Published online May 26, 2023. doi: 10.4252/wjsc.v15.i5.466
Peer-review started: December 30, 2022
First decision: March 14, 2023
Revised: March 28, 2023
Accepted: April 17, 2023
Article in press: April 17, 2023
Published online: May 26, 2023
Processing time: 147 Days and 5.8 Hours
The corneal epithelium is composed of stratified squamous epithelial cells on the outer surface of the eye, which acts as a protective barrier and is critical for clear and stable vision. Its continuous renewal or wound healing depends on the proliferation and differentiation of limbal stem cells (LSCs), a cell population that resides at the limbus in a highly regulated niche. Dysfunction of LSCs or their niche can cause limbal stem cell deficiency, a disease that is manifested by failed epithelial wound healing or even blindness. Nevertheless, compared to stem cells in other tissues, little is known about the LSCs and their niche. With the advent of single-cell RNA sequencing, our understanding of LSC characteristics and their microenvironment has grown considerably. In this review, we summarized the current findings from single-cell studies in the field of cornea research and focused on important advancements driven by this technology, including the heterogeneity of the LSC population, novel LSC markers and regulation of the LSC niche, which will provide a reference for clinical issues such as corneal epithelial wound healing, ocular surface reconstruction and interventions for related diseases.
Core Tip: Limbal stem cells (LSCs), a cell population that resides at the limbus in a highly regulated niche. With the advent of single-cell RNA sequencing, our understanding of LSC characteristics and their microenvironment has grown considerably. This review focuses on the current research on single cell sequencing in LSCs. We highlight the heterogeneity, novel specific markers and niche regulation of LSCs.
- Citation: Sun D, Shi WY, Dou SQ. Single-cell RNA sequencing in cornea research: Insights into limbal stem cells and their niche regulation. World J Stem Cells 2023; 15(5): 466-475
- URL: https://www.wjgnet.com/1948-0210/full/v15/i5/466.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v15.i5.466
The cornea is a unique transparent tissue in the human body exposed to the external environment and is the window for sight[1,2]. Specifically, the corneal epithelium acts as a protective barrier on the ocular surface and is constantly regenerating. This unique property of the corneal epithelium is dependent on self-renewing epithelial stem cells located at the limbus, known as limbal stem cells (LSCs)[3-5]. LSCs reside in the “palisades of Vogt” (also known as limbal epithelial crypts) and are critical for corneal epithelial regeneration and wound healing. LSCs respond to corneal epithelial cell renewal or wound healing by differentiating to produce limbal progenitor cells (LPCs) and transient amplifying cells (TACs), which then migrate to the central cornea to replenish the corneal epithelium[6-9]. This process was summarized as the XYZ hypothesis[10] and explained the balance of cell numbers and homeostasis in the corneal epithelium maintained by LSCs.
Like the stem cells in other tissues, the surrounding microenvironment or limbal niche strictly supports and regulates the functional behaviors of LSCs[11,12]. The limbal niche has unique characteristics and components, including mesenchymal cells, immune cells, melanocytes, vascular cells, extracellular matrix and signaling molecules (e.g., growth factors and cytokines)[13-16]. Significant pathology involving any component of the limbal niche can lead to the dysfunction of LSCs or even result in limbal stem cell deficiency (LSCD), a disease that is characterized by impaired wound healing or blindness[17,18].
Various studies have identified numerous markers of LSCs but identifying definitive molecular signatures to distinguish LSCs and other corneal epithelial cells is still challenging. The unclear internal heterogeneity of the LSC population can increase the difficulty in efficiently isolating pure LSCs for clinical transplantation. In addition, emerging evidence supports that reconstruction of the limbal niche may be introduced to treat LSCD. Therefore, understanding the function and niche regulation of LSCs is needed to discover novel therapies for ocular surface disease.
