligands, benzo[a]pyrene (B[a]P) and FICZ, were analyzed in mice temporomandibular joint osteoarthritis[15]. The above study demonstrated that B[a]P induced mandibular subchondral bone resorption in an AhR-dependent manner. However, FICZ exerted a therapeutic effect and rescued the bone loss in vivo at both low (100mg/kg) and high (100mg/kg) concentrations. Moreover, 200ng/mL FICZ promoted the osteogenic differentiation on MC3T3 E1 cells in vitro, resulting in more obvious ALP staining and ARS, and increased mRNA expressions of osteogenic markers, including ALP, osteocalcin (OCN) and collagen type I alpha 1. Consistently, in a previous study by our group, 500 nM FICZ enhanced the ALP staining and ARS of rat BMSCs after osteogenic induction[11]. In addition to ligand-activated AhR, its role in bone ossification was investigated in transgenic mice. In the semistable fracture healing model in mice, the expression of AhR in the healing callus tissue was more than 2-fold higher on the seventh day after fracture than in uninjured samples. On the 14th day after fracture, AhR expression had increased by 10-fold in callus tissue. To determine whether the loss of AhR affects bone healing, the researchers established a tibial fracture model in wild type (WT) mice and AhR knockout (KO) mice and performed micro-computed tomography (micro-CT) scan analysis two weeks later. Mineralized callus tissue in the fracture gap was observed in the former but not the latter mice. At the third week, micro-CT analysis also showed that AhR KO mice had less bone formation than WT mice[7]. In another study[16], the ALP and OCN mRNA expression levels of BMSCs obtained from AhR KO mice were lower than those of WT mice after 8-10 d of osteogenic induction. Interestingly, the ALP and OCN expression patterns in the BMSCs of AhR KO mice after osteogenic induction were rather parallel to the TCDD-suppressed responses in BMSCs from WT mice. The results of present study were consistent with above literatures.
Additionally, AhR has been proven to be involved in modulating immune/inflammatory disease by targeting specific gene expression and altering immune differentiation[8]. Similarly, complex ligand interactions that control AhR function might result in diverse immunologic effects including immunosuppressive or pro-inflammatory downstream functions[17]. In another study, peritoneal macrophages from WT and AhR-null mice were polarized toward the M1 or M2 phenotype by stimulation with lipopolysaccharide/interferon-g or interleukin (IL)-4[18]. The results indicated that AhR-null macrophages presented higher levels of M1 markers including IL-1b, IL-6, IL-12 and tumor necrosis factor-a, and lower levels of M2 markers, including chitinase-like 3 (or called Ym1) and IL-10. It was found that the binding of AhR to the promoters of IL-10 and arginase-1 was increased in macrophages after uptake of apoptotic cells to promote M2 polarization[19]. AhR not only affects the phenotype of macrophages themselves but also influences the results of other cells in regulating macrophage polarization. Treatment with the AhR ligand FICZ attenuated calcium oxalate nephrocalcinosis in a mouse model. Bone marrow-derived macrophages (BMDMs) and calcium oxalate monohydrate (100mg/mL)-treated renal tubular epithelial cells were cocultured in transwell system. FICZ supplement in the system promoted the expression of M2 markers and diminished the expression of M1 markers in BMDMs. The molecular mechanism was that AhR directly targeted downstream microRNA-142a-3p, which suppressed interferon regulatory factor 1 and hypoxia inducible factor 1 alpha by binding to their 3’ untranslated region[20]. In another study, another AhR ligand, TCDD, was added to a coculture system of mBMSCs and macrophages. Treatment of BMSCs with TCDD resulted in a significant increase in M2 markers and a decrease in M1 markers in macrophages. The AhR antagonist CH223191 alleviated the macrophage-modulating effect[21]. In the present study, AhR overexpression in mBMSCs promoted its ability of polarizing macrophages into M2-like phenotype.