With the development and maturity of sequencing technology, more and more genomic, trans
LSCs are located in the basal layer of the corneal epithelium. As previously mentioned, they are characterized by a high proliferative potential, small size, high nucleoplasmic ratio and slow cell cycle[41,42]. LSCs are scarce, and finding bona-fide markers to distinguish them from other basal epithelial cells is challenging. In addition, few studies have investigated the heterogeneity and hierarchy of LSCs. Understanding the heterogeneity of LSCs is important for comprehending the function to effectively isolate them for clinical transplantation.
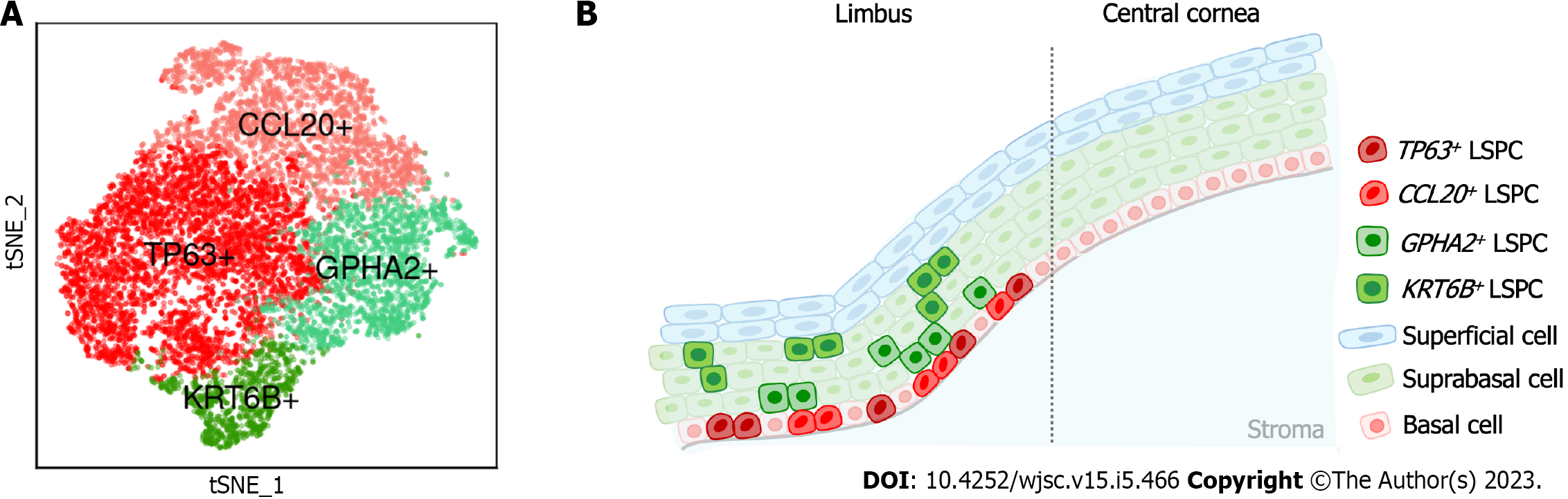
Dou et al[43] performed scRNA-Seq on human limbal tissues and identified four subclusters of stem/progenitor cells after single-cell transcriptome analysis. In this study, the authors annotated eight cell types, including prominent limbal epithelial cells, stromal cells and other rare cell populations. The authors then subclustered limbal epithelial cells and resolved their heterogeneity, including limbal stem/progenitor cells (LSPCs), limbal suprabasal cells and limbal superficial cells. To further explore the LSC population, the authors then subclustered LSPCs and obtained four subpopulations (Figure 1) including: (1) A subpopulation with the classical LSC marker TP63[44]; (2) A subpopulation with high expression of CCL20, which is a chemokine that can induce cell migration and proliferation[45]; (3) A subpopulation with specific expression of GPHA2, a marker recently identified in quiescent LSCs (qLSCs) from humans and mice[46,47]; and (4) A subpopulation with high expression of KRT6B, which is associated with rapid keratinocyte division and contributes to inhibiting the migration of mitotic cell populations from the basal layer[48]. The authors then investigated the differences in stemness and differentiation status and observed that TP63+ and CCL20+ cells presented a high stemness state, whereas GPHA2+ and KRT6B+ showed a high differentiation state.
Another study by Li et al[49] annotated five subtypes from the limbal basal epithelium of the human cornea. They characterized terminally differentiated cells (TDCs), post-mitotic cells, TACs, LPCs and LSCs. Furthermore, the authors discovered that these five subtypes represented the major stages and trajectories of human LSC proliferation and differentiation (from LSCs, LPCs, TACs and post-mitotic cells to TDCs), and they were spatially situated in different regions from the limbus to the central cornea. In TDCs, corneal epithelium-specific differentiation markers and keratinocyte keratinization markers were expressed at the highest levels, while the LSC differentiation markers had the lowest expression.
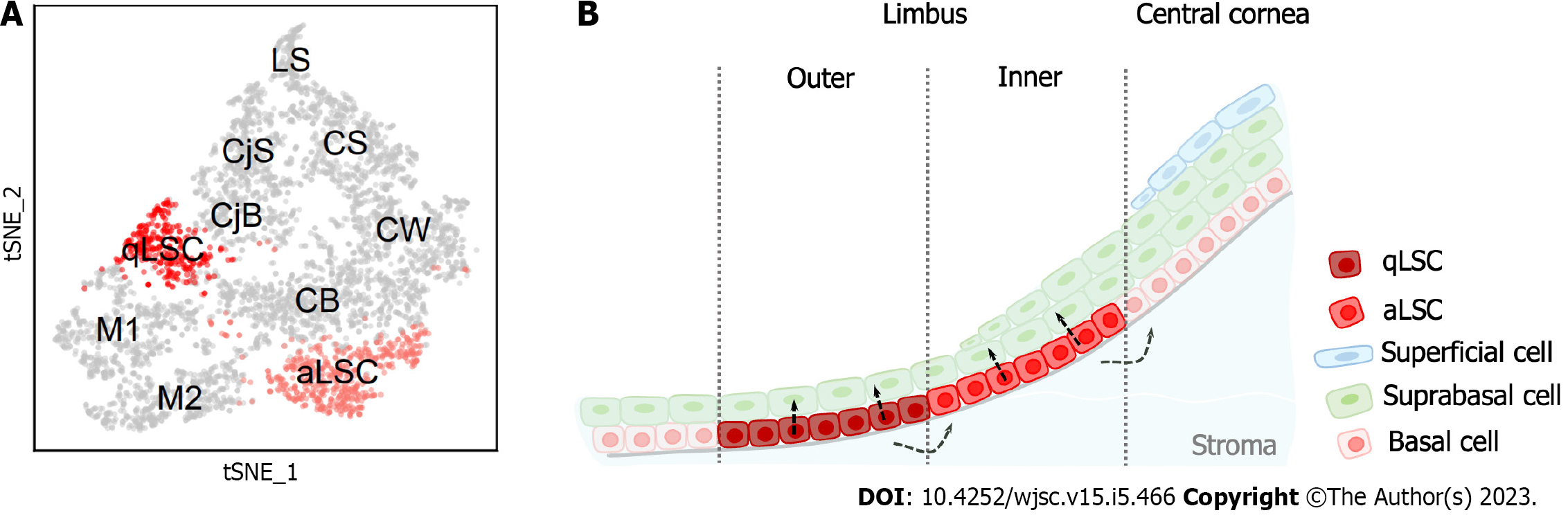
LSCs in mice are also heterogeneous and behave differently than human LSCs. Altshuler et al[46] combined scRNA-Seq and quantitative lineage tracing for in-depth analysis of mouse limbal epithelium. The authors revealed the presence of two distinct subpopulations of mouse LSCs that were in separate and well-defined spatial locations called the “inner” and “outer” limbus (Figure 2). The inner limbus contains active LSCs, which maintain the homeostasis of the corneal epithelium. The outer limbus contains qLSCs that have a significantly lower rate of division and are involved in wound healing and border formation. Spectral tracking experiments displayed that qLSCs can quickly exit the dormant state and enter the cell cycle in response to injury, suggesting that qLSCs are a reservoir for tissue regeneration. In addition, their circumferentially extended clonal growth model and continuous localization on the border highly indicates that these cells play a crucial role in border maintenance. Notably, this finding was also confirmed by a study utilizing the two-photon live imaging approach[50]. Collectively, LSCs are highly heterogeneous in both humans and mice, unlike stem cells in other tissues. Further studies are needed to investigate the self-renewal and differentiation mechanisms of LSCs.