It was reported that AhR signaling exhibits considerable crosstalk with other transcription factors, such as those in the nuclear factor-kB family and signal transducer and activator of transcription family[17,22]. AhR can bind to other molecules to undergo a conformational change exposing a nuclear localization signal[17]. STAT3 is critical in regulating immune responses[23] and osteogenic differentiation processes[24]. Our previous study showed that FICZ-stimulated AhR alleviated the inflammatory response in periodontal ligament cells by increasing the phosphorylation of STAT3[10]. The interplay between AhR and STAT3 was explored in various cell types. In A549 cells the interaction of AhR and STAT3 was detected via Co-IP. Increased AhR by reduning upregulated the expression of STAT3 and the downstream IL-10, which alleviate severe pneumonia[25]. Lactobacillus johnsonii N6.2-derived nano-sized vesicles led to the nuclear translocation of AhR in pancreatic b cells and enhanced the phosphorylation of STAT3 and expression of IL-10, which reduced the apoptosis and improved the expression of genes related to glucose transport[26]. In the present study, the direct AhR and STAT3 interaction was also observed in mBMSCs. Moreover, AhR overexpression upregulated the phosphorylation of STAT3. However, the further research is needed to fully understand the precise molecular mechanisms underlying the AhR-STAT3 interaction.
In conclusion, AhR plays a promotive role in the regenerative potential of mBMSCs, including osteogenic differentiation and polarizing macrophages to an anti-inflammatory phenotype. Mechanistically, AhR can interact with STAT3, thereby increasing the phosphorylation level of STAT3. Inhibition of STAT3 partially counteracted the beneficial effect of AhR. Hence, AhR might be a target for achieving optimal bone regeneration in mBMSCs-based tissue engineering.
Bone marrow mesenchymal stromal cells (BMSCs) are one of the most commonly used seed cells in bone tissue engineering. Aryl hydrocarbon receptor (AhR) has been recognized as a nuclear receptor that modulates bone turnover. However, the function of constitutive AhR in BMSCs remains unclear.
To explore whether AhR is involved in the regenerative potential of mouse BMSCs (mBMSCs).
To investigate the role of AhR in the osteogenic and macrophage-modulating potential of mBMSCs and the underlying mechanism.
Immunochemistry and immunofluorescent staining were used to observe the expression of AhR in mouse bone marrow tissue and mBMSCs. The overexpression or knockdown of AhR was achieved by lentivirus-mediated plasmid. The osteogenic potential was observed by alkaline phosphatase and alizarin red staining. The mRNA and protein levels of osteogenic markers were detected by quantitative polymerase chain reaction and western blot. After coculture with different mBMSCs, the cluster of differentiation (CD) 86 and CD206 expressions levels in RAW 264.7 cells were analyzed by flow cytometry. To explore the underlying molecular mechanism, the interaction of AhR with signal transducer and activator of transcription 3 (STAT3) was observed by co-immunoprecipitation and phosphorylation of STAT3 was detected by western blot.
AhR expressions in mouse bone marrow tissue and isolated mBMSCs were detected. AhR overexpression enhanced the osteogenic potential of mBMSCs while AhR knockdown suppressed it. The ratio of CD86+ RAW 264.7 cells cocultured with AhR-overexpressed mBMSCs was reduced and that of CD206+ cells was increased. AhR directly interacted with STAT3. AhR overexpression increased the phosphorylation of STAT3. After inhibition of STAT3 via stattic, the promotive effects of AhR overexpression on the osteogenic differentiation and macrophage-modulating were partially counteracted.
AhR plays a beneficial role in the regenerative potential of mBMSCs partially by increasing phosphorylation of STAT3.
This study suggested that AhR and its interaction with STAT3 might be a potential candidate target for achieving optimal bone regeneration in mBMSCs-based tissue engineering.
| 1. | Yousefi AM, James PF, Akbarzadeh R, Subramanian A, Flavin C, Oudadesse H. Prospect of Stem Cells in Bone Tissue Engineering: A Review. Stem Cells Int. 2016;2016:6180487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 126] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 2. | Zhang Y, Du Z, Li D, Wan Z, Zheng T, Zhang X, Yu Y, Yang X, Cai Q. Catalpol modulating the crosstalking between mesenchymal stromal cells and macrophages via paracrine to enhance angiogenesis and osteogenesis. Exp Cell Res. 2022;418:113269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 3. | Tasso R, Ulivi V, Reverberi D, Lo Sicco C, Descalzi F, Cancedda R. In vivo implanted bone marrow-derived mesenchymal stem cells trigger a cascade of cellular events leading to the formation of an ectopic bone regenerative niche. Stem Cells Dev. 2013;22:3178-3191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 4. | Mahon OR, Browe DC, Gonzalez-Fernandez T, Pitacco P, Whelan IT, Von Euw S, Hobbs C, Nicolosi V, Cunningham KT, Mills KHG, Kelly DJ, Dunne A. Nano-particle mediated M2 macrophage polarization enhances bone formation and MSC osteogenesis in an IL-10 dependent manner. Biomaterials. 2020;239:119833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 292] [Article Influence: 48.7] [Reference Citation Analysis (0)] |
| 5. | Qi Y, Zhu T, Zhang T, Wang X, Li W, Chen D, Meng H, An S. M1 macrophage-derived exosomes transfer miR-222 to induce bone marrow mesenchymal stem cell apoptosis. Lab Invest. 2021;101:1318-1326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 6. | Mulero-Navarro S, Fernandez-Salguero PM. New Trends in Aryl Hydrocarbon Receptor Biology. Front Cell Dev Biol. 2016;4:45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 152] [Cited by in RCA: 201] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 7. | Izawa T, Arakaki R, Mori H, Tsunematsu T, Kudo Y, Tanaka E, Ishimaru N. The Nuclear Receptor AhR Controls Bone Homeostasis by Regulating Osteoclast Differentiation via the RANK/c-Fos Signaling Axis. J Immunol. 2016;197:4639-4650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 8. | Neavin DR, Liu D, Ray B, Weinshilboum RM. The Role of the Aryl Hydrocarbon Receptor (AHR) in Immune and Inflammatory Diseases. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 223] [Article Influence: 27.9] [Reference Citation Analysis (2)] |
| 9. | Safe S, Jin UH, Park H, Chapkin RS, Jayaraman A. Aryl Hydrocarbon Receptor (AHR) Ligands as Selective AHR Modulators (SAhRMs). Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 94] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 10. | Huang J, Cai X, Ou Y, Fan L, Zhou Y, Wang Y. Protective roles of FICZ and aryl hydrocarbon receptor axis on alveolar bone loss and inflammation in experimental periodontitis. J Clin Periodontol. 2019;46:882-893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 11. | Huang J, Wang Y, Zhou Y. Beneficial roles of the AhR ligand FICZ on the regenerative potentials of BMSCs and primed cartilage templates. RSC Adv. 2022;12:11505-11516. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 12. | Miki Y, Hata S, Saito R, Ono K, Sasano H, Kumamoto H. Expression of Aryl Hydrocarbon Receptor in Bone Tissues. In: Sasaki K, Suzuki O, Takahashi N, editors. Interface Oral Health Science. 2011. Tokyo: Springer, 2021:134–136. |
| 13. | Rothhammer V, Quintana FJ. The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease. Nat Rev Immunol. 2019;19:184-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 409] [Cited by in RCA: 893] [Article Influence: 127.6] [Reference Citation Analysis (0)] |