Since 1989, when LSCs were discovered[4], a series of markers have been found to identify these cells, such as TP63, KRT3, KRT12. However, the marker pattern typically labels the broad limbal basal cell population. Accurately distinguishing LSCs from other epithelial cells is still challenging and is still an active area of research. Altshuler et al[46] discovered a novel set of markers to accurately identify LSCs. They applied in situ hybridization probes for Krt4 and Krt12 to label mouse conjunctival and corneal basal and suprabasal cells, respectively. Gpha2 staining could obviously demarcate the outer LSCs (also known as qLSCs), while the inner LSCs (also known as active LSCs) were labeled as Atf3+. Then, they used immunofluorescence staining to confirm that the outer limbal epithelial basal cells were Krt15+/Ifitm3+/Cd63+, and the inner limbal epithelial basal cells were Atf3+/Mt1-2+. Next, the authors explored the correlation between mouse and human LSC markers. Immunofluorescence images revealed that KRT15, IFITM3 and GPHA2 were expressed in human limbus epithelial basal cells. Ifitm3 was found to be restricted to cellular vesicles in the cytoplasm of undifferentiated limbal cells, which was consistent with a previous study’s findings[51]. Ifitm3 knockdown led to a differentiation phenotype and a reduced colony-forming capacity. These experiments suggest that Ifitm3 and Gpha2 can be used to identify LSCs, and Ifitm3 mediates the undifferentiated state.
Gpha2 has been frequently studied in human LSCs. Dou et al[43] explored the four subclusters of LSPCs, which were identified by TP63, CCL20, GPHA2 and KRT6B. Collin et al[47] identified several novel genes, one of which was GPHA2, using an unbiased approach to recognize marker genes that were highly expressed in human LSCs relative to other corneal epithelial cells. High and specific expression of GPHA2 was observed in the limbus crypts, which was consistent with the findings of Altshuler et al[46]. Moreover, the authors used RNA interference (RNAi) to downregulate GPHA2 and observed a significant reduction in cell proliferation and differentiation efficiency, indicating an important role of GPHA2 in maintaining the undifferentiated state of human LSCs. The authors also performed flow activated cell sorting analysis with colony forming efficiency assays to confirm the RNAi data.
Other LSC markers have also been identified. Li et al[49] identified TSPAN7+ and SOX17+ cells distributed in a scattered pattern in human limbus epithelium basal cells. The authors established an in vitro model of epithelial cells and discovered TSPAN7 and SOX17 were not strongly expressed in human limbal epithelial cells. However, mRNA and protein expression levels were significantly activated after injury, especially during cell migration and growth. The authors also utilized RNAi to downregulate TSPAN7 and SOX17 and observed inhibited cell proliferation and significantly delayed epithelial regeneration during wound healing. Overall, the discovery of novel markers of LSCs (Table 1) can help us to better distinguish LSCs from other cells to further understand the function and state of LSCs and provide a more effective strategy for the isolation, culture and clinical application of LSCs.
| LSC subtype | Marker | Species | Ref. |
| LSPC with high stemness | TP63, CCL20 | Human | [43] |
| LSPC with high differentiation | GPHA2, KRT6B | Human | |
| LSC | TSPAN7, SOX17, SELE, ECSCR, RAMP3, RNASE1, NPCD1, NNMT, SLC2A3, KLF2, PDK4 | Human | [49] |
| Limbal progenitor cell | DCN, PLIN2, DEGS1, MMP10, IFITM3, SLC6A6, LTB4R, SLP1 | Human | |
| qLSC | Gpha2, Cd63, Ifitm3 | Mouse | [46] |
| aLSC | Atf3, Socs3, Mt1, Prdm1 | Mouse |
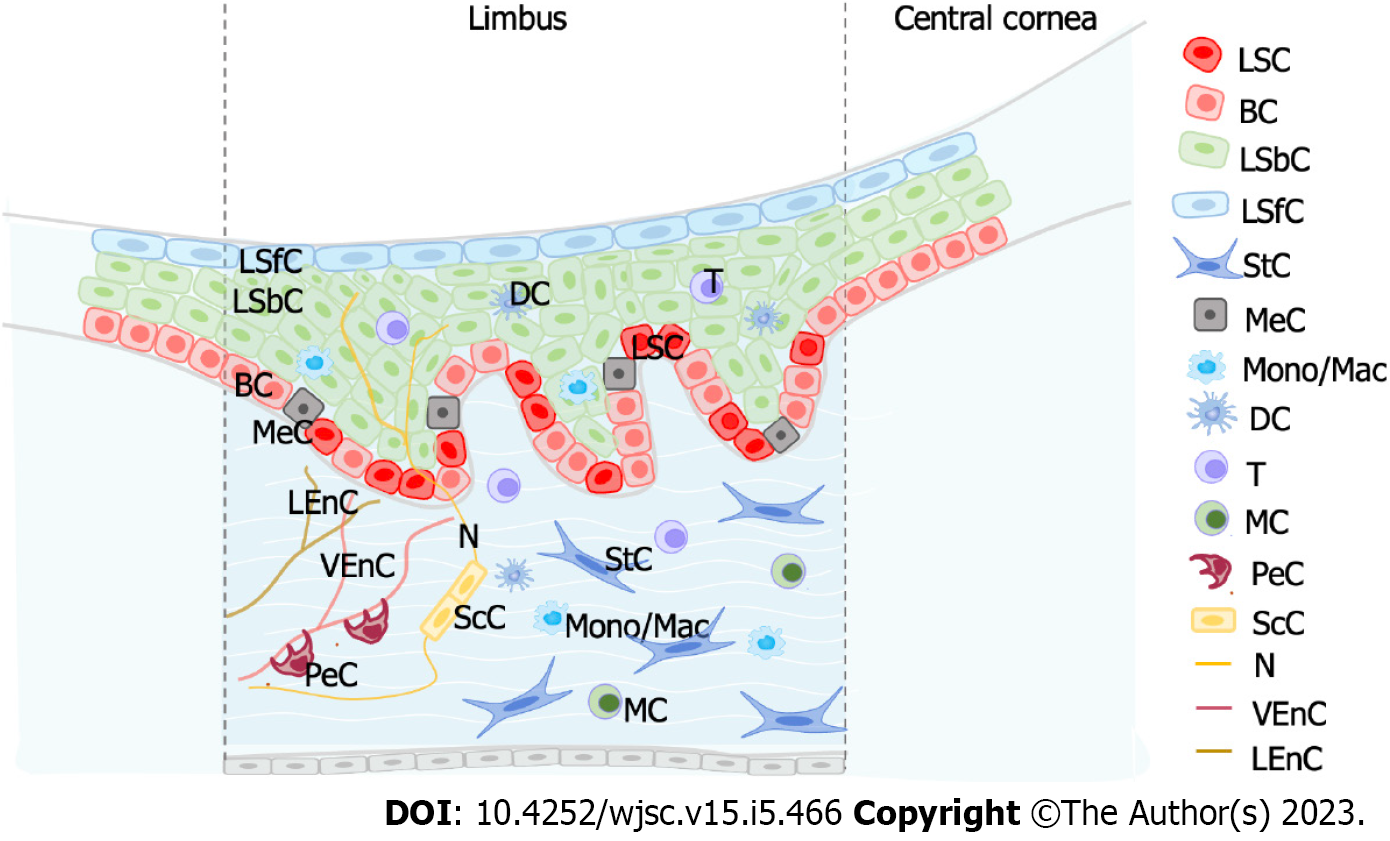
LSC proliferation, migration and differentiation are inseparable from the regulation of the limbal niche microenvironment. The stem cell niche is the local microenvironment directly promoting or protecting stem cell populations[52-54]. The LSC niche provides a sheltered environment that protects LSCs from stimulation[55-58]. If the LSC niche is involved in pathological damage, then LSC dysfunction can occur. Therefore, the study of the LSC niche is essential.
Collin et al[47] investigated the interaction between LSCs and the limbal niche by single-cell analysis. The authors combined scRNA-Seq and ATAC-Seq and performed CellPhoneDB analysis[59]. They identified multiple significant interactions between human LSCs and regulatory factors of immune cells such as proinflammatory cytokines [tumor necrosis factor, interleukin (IL)-1β, IL-6, IL-17A, interferon γ, and oncostatin M], proinflammatory cell surface receptor (triggering receptor expressed on myeloid cells 1), proinflammatory cytokine expression (adaptor complexes 1) and regulators of inflammatory responses (nuclear factor kappa B, RELA, colony-stimulating factor 2, phosphoinositide 3-kinase, extracellular signal-regulated kinase 1/2, and F2). The authors verified that limbal epithelial cells were significantly reduced in cell culture medium containing tumor necrosis factor-α and IL-1β. This suggested that proinflammatory cytokines produced by immune cells were involved in the apoptosis of limbal epithelial cells[60], thus mimicking the central corneal defect and stimulating the proliferation of LSCs[61]. This was also consistent with other reports showing that the addition of proinflammatory factors to limbal epithelial cell cultures can directly affect the expression of LSC markers and their colony forming efficiency capacity[60,62-64].
Dou et al[43] systematically explored intercellular communication between LSPCs and other cell populations based on ligand-receptor analysis. By correlating the corresponding receptor-ligands in human LSPCs and their niche cells, the authors observed that LSPCs were regulated by the limbal niche as well as by other cells in the limbal niche. The Notch signaling pathway was also involved in cell-cell interaction between LSPCs and their niche cells. NOTCH1-4 receptors were expressed in LSPCs, and their relevant ligands were primarily identified in niche cells, such as Schwann cells, stromal cells, pericytes and LSPCs. Likewise, the WNT7A, WNT7B and WNT5A ligands, which participate in the Wnt/β-catenin signaling pathway, were detected on LSPCs. Their corresponding receptors were primarily detected on limbal epithelial cells, stromal cells, immune cells, Schwann cells and LSPCs. The presence of multiple chemokines, such as CCL4, CCL4L2, IL-1β and IL-24, on LSPCs and their paired receptors indicated that immune cell interactions may potentially regulate LSPCs.
Altshuler et al[46] revealed that T cells acted as niche cells and served its function in the maintenance of quiescence, epithelial thickness control and wound healing. By studying the limbus of the severe combined immunodeficiency (SCID) and non-obese diabetic SCID mice, which are unable to make mature T and B lymphocytes, it was observed that the GPHA2 protein was substantially decreased to almost undetectable levels. In contrast, the expression of Ifitm3 did not rely on the existence of immune cells, implying that it was regulated by other niche cells. When T cells were inhibited by topical application of the corticosteroid dexamethasone, LSCs showed a dramatic reduction in Cd63 and Gpha2 expression levels and increased cell proliferation, demonstrating that T cells played a crucial role in regulating qLSCs. Finally, corneal epithelial debridement followed by epithelial closure by fluorescein dye infiltration revealed delayed epithelial wound healing in mice lacking T cells.
In addition, other niche cells were determined to be important for the microenvironment regulation of LSCs. Oxidative stress can lead to a variety of eye diseases, such as keratitis, cataracts and retinal diseases, which are subject to varying degrees of oxidative damage[65,66]. Recently, studies found that melanocytes in the limbal niche (as antioxidant systems) protected LSPCs from UV-induced oxidative damage and reduced oxidative stress through the transfer of melanosomes[67,68]. Moreover, by ligand analysis, Dou et al[43] identified the intercellular communications between melanocytes and LSCs. NAMPT, as a ligand, was highly expressed in melanocytes and had been reported to act as a critical switch in melanoma cells. CD44 acted as a receptor and was also highly enriched in melanocytes.
Vascular endothelial cells are also one of the important niche cells of LSCs. It has been reported that vascular endothelial cells were highly correlated with the classic Wnt signaling pathway involved in the regulation of the corneal limbal niche[69,70]. Furthermore, Dou et al[43] performed a differential expression analysis with the integration of the scRNA-Seq datasets from the limbus and the skin and observed that the vascular endothelial cells from the limbus highly expressed anti-vascular factors compared to that from the skin, consistent with characteristics of corneal angiogenic privilege. Above all, these studies have shown that the regulation of the LSC niche (Figure 3) occupies a key role in the growth, development, proliferation and differentiation of LSCs.