| 14. | Watson ATD, Nordberg RC, Loboa EG, Kullman SW. Evidence for Aryl hydrocarbon Receptor-Mediated Inhibition of Osteoblast Differentiation in Human Mesenchymal Stem Cells. Toxicol Sci. 2019;167:145-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 15. | Yoshikawa Y, Izawa T, Hamada Y, Takenaga H, Wang Z, Ishimaru N, Kamioka H. Roles for B[a]P and FICZ in subchondral bone metabolism and experimental temporomandibular joint osteoarthritis via the AhR/Cyp1a1 signaling axis. Sci Rep. 2021;11:14927. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 16. | Korkalainen M, Kallio E, Olkku A, Nelo K, Ilvesaro J, Tuukkanen J, Mahonen A, Viluksela M. Dioxins interfere with differentiation of osteoblasts and osteoclasts. Bone. 2009;44:1134-1142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 75] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 17. | Lamorte S, Shinde R, McGaha TL. Nuclear receptors, the aryl hydrocarbon receptor, and macrophage function. Mol Aspects Med. 2021;78:100942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 18. | Climaco-Arvizu S, Domínguez-Acosta O, Cabañas-Cortés MA, Rodríguez-Sosa M, Gonzalez FJ, Vega L, Elizondo G. Aryl hydrocarbon receptor influences nitric oxide and arginine production and alters M1/M2 macrophage polarization. Life Sci. 2016;155:76-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 63] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 19. | Shinde R, Hezaveh K, Halaby MJ, Kloetgen A, Chakravarthy A, da Silva Medina T, Deol R, Manion KP, Baglaenko Y, Eldh M, Lamorte S, Wallace D, Chodisetti SB, Ravishankar B, Liu H, Chaudhary K, Munn DH, Tsirigos A, Madaio M, Gabrielsson S, Touma Z, Wither J, De Carvalho DD, McGaha TL. Apoptotic cell-induced AhR activity is required for immunological tolerance and suppression of systemic lupus erythematosus in mice and humans. Nat Immunol. 2018;19:571-582. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 163] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 20. | Yang X, Liu H, Ye T, Duan C, Lv P, Wu X, Liu J, Jiang K, Lu H, Yang H, Xia D, Peng E, Chen Z, Tang K, Ye Z. AhR activation attenuates calcium oxalate nephrocalcinosis by diminishing M1 macrophage polarization and promoting M2 macrophage polarization. Theranostics. 2020;10:12011-12025. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 84] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 21. | Cui Z, Feng Y, Li D, Li T, Gao P, Xu T. Activation of aryl hydrocarbon receptor (AhR) in mesenchymal stem cells modulates macrophage polarization in asthma. J Immunotoxicol. 2020;17:21-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 22. | Øvrevik J, Låg M, Lecureur V, Gilot D, Lagadic-Gossmann D, Refsnes M, Schwarze PE, Skuland T, Becher R, Holme JA. AhR and Arnt differentially regulate NF-κB signaling and chemokine responses in human bronchial epithelial cells. Cell Commun Signal. 2014;12:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 71] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 23. | Hillmer EJ, Zhang H, Li HS, Watowich SS. STAT3 signaling in immunity. Cytokine Growth Factor Rev. 2016;31:1-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 574] [Article Influence: 57.4] [Reference Citation Analysis (0)] |
| 24. | Zhou S, Dai Q, Huang X, Jin A, Yang Y, Gong X, Xu H, Gao X, Jiang L. STAT3 is critical for skeletal development and bone homeostasis by regulating osteogenesis. Nat Commun. 2021;12:6891. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 25. | Luo S, Gan L, Liu S, Zhong L, Chen M, Zhang H, Li J, Huang L, Lv C. The synergistic Reduning and cefmetazole sodium treatment of severe pneumonia is mediated by the AhR-Src-STAT3 pathway. J Thorac Dis. 2022;14:474-493. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 26. | Teixeira LD, Harrison NA, da Silva DR, Mathews CE, Gonzalez CF, Lorca GL. Nanovesicles From Lactobacillus johnsonii N6.2 Reduce Apoptosis in Human Beta Cells by Promoting AHR Translocation and IL10 Secretion. Front Immunol. 2022;13:899413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cell and tissue engineering
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Jaing TH, Taiwan; Kode JA, India; Stogov MV, Russia; Tanabe S, Japan S-Editor: Cong Lin L-Editor: Lin C P-Editor: Zhang XD