The first Drop-Seq experiments were performed on mouse retina in 2015[23]. Since this revolutionary experiment, single-cell sequencing technology has been widely used in many fields, including ophthalmology, and gene expression has been studied at an unprecedented resolution in multiple ocular tissues. Corneal transparency is essential for normal vision; thus, comprehension of the mechanisms related to corneal wound healing and regeneration is crucial for the treatment of patients suffering from corneal disease. Currently, corneal epithelial regeneration is a relatively satisfactory approach and has the potential to treat corneal superficial scars. However, for multiple corneal basal scars or endothelial disease, corneal transplantation remains the only option to restore clear vision[71-73]. Unfortunately, corneal clouding remains one of the leading causes of blindness worldwide due to the lack of corneal donor tissue or the limited availability of corneal surgery[74,75]. Although most studies support corneal regeneration through LSC therapies[76,77], the study of LSCs is particularly important.
This review focused on the current research on single-cell sequencing in LSCs. We highlighted the heterogeneity of LSCs and presented several novel specific markers of LSCs and the role of niche regulation of LSCs. LSCs can be identified in both humans and mice, and several markers, such as GHPA2 and IFITM3, can be highly and specifically expressed on LSCs. Moreover, both T cell regulation in mice studied by Altshuler et al[46] and immune cell regulation in humans studied by Collin et al[47] and Dou et al[43] suggest that niche regulation is of vital importance for LSCs.
Future research can still benefit from RNA-Seq technology as it can aid in acquisition of further knowledge on the functions and characteristics of LSCs, including in the discovery of more novel highly specific expression markers and more niche regulated components that can promote or inhibit the proliferation and differentiation of LSCs. These discoveries should be translated into better prevention and treatment strategies to treat blindness and improve the clinical prognosis of patients with LSCD and other LSC-related diseases.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Ophthalmology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C, C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Das Mohapatra SS, India; Jiang FX, Australia; Mariappan I, India; Salvadori M, Italy; Sidoti A, Italy; Ventura C, Italy S-Editor: Wang JJ L-Editor: A P-Editor: Zhang XD
| 1. | Sridhar MS. Anatomy of cornea and ocular surface. Indian J Ophthalmol. 2018;66:190-194. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 343] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 2. | Secker GA, Daniels JT. Corneal epithelial stem cells: deficiency and regulation. Stem Cell Rev. 2008;4:159-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 44] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 3. | Bonnet C, González S, Roberts JS, Robertson SYT, Ruiz M, Zheng J, Deng SX. Human limbal epithelial stem cell regulation, bioengineering and function. Prog Retin Eye Res. 2021;85:100956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 4. | Cotsarelis G, Cheng SZ, Dong G, Sun TT, Lavker RM. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: implications on epithelial stem cells. Cell. 1989;57:201-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 977] [Cited by in RCA: 990] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 5. | Davanger M, Evensen A. Role of the pericorneal papillary structure in renewal of corneal epithelium. Nature. 1971;229:560-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 425] [Cited by in RCA: 405] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 6. | Collinson JM, Morris L, Reid AI, Ramaesh T, Keighren MA, Flockhart JH, Hill RE, Tan SS, Ramaesh K, Dhillon B, West JD. Clonal analysis of patterns of growth, stem cell activity, and cell movement during the development and maintenance of the murine corneal epithelium. Dev Dyn. 2002;224:432-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 111] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 7. | Nagasaki T, Zhao J. Centripetal movement of corneal epithelial cells in the normal adult mouse. Invest Ophthalmol Vis Sci. 2003;44:558-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 101] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 8. | Pellegrini G, Golisano O, Paterna P, Lambiase A, Bonini S, Rama P, De Luca M. Location and clonal analysis of stem cells and their differentiated progeny in the human ocular surface. J Cell Biol. 1999;145:769-782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 530] [Cited by in RCA: 514] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 9. | Zhou M, Li XM, Lavker RM. Transcriptional profiling of enriched populations of stem cells versus transient amplifying cells. A comparison of limbal and corneal epithelial basal cells. J Biol Chem. 2006;281:19600-19609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 55] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 10. | Mathers WD, Lemp MA. Morphology and movement of corneal surface cells in humans. Curr Eye Res. 1992;11:517-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 11. | Arwert EN, Hoste E, Watt FM. Epithelial stem cells, wound healing and cancer. Nat Rev Cancer. 2012;12:170-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 322] [Cited by in RCA: 365] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 12. | Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415-418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2119] [Cited by in RCA: 1990] [Article Influence: 132.7] [Reference Citation Analysis (0)] |
| 13. | Grieve K, Ghoubay D, Georgeon C, Thouvenin O, Bouheraoua N, Paques M, Borderie VM. Three-dimensional structure of the mammalian limbal stem cell niche. Exp Eye Res. 2015;140:75-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 77] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 14. | Massie I, Dziasko M, Kureshi A, Levis HJ, Morgan L, Neale M, Sheth R, Tovell VE, Vernon AJ, Funderburgh JL, Daniels JT. Advanced imaging and tissue engineering of the human limbal epithelial stem cell niche. Methods Mol Biol. 2015;1235:179-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 15. | Parfitt GJ, Kavianpour B, Wu KL, Xie Y, Brown DJ, Jester JV. Immunofluorescence Tomography of Mouse Ocular Surface Epithelial Stem Cells and Their Niche Microenvironment. Invest Ophthalmol Vis Sci. 2015;56:7338-7344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 16. | Ramírez BE, Victoria DA, Murillo GM, Herreras JM, Calonge M. In vivo confocal microscopy assessment of the corneoscleral limbal stem cell niche before and after biopsy for cultivated limbal epithelial transplantation to restore corneal epithelium. Histol Histopathol. 2015;30:183-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 17. | Kim BY, Riaz KM, Bakhtiari P, Chan CC, Welder JD, Holland EJ, Basti S, Djalilian AR. Medically reversible limbal stem cell disease: clinical features and management strategies. Ophthalmology. 2014;121:2053-2058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 79] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 18. | Notara M, Refaian N, Braun G, Steven P, Bock F, Cursiefen C. Short-term uvb-irradiation leads to putative limbal stem cell damage and niche cell-mediated upregulation of macrophage recruiting cytokines. Stem Cell Res. 2015;15:643-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 19. | Khan SY, Ali M, Kabir F, Na CH, Delannoy M, Ma Y, Qiu C, Costello MJ, Hejtmancik JF, Riazuddin SA. The role of FYCO1-dependent autophagy in lens fiber cell differentiation. Autophagy. 2022;18:2198-2215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 20. | Hata M, Hata M, Andriessen EM, Juneau R, Pilon F, Crespo-Garcia S, Diaz-Marin R, Guber V, Binet F, Fournier F, Buscarlet M, Grou C, Calderon V, Heckel E, Melichar HJ, Joyal JS, Wilson AM, Sapieha P. Early-life peripheral infections reprogram retinal microglia and aggravate neovascular age-related macular degeneration in later life. J Clin Invest. 2023;133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 21. | Donato L, Alibrandi S, Scimone C, Rinaldi C, Dascola A, Calamuneri A, D'Angelo R, Sidoti A. The impact of modifier genes on cone-rod dystrophy heterogeneity: An explorative familial pilot study and a hypothesis on neurotransmission impairment. PLoS One. 2022;17:e0278857. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 37] [Reference Citation Analysis (0)] |
| 22. | Cowan CS, Renner M, De Gennaro M, Gross-Scherf B, Goldblum D, Hou Y, Munz M, Rodrigues TM, Krol J, Szikra T, Cuttat R, Waldt A, Papasaikas P, Diggelmann R, Patino-Alvarez CP, Galliker P, Spirig SE, Pavlinic D, Gerber-Hollbach N, Schuierer S, Srdanovic A, Balogh M, Panero R, Kusnyerik A, Szabo A, Stadler MB, Orgül S, Picelli S, Hasler PW, Hierlemann A, Scholl HPN, Roma G, Nigsch F, Roska B. Cell Types of the Human Retina and Its Organoids at Single-Cell Resolution. Cell. 2020;182:1623-1640.e34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 456] [Article Influence: 91.2] [Reference Citation Analysis (0)] |
| 23. | Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202-1214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5595] [Cited by in RCA: 5067] [Article Influence: 460.6] [Reference Citation Analysis (0)] |
| 24. | Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, McCarroll SA, Cepko CL, Regev A, Sanes JR. Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell. 2016;166:1308-1323.e30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 788] [Cited by in RCA: 816] [Article Influence: 81.6] [Reference Citation Analysis (0)] |
| 25. | Tasic B. Single cell transcriptomics in neuroscience: cell classification and beyond. Curr Opin Neurobiol. 2018;50:242-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 66] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 26. | Altschuler SJ, Wu LF. Cellular heterogeneity: do differences make a difference? Cell. 2010;141:559-563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1031] [Cited by in RCA: 828] [Article Influence: 51.8] [Reference Citation Analysis (0)] |
| 27. | Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58:610-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 720] [Cited by in RCA: 973] [Article Influence: 88.5] [Reference Citation Analysis (1)] |
| 28. | Liu S, Trapnell C. Single-cell transcriptome sequencing: recent advances and remaining challenges. F1000Res. 2016;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 163] [Cited by in RCA: 161] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 29. | Svensson V, Vento-Tormo R, Teichmann SA. Exponential scaling of single-cell RNA-seq in the past decade. Nat Protoc. 2018;13:599-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 505] [Cited by in RCA: 624] [Article Influence: 78.0] [Reference Citation Analysis (0)] |
| 30. | Heng JS, Hackett SF, Stein-O'Brien GL, Winer BL, Williams J, Goff LA, Nathans J. Comprehensive analysis of a mouse model of spontaneous uveoretinitis using single-cell RNA sequencing. Proc Natl Acad Sci U S A. 2019;116:26734-26744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 31. | Hu Y, Wang X, Hu B, Mao Y, Chen Y, Yan L, Yong J, Dong J, Wei Y, Wang W, Wen L, Qiao J, Tang F. Dissecting the transcriptome landscape of the human fetal neural retina and retinal pigment epithelium by single-cell RNA-seq analysis. PLoS Biol. 2019;17:e3000365. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 112] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 32. | Peng YR, Shekhar K, Yan W, Herrmann D, Sappington A, Bryman GS, van Zyl T, Do MTH, Regev A, Sanes JR. Molecular Classification and Comparative Taxonomics of Foveal and Peripheral Cells in Primate Retina. Cell. 2019;176:1222-1237.e22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 353] [Article Influence: 50.4] [Reference Citation Analysis (0)] |
| 33. | Lukowski SW, Lo CY, Sharov AA, Nguyen Q, Fang L, Hung SS, Zhu L, Zhang T, Grünert U, Nguyen T, Senabouth A, Jabbari JS, Welby E, Sowden JC, Waugh HS, Mackey A, Pollock G, Lamb TD, Wang PY, Hewitt AW, Gillies MC, Powell JE, Wong RC. A single-cell transcriptome atlas of the adult human retina. EMBO J. 2019;38:e100811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 227] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 34. | Clark BS, Stein-O'Brien GL, Shiau F, Cannon GH, Davis-Marcisak E, Sherman T, Santiago CP, Hoang TV, Rajaii F, James-Esposito RE, Gronostajski RM, Fertig EJ, Goff LA, Blackshaw S. Single-Cell RNA-Seq Analysis of Retinal Development Identifies NFI Factors as Regulating Mitotic Exit and Late-Born Cell Specification. Neuron. 2019;102:1111-1126.e5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 460] [Cited by in RCA: 354] [Article Influence: 50.6] [Reference Citation Analysis (0)] |
| 35. | Gautam P, Hamashima K, Chen Y, Zeng Y, Makovoz B, Parikh BH, Lee HY, Lau KA, Su X, Wong RCB, Chan WK, Li H, Blenkinsop TA, Loh YH. Multi-species single-cell transcriptomic analysis of ocular compartment regulons. Nat Commun. 2021;12:5675. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 36. | van Zyl T, Yan W, McAdams AM, Monavarfeshani A, Hageman GS, Sanes JR. Cell atlas of the human ocular anterior segment: Tissue-specific and shared cell types. Proc Natl Acad Sci U S A. 2022;119:e2200914119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 96] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 37. | Lehmann GL, Hanke-Gogokhia C, Hu Y, Bareja R, Salfati Z, Ginsberg M, Nolan DJ, Mendez-Huergo SP, Dalotto-Moreno T, Wojcinski A, Ochoa F, Zeng S, Cerliani JP, Panagis L, Zager PJ, Mullins RF, Ogura S, Lutty GA, Bang J, Zippin JH, Romano C, Rabinovich GA, Elemento O, Joyner AL, Rafii S, Rodriguez-Boulan E, Benedicto I. Single-cell profiling reveals an endothelium-mediated immunomodulatory pathway in the eye choroid. J Exp Med. 2020;217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 38. | Wu H, Chen W, Zhao F, Zhou Q, Reinach PS, Deng L, Ma L, Luo S, Srinivasalu N, Pan M, Hu Y, Pei X, Sun J, Ren R, Xiong Y, Zhou Z, Zhang S, Tian G, Fang J, Zhang L, Lang J, Wu D, Zeng C, Qu J, Zhou X. Scleral hypoxia is a target for myopia control. Proc Natl Acad Sci U S A. 2018;115:E7091-E7100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 354] [Article Influence: 44.3] [Reference Citation Analysis (0)] |
| 39. | Dou S, Wang Q, Zhang B, Wei C, Wang H, Liu T, Duan H, Jiang H, Liu M, Qi X, Zhou Q, Xie L, Shi W, Gao H. Single-cell atlas of keratoconus corneas revealed aberrant transcriptional signatures and implicated mechanical stretch as a trigger for keratoconus pathogenesis. Cell Discov. 2022;8:66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 67] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 40. | Li JM, Kim S, Zhang Y, Bian F, Hu J, Lu R, Pflugfelder SC, Chen R, Li DQ. Single-Cell Transcriptomics Identifies a Unique Entity and Signature Markers of Transit-Amplifying Cells in Human Corneal Limbus. Invest Ophthalmol Vis Sci. 2021;62:36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 41. | Gonzalez G, Sasamoto Y, Ksander BR, Frank MH, Frank NY. Limbal stem cells: identity, developmental origin, and therapeutic potential. Wiley Interdiscip Rev Dev Biol. 2018;7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 92] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 42. | Yoon JJ, Ismail S, Sherwin T. Limbal stem cells: Central concepts of corneal epithelial homeostasis. World J Stem Cells. 2014;6:391-403. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 93] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 43. | Dou S, Wang Q, Qi X, Zhang B, Jiang H, Chen S, Duan H, Lu Y, Dong J, Cao Y, Xie L, Zhou Q, Shi W. Molecular identity of human limbal heterogeneity involved in corneal homeostasis and privilege. Ocul Surf. 2021;21:206-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (1)] |
| 44. | Claudinot S, Sakabe JI, Oshima H, Gonneau C, Mitsiadis T, Littman D, Bonfanti P, Martens G, Nicolas M, Rochat A, Barrandon Y. Tp63-expressing adult epithelial stem cells cross lineages boundaries revealing latent hairy skin competence. Nat Commun. 2020;11:5645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 45. | Wang B, Shi L, Sun X, Wang L, Wang X, Chen C. Production of CCL20 from lung cancer cells induces the cell migration and proliferation through PI3K pathway. J Cell Mol Med. 2016;20:920-929. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 76] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 46. | Altshuler A, Amitai-Lange A, Tarazi N, Dey S, Strinkovsky L, Hadad-Porat S, Bhattacharya S, Nasser W, Imeri J, Ben-David G, Abboud-Jarrous G, Tiosano B, Berkowitz E, Karin N, Savir Y, Shalom-Feuerstein R. Discrete limbal epithelial stem cell populations mediate corneal homeostasis and wound healing. Cell Stem Cell. 2021;28:1248-1261.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 116] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 47. | Collin J, Queen R, Zerti D, Bojic S, Dorgau B, Moyse N, Molina MM, Yang C, Dey S, Reynolds G, Hussain R, Coxhead JM, Lisgo S, Henderson D, Joseph A, Rooney P, Ghosh S, Clarke L, Connon C, Haniffa M, Figueiredo F, Armstrong L, Lako M. A single cell atlas of human cornea that defines its development, limbal progenitor cells and their interactions with the immune cells. Ocul Surf. 2021;21:279-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 156] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 48. | Finnegan A, Cho RJ, Luu A, Harirchian P, Lee J, Cheng JB, Song JS. Single-Cell Transcriptomics Reveals Spatial and Temporal Turnover of Keratinocyte Differentiation Regulators. Front Genet. 2019;10:775. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 49. | Li DQ, Kim S, Li JM, Gao Q, Choi J, Bian F, Hu J, Zhang Y, Li J, Lu R, Li Y, Pflugfelder SC, Miao H, Chen R. Single-cell transcriptomics identifies limbal stem cell population and cell types mapping its differentiation trajectory in limbal basal epithelium of human cornea. Ocul Surf. 2021;20:20-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 87] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 50. | Farrelly O, Suzuki-Horiuchi Y, Brewster M, Kuri P, Huang S, Rice G, Bae H, Xu J, Dentchev T, Lee V, Rompolas P. Two-photon live imaging of single corneal stem cells reveals compartmentalized organization of the limbal niche. Cell Stem Cell. 2021;28:1233-1247.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 51. | Wu X, Dao Thi VL, Huang Y, Billerbeck E, Saha D, Hoffmann HH, Wang Y, Silva LAV, Sarbanes S, Sun T, Andrus L, Yu Y, Quirk C, Li M, MacDonald MR, Schneider WM, An X, Rosenberg BR, Rice CM. Intrinsic Immunity Shapes Viral Resistance of Stem Cells. Cell. 2018;172:423-438.e25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 306] [Cited by in RCA: 292] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 52. | Nishikawa SI, Osawa M. What is a stem cell niche? Ernst Schering Res Found Workshop. 2006;1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 53. | Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598-611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1612] [Cited by in RCA: 1451] [Article Influence: 80.6] [Reference Citation Analysis (0)] |
| 54. | Li L, Xie T. Stem cell niche: structure and function. Annu Rev Cell Dev Biol. 2005;21:605-631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 854] [Cited by in RCA: 823] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 55. | Yazdanpanah G, Haq Z, Kang K, Jabbehdari S, Rosenblatt ML, Djalilian AR. Strategies for reconstructing the limbal stem cell niche. Ocul Surf. 2019;17:230-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 72] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 56. | Tseng SC, He H, Zhang S, Chen SY. Niche Regulation of Limbal Epithelial Stem Cells: Relationship between Inflammation and Regeneration. Ocul Surf. 2016;14:100-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 75] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 57. | Li W, Hayashida Y, Chen YT, Tseng SC. Niche regulation of corneal epithelial stem cells at the limbus. Cell Res. 2007;17:26-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 225] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 58. | Guo P, Sun H, Zhang Y, Tighe S, Chen S, Su CW, Liu Y, Zhao H, Hu M, Zhu Y. Limbal niche cells are a potent resource of adult mesenchymal progenitors. J Cell Mol Med. 2018;22:3315-3322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 59. | Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15:1484-1506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 835] [Cited by in RCA: 2213] [Article Influence: 368.8] [Reference Citation Analysis (0)] |
| 60. | Yang L, Zhang S, Duan H, Dong M, Hu X, Zhang Z, Wang Y, Zhang X, Shi W, Zhou Q. Different Effects of Pro-Inflammatory Factors and Hyperosmotic Stress on Corneal Epithelial Stem/Progenitor Cells and Wound Healing in Mice. Stem Cells Transl Med. 2019;8:46-57. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 61. | Puri S, Sun M, Mutoji KN, Gesteira TF, Coulson-Thomas VJ. Epithelial Cell Migration and Proliferation Patterns During Initial Wound Closure in Normal Mice and an Experimental Model of Limbal Stem Cell Deficiency. Invest Ophthalmol Vis Sci. 2020;61:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 62. | Notara M, Shortt AJ, Galatowicz G, Calder V, Daniels JT. IL6 and the human limbal stem cell niche: a mediator of epithelial-stromal interaction. Stem Cell Res. 2010;5:188-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 66] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 63. | Wang W, Li S, Xu L, Jiang M, Li X, Zhang Y, Tighe S, Zhu Y, Li G. Differential Gene Expression between Limbal Niche Progenitors and Bone Marrow Derived Mesenchymal Stem Cells. Int J Med Sci. 2020;17:549-557. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 64. | Veréb Z, Albert R, Póliska S, Olstad OK, Akhtar S, Moe MC, Petrovski G. Comparison of upstream regulators in human ex vivo cultured cornea limbal epithelial stem cells and differentiated corneal epithelial cells. BMC Genomics. 2013;14:900. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 65. | Wakamatsu TH, Dogru M, Tsubota K. Tearful relations: oxidative stress, inflammation and eye diseases. Arq Bras Oftalmol. 2008;71:72-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 101] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 66. | Donato L, Scimone C, Alibrandi S, Scalinci SZ, Rinaldi C, D'Angelo R, Sidoti A. Epitranscriptome Analysis of Oxidative Stressed Retinal Epithelial Cells Depicted a Possible RNA Editing Landscape of Retinal Degeneration. Antioxidants (Basel). 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 30] [Reference Citation Analysis (0)] |
| 67. | Polisetti N, Gießl A, Zenkel M, Heger L, Dudziak D, Naschberger E, Stich L, Steinkasserer A, Kruse FE, Schlötzer-Schrehardt U. Melanocytes as emerging key players in niche regulation of limbal epithelial stem cells. Ocul Surf. 2021;22:172-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 68. | Bose B, Najwa AR, Shenoy P S. Oxidative Damages to Eye Stem Cells, in Response to, Bright and Ultraviolet Light, Their Associated Mechanisms, and Salvage Pathways. Mol Biotechnol. 2019;61:145-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 69. | Scimone C, Donato L, Marino S, Alafaci C, D'Angelo R, Sidoti A. Vis-à-vis: a focus on genetic features of cerebral cavernous malformations and brain arteriovenous malformations pathogenesis. Neurol Sci. 2019;40:243-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 70. | Scimone C, Donato L, Alibrandi S, Esposito T, Alafaci C, D'Angelo R, Sidoti A. Transcriptome analysis provides new molecular signatures in sporadic Cerebral Cavernous Malformation endothelial cells. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 71. | Wilson SE, Torricelli AAM, Marino GK. Corneal epithelial basement membrane: Structure, function and regeneration. Exp Eye Res. 2020;194:108002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 72. | Mobaraki M, Abbasi R, Omidian Vandchali S, Ghaffari M, Moztarzadeh F, Mozafari M. Corneal Repair and Regeneration: Current Concepts and Future Directions. Front Bioeng Biotechnol. 2019;7:135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 122] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 73. | Mimura T, Yamagami S, Amano S. Corneal endothelial regeneration and tissue engineering. Prog Retin Eye Res. 2013;35:1-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 90] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 74. | Garg P, Krishna PV, Stratis AK, Gopinathan U. The value of corneal transplantation in reducing blindness. Eye (Lond). 2005;19:1106-1114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 95] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 75. | Congdon NG, Friedman DS, Lietman T. Important causes of visual impairment in the world today. JAMA. 2003;290:2057-2060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 479] [Cited by in RCA: 494] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 76. | Hsu CC, Peng CH, Hung KH, Lee YY, Lin TC, Jang SF, Liu JH, Chen YT, Woung LC, Wang CY, Tsa CY, Chiou SH, Chen SJ, Chang YL. Stem Cell Therapy for Corneal Regeneration Medicine and Contemporary Nanomedicine for Corneal Disorders. Cell Transplant. 2015;24:1915-1930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 77. | Nurković JS, Vojinović R, Dolićanin Z. Corneal Stem Cells as a Source of Regenerative Cell-Based Therapy. Stem Cells Int. 2020;2020:8813447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